Abstract
Spotted fever group rickettsiae (SFG) are a neglected group of bacteria, belonging to the genus Rickettsia, that represent a large number of new and emerging infectious diseases with a worldwide distribution. The diseases are zoonotic and are transmitted by arthropod vectors, mainly ticks, fleas and mites, to hosts such as wild animals. Domesticated animals and humans are accidental hosts. In Asia, local people in endemic areas as well as travellers to these regions are at high risk of infection. In this review we compare SFG molecular and serological diagnostic methods and discuss their limitations. While there is a large range of molecular diagnostics and serological assays, both approaches have limitations and a positive result is dependent on the timing of sample collection. There is an increasing need for less expensive and easy-to-use diagnostic tests. However, despite many tests being available, their lack of suitability for use in resource-limited regions is of concern, as many require technical expertise, expensive equipment and reagents. In addition, many existing diagnostic tests still require rigorous validation in the regions and populations where these tests may be used, in particular to establish coherent and worthwhile cut-offs. It is likely that the best strategy is to use a real-time quantitative polymerase chain reaction (qPCR) and immunofluorescence assay in tandem. If the specimen is collected early enough in the infection there will be no antibodies but there will be a greater chance of a PCR positive result. Conversely, when there are detectable antibodies it is less likely that there will be a positive PCR result. It is therefore extremely important that a complete medical history is provided especially the number of days of fever prior to sample collection. More effort is required to develop and validate SFG diagnostics and those of other rickettsial infections.
Key words: Asia, diagnosis, rickettsial infection, SFG, spotted fever rickettsia
Introduction
Rickettsioses are infectious diseases caused by obligate intracellular gram-negative bacteria. They belong to the order Rickettsiales and reside in a wide range of arthropod vectors such as fleas, ticks and mites [1]. These vectors can transmit pathogens to humans at the bite site, who may or may not subsequently develop disease. Rickettsial diseases have been reported to be the second most common cause of non-malarial febrile illness in the Southeast Asia region after dengue infection [2].
Pathogenic members of the Rickettsia are classified into two major groups: spotted fever group (SFG) and typhus group (TG) rickettsiae and additional, Orientia tsutsugamushi and O. chuto are classified as scrub typhus group (STG) [3]. Although rickettsial diseases have worldwide distribution, there are endemic and hyper-endemic areas. TG and STG rickettsiae are widely diagnosed in Southeast Asia [1, 4, 5]. In Asia, TG infections are mainly caused by Rickettsia typhi [6, 7] which is the etiologic agent of murine typhus (or endemic typhus). Rickettsia prowazekii, also a TG rickettsiae and responsible for louse-borne typhus (epidemic typhus), is rarely detected. Scrub typhus is caused by O. tsutsugamushi along with the related O. chuto [8, 9]. O. tsutsugamushi is widespread in the Asia-Pacific (and northern Australia) however, when O. chuto is included, the geographical range is extended to include the Middle-east, Africa and South America [9, 10]. The SFG consists of over 30 species that can be found worldwide. The one of the most studied is R. rickettsii which causes Rocky Mountain spotted fever (RMSF) in North America [11]. Other species such as R. australis and R. honei are prevalent in northern Australia [12]. Rickettsia conorii is responsible for Mediterranean spotted fever (MSF) in several parts of Europe, Africa and Asia [13–15]. Rickettsia felis (known as the causative agent of flea-borne spotted fever) is seen as an emerging infectious disease. First identified in the USA and now seen worldwide, it is also responsible for many cases of febrile illness in Africa [16].
The main arthropod vectors of SFG are hard ticks (Ixodidae), although soft ticks (Argasidae) are also implicated in a number of SFG [1, 17]. Other SFG such as R. felis, may be transmitted by fleas, whilst more recently, mosquitoes have also been demonstrated to be competent vectors [18]. Dependent on the vector, transmission of SFG occurs via salivary products produced during feeding or through inoculation with the faeces of infected vectors on the wound, mucosal surfaces and via inhalation. Rickettsial infections occur following infection of the endothelial cell lining of blood vessels (microvascular endothelium in the case of infection by R. conorii and both microvascular and macrovascular endothelium in the case of R. rickettsii) [19, 20]. Symptoms at clinical presentation are variable and are often similar to many other acute febrile illnesses. Careful clinical observation by physicians and reliable laboratory diagnosis can lead to early appropriate administration of antibiotic therapy and patient management and thereby reduce patient mortality or clinical complications. The purpose of this article is to compare and contrast molecular and serological identification methods including limitations for the diagnosis of SFG infections with reference to the Asian perspective especially in low- and middle-income countries (LMICs)
SFG infection and diagnosis
Infection and clinical presentation
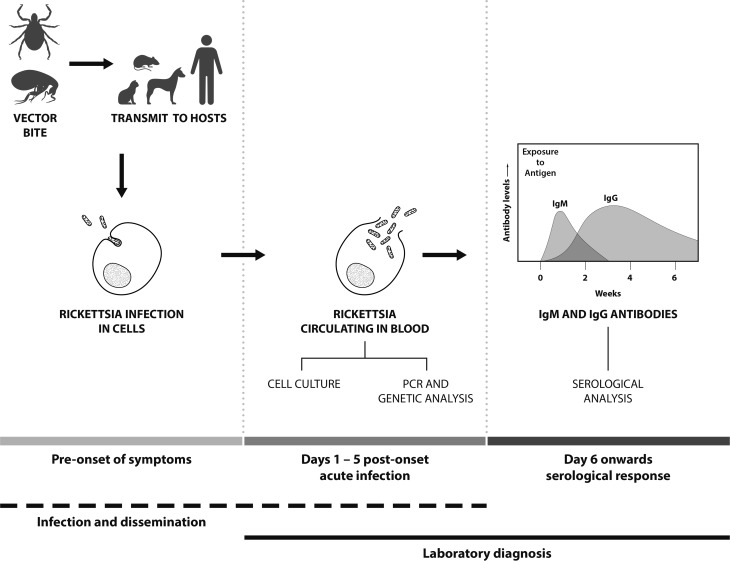
The infection cycle of SFG in humans starts with the arthropod bite (normally ticks or fleas) followed by an incubation period of up to fifteen days prior to the onset of clinical symptoms (Fig. 1). Clinical symptoms of SFG infection can vary, ranging from mild to life-threatening. Patients suspected of SFG infection normally present with fever, nausea, vomiting, maculopapular rash and occasionally eschars at the site of inoculation [21]. SFG infection can result in variable and often severe clinical symptoms in individual patients and the lack of eschar evidence can lead to misdiagnosis and often delays in commencing antibiotic treatment. For instance, in Hong Kong, treatment was delayed due to misdiagnosis of a case of MSF infection with meningitis symptoms; this resulted in inappropriate treatment and a fatal outcome [22]. In addition, some MSF patients have shown complications such as hearing loss [23], acute myocarditis [24] and cerebral infarction [25]. In Japan, severe Japanese spotted fever cases (R. japonica) have been reported with acute respiratory failure complications, including acute respiratory distress syndrome [26, 27]. In other cases, complications from SFG infections can include renal failure, purpura fulminans and severe pneumonia [28]. Therefore, the availability of diagnostic techniques for SFG infections is important, as well as SFG-awareness of physicians to enable the early administration of appropriate antibiotic treatment.
Fig. 1.
Features and temporal aspects of SFG Rickettsia infection and diagnosis.
SFG laboratory diagnosis
Molecular detection
Polymerase chain reaction (PCR)-based detection is the primary method to detect SFG, especially for the early detection of infection before the development of detectable antibodies [29] (Fig. 1). Molecular-based techniques need to be sensitive, and the sample type and assay used determine the success of detection. PCRs are used for both epidemiological and diagnostic purposes and SFG DNA may be isolated from arthropods, animal hosts and human clinical samples including whole blood, buffy coat, serum, tissue biopsies (such as skin), eschar scrapings and swabs [30]. For human clinical diagnostic purposes, whole blood or buffy coat is the preferred sample type, as SFG are intracellular (the cellular component being concentrated in the buffy coat fraction, increasing the sensitivity of detection) and the sample type being easily collected. Concentration of the sample type and maximizing the sensitivity of assays are a very important consideration for the detection of rickettsiae. Although little is known of the quantities of SFG in blood, quantities of other rickettsial organisms in the blood of an infected patient are variable. In RMSF with nonfatal outcomes, R. rickettsii copy numbers ranged from 8.40 × 101 to 3.95 × 105 copies/ml of blood, whilst in patients with fatal infections copy numbers ranged from 1.41 × 103 to 2.05 × 106 copies/ml of blood [31]. By comparison, the STG O. tsutsugamushi may be found at a median density of 13 genome copies/ml of blood (IQR: 0–334) [32]. Serum or plasma may be used for the PCR, but these samples are less than optimal as there will be fewer patient cells (meaning lower rickettsiae concentration). In addition, serum may have increased concentrations of blood fibrinogen and fibrin materials which can bind to Rickettsia DNA decreasing availability of DNA target for PCRs [33]. Eschars are a suitable sample type for the PCR (as well as culture techniques) and may be sampled as either scrapings, swabs or biopsied specimen [34]. As with any sample type, the specimen needs to be maintained at optimal temperatures or preserved prior to detection and/or isolation. Formalin-fixed and paraffin-embedded tissues can also be used, but fixation using formalin causes nucleic acid fragmentation and reduces the quality and quantity of nucleic acids as well as limiting the length of PCR products [35].
Selection of an appropriate gene target is important. Conserved gene targets enable a broad Rickettsia genus- or SFG-level of detection. The use of gene targets such as the citrate synthase (gltA), 16S rRNA and 17 kDa lipoprotein outer membrane antigens (17 kDa) generally confirms the presence of SFG or TG rickettsiae [36–39]. The use of Outer Membrane Protein A (190 kDa) (ompA) and B (ompB) genes appear to be more specific and discriminating for SFG in both patient and animal samples [29, 38].With all of these gene targets, down-stream sequencing of the PCR product will discern, in most cases, specific species. It is strongly recommended that multiple gene targets are used to gain an accurate identification.
A variety of different PCR-based assays are available for the diagnosis of SFG infections. Table 1 lists the most commonly used conventional (cPCR), nested (nPCR) and quantitative PCRs (qPCRs) that have been developed. Many of the primer sets have been mixed-and-matched and optimized in different studies to maximize the identification of SFG infections. cPCR assays have targeted most of the key genes discussed. nPCRs (which use two sets of primers) have been developed to increase the detection sensitivity over cPCRs although, as with other PCR assays, there may still be difficulty in differentiating closely-related SFG species, such as R. conorii and R. sibirica [40]. Several real-time or quantitative PCR (qPCR) assays developed for rickettsial detection have largely replaced the use of nPCRs due to a greater sensitivity and shorter run-time (Table 1). Furthermore, multiplexed qPCR is an efficient method that demonstrates greater analytical power, as multiple primers and probes are combined into a single assay, either targeting a number of different species or different gene targets for a single species at the same time [35].
Table 1.
Summary of common SFG PCRs used for diagnosis and sequencing for species identification purposes
| Target | Oligo name | Oligo combinations | PCR type | Original reference | PCR use | Further references | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 16S rDNA | 16SU17Fa | 1 | 2 | (1) cPCR (2) nPCR |
(1) [41] (2) [42] |
VD/HD/S | [29] | |||||||
| Rc16S.452n | 1 | – | ||||||||||||
| 16SU1592R | – | 2 | ||||||||||||
| 16sR34F | – | 2 | ||||||||||||
| 16sOR1198R | – | 2 | ||||||||||||
| 17 kDa | R17K249R | 1 | (1) qPCRb | [43] | VD/HD | |||||||||
| R17K135F | 1 | |||||||||||||
| R17Kbprobe | 1 | |||||||||||||
| R17K128F2 | 1 | (1) qPCRc | [44] | VD | [42,45, 46] | |||||||||
| R17K238R | 1 | |||||||||||||
| R17K202TaqP | 1 | |||||||||||||
| R17–122 | 1 | – | 3 | 4 | (1) cPCR (2) cPCR (3) nPCR (4) nPCR |
(1/4) [47] (2) [48] |
HD | [11, 38, 49] | ||||||
| R17–500 | 1 | – | 3 | 4 | ||||||||||
| Tz-16–20 | – | 2 | 3 | – | ||||||||||
| Tz-15–19 | – | 2 | 3 | – | ||||||||||
| RP1D | – | – | – | 4 | ||||||||||
| RP2 | – | – | – | 4 | ||||||||||
| R17kM61F | – | – | 3 | (1) cPCR (2) nPCR (3) nPCR |
[42] | VD/HD/S | [29, 45, 50] | |||||||
| R17k31Fd | 1 | 2 | 3 | |||||||||||
| R17k469R | – | 2 | 3 | |||||||||||
| Rr17k2608RNe | 1 | 2 | – | |||||||||||
| gltA | CS1dF | 1 | 2 | (1) cPCR (2) nPCR |
(1) [51] (2) [52] |
VD/S | [45] | |||||||
| CS1273R | 1 | 2 | ||||||||||||
| CS1234R | – | 2 | ||||||||||||
| RpECS1258nf | 1 | 2 | (1) cPCR (2) nPCR |
(1) [51, 53] (2) [40] |
VD/HD/S | [29, 38, 45, 49] | ||||||||
| RpCS.877 pg | 1 | 2 | ||||||||||||
| RpCS.1233n | – | 2 | ||||||||||||
| RpCS.896p | – | 2 | ||||||||||||
| CS-78 | 1 | (1) cPCR | [54] | VD/HD | [29] | |||||||||
| CS-323 | 1 | |||||||||||||
| CS-F | 1 | (1) qPCR | [55] | VD/HD | [42] | |||||||||
| CS-R | 1 | |||||||||||||
| CS-P | 1 | |||||||||||||
| ompA | RompA642R | 1 | (1) cPCR | [42] | VD/S | [45] | ||||||||
| RompAM50F | 1 | |||||||||||||
| RR190-70 | 1 | 2 | 3 | 4 | – | – | – | – | – | (1) cPCR (2) cPCR (3) nPCR (4) cPCR (5) nPCR (6) cPCR (7) nPCR (8) cPCR (9) qPCR |
(1) [49] (2) [53] (4) [56] (7) [36] (8) [57] |
VD/HD/S | [11, 29, 45, 58] | |
| RR190–701 | 1 | − | 3 | 4 | 5 | 6 | – | – | 9 | |||||
| 190-RN1 | 1 | – | – | – | – | – | – | – | – | |||||
| 190-FN1 | 1 | – | – | – | – | – | – | – | – | |||||
| Rr190k.720n | – | – | – | – | – | – | 7 | – | – | |||||
| Rr190k.71p | – | – | – | – | – | – | 7 | – | – | |||||
| RR190–602 | – | 2 | 3 | – | – | 6 | 7 | – | – | |||||
| RR190.547F | – | – | – | – | 5 | – | – | 8 | 9 | |||||
| RR190.701R | – | – | − | – | 5 | – | – | 8 | – | |||||
| ompB | Rf1524R | 1 | (1) cPCR | [42] | VD/S | |||||||||
| ompB1570R | 1 | |||||||||||||
| 120–607F | 1 | (1) nPCR | [39, 59] (#237) | VD/S | [45] | |||||||||
| RompB11F | 1 | |||||||||||||
| RompB1902R | 1 | |||||||||||||
| RAK1009F | 1 | (1) cPCR | [59] (#237) | VD/S | [45] | |||||||||
| RAK1452R | 1 | |||||||||||||
| Rc.rompB.4836n | 1 | (1) nPCR | [40] | HD | [38] | |||||||||
| Rc.rompB.4362p | 1 | |||||||||||||
| Rc.rompB.4762n | 1 | |||||||||||||
| Rc.rompB.4496p | 1 | |||||||||||||
| rompB OF | 1 | (1) nPCR | [40] | VD/HD | [29] | |||||||||
| rompB OR | 1 | |||||||||||||
| rompB SFG IF | 1 | |||||||||||||
| rompB SFG IR | 1 | |||||||||||||
| rplP | PanR8-F | 1 | (1) qPCR | [11] | HD | |||||||||
| PanR8-P | 1 | |||||||||||||
| PanR8-R | 1 | |||||||||||||
| sca4 | RrD928R | 1 | (1) nPCR | [59](#237) | VD/S | [45] | ||||||||
| RrD1826R | 1 | |||||||||||||
| RrD749F | 1 | |||||||||||||
Species-specific PCRs excluded. PCR type: cPCR, conventional; nPCR, nested (including semi-nested); qPCR, quantitative/real-time PCR. PCR use: VD, diagnostics in arthropod vectors; HD, diagnostics in human samples; S, used for sequencing purposes.
aka: fD1.
aka: Rick17 assay.
aka: Rick17b assay (developed from Rick17 assay).
aka: 17 kDa-2/Rr2608Rnew/Primer2; base change G20A in references [29,42,45]; base change T7C in references [42,45]; base change A22C in reference [38].
Base change A22C in reference [38].
aka: CS1273R; base change G5A in reference [38].
An alternative to the standard PCR assays, suitable for more field-based diagnosis, is the loop-mediated isothermal amplification assay (LAMP). This has a potential for application as a simple and rapid molecular SFG detection technique, employing an isothermal (constant temperature) nucleic acid technique for the amplification of DNA. The amplification is performed at 60–65 °C and the stop reaction at 80 °C [60]. The LAMP has been reported to be more sensitive than the nPCR to detect ompB from SFG in China, with 73% sensitivity and 100% specificity compared with the nPCR which gave negative results [60].
As with all PCR assays, it is also possible that a vector may carry more than one rickettsial species resulting in multiple PCR products being obtained if using broad range primers (not species-specific). This can cause difficulties in interpreting results, and may even result in misdiagnosing of the actual agent causing disease. Therefore, if the PCR results show positivity for SFG but the assay cannot differentiate between species, the amplicons should be sequenced for further species identification which provides useful epidemiological information.
Serological detection
The genus Rickettsia and Orientia can be characterized into three major antigenic groups: SFG, TG and STG [37]. There are serological cross reactions between SFG and TG antibodies [17, 30, 37] as well amongst antibodies of Rickettsia spp. within SFG and therefore serological diagnosis is normally only made to the antigenic (serogroup) group level. Discrimination to the species level within SFG using serological techniques is extremely difficult and only possible following cross-absorption using western blot techniques [30, 61].
In many cases, rickettsial infection is confirmed by the use of serological techniques such as indirect immunofluorescence assay (IFA), indirect immunoperoxidase (IIP) test, enzyme-linked immunosorbent assay (ELISA) and the Weil–Felix agglutination test (WFT). Immunoglobulin M (IgM) antibodies are present in the early phase of an infection and reduces afterwards until they are undetectable after a few weeks (Fig. 1). In contrast, IgG increases after the second week of illness and persists at low titers for years in some cases (30% found after one year) [62]. The WFT was developed in 1916 and primarily used in mid-1940's for the identification of TG rickettsiae infections [63]. The WFT is an inexpensive test, principally based on the antibodies against Gram-negative Proteus spp. antigens of OX (O-specific polysaccharide chain of outer membrane lipopolysaccharide) which cross-reacts with Rickettsia antigens OX-2 and OX-19 with the former more specific to SFG [64]. However, the WFT demonstrates low sensitivity in the acute phase of infection as demonstrated by 47% sensitivity in RMSF [65] and 33% sensitivity for SFG in Sri Lanka [66] and is rarely use nowadays to this shortcoming.
In many Asian countries, IFA and IIP are recognized as standard methods for routine diagnosis. The sensitivity of IIP and IFA often depends on the timing of serum collection due to the lack of antibodies in the first week of illness prior to complete seroconversion [67] although it can demonstrate high sensitivity (83–100%) and specificity (99–100%) (Table 2) [37]. A seroconversion or a four-fold difference in antibodies from acute to convalescent phase IgM and IgG antibodies is considered to be significant [62, 68] however, the diagnostic accuracy of such tests is also dependent on the cut-offs applied and therefore a understanding of the background immunity in endemic and non-endemic populations is essential with higher cut-offs often used in endemic settings. Furthermore, IFA and IIP results are dependent on appropriate antigenic types (often R. honei and R. conorii in Asia), well-trained personnel for the interpretation of the results, and requires a fluorescence microscope, which is often expensive and difficult to maintain.
Table 2.
The comparison of sensitivity and specificity of serological techniques for SFG infection
| Year | Location | Method | Sample type | Samples | % Sens | % Spec | Rickettsial species | Reference |
|---|---|---|---|---|---|---|---|---|
| 1975 | USA | IFA | Serum | 571 | 85–97 | 99–100 | RMSF | [69] |
| 1975 | USA | IFA | Serum | 42 | 86 | NA | SFG | [70] |
| 1983 | USA | IFA-IgG/M | Serum | 50 | 83–100 | 100 | RMSF | [71] |
| 1981–1984 | Georgia | IFA | Serum | 1774 | 94 | NA | RMSF | [65] |
| 1983 | USA | ELISA-IgG ELISA-IgM |
Serum | 50 | 93–100 100 |
100 100 |
RMSF | [71] |
| 2009 | India | ELISA-IgM | Blood | 161 | 91 | 100 | SFG & STG | [72] |
| 2010–2012 | India | ELISA-IgG/M | Serum | 103 | 85 | 100 | SFG | [73] |
| 1975 | USA | CF | Serum | 571 | 85.4 | 100 | RMSF | [69] |
| 1975 | USA | CF | Serum | 42 | 62 | NA | SFG | [70] |
| 1981–1984 | Georgia | CF | Serum | 1774 | 63 | NA | RMSF | [65] |
| 1975 | USA | WF | Serum | 42 | 21 | NA | SFG | [70] |
| 1981–1984 | Georgia | WF (OX-19) WF (OX-2) |
Serum | 1774 | 70 47 |
NA NA |
RMSF | [65] |
| 2001 | Sri Lanka | WF | Serum | 30 | 33 | 46 | SFG, STG & TG | [66] |
| 2009 | Central India | WF | Serum | 157 | 49 | 96 | SFG & STG | [72] |
| 1975 | USA | MA | Serum | 571 | 56.1 | 99 | RMSF | [69] |
NA, not available; ST, scrub typhus; MT, murine typhus; Sens, sensitivity; Spec, specificity; WF, Weil–Felix; CF, complement fixation; IFA, immunofluorescence assay; RMSF, Rocky Mountain spotted fever; SFG, spotted fever group Rickettsia; STG, scrub typhus group; TG, typhus group.
Enzyme-linked immunosorbent assays (ELISAs) are widely used and demonstrate high sensitivity and specificity. In an Indian study, researchers used a commercial R. conorii ELISA IgG/IgM kit that demonstrated 85% sensitivity and 100% specificity [73] (Table 2). The advantage of ELISA methodologies is that they allow screening of large batches of samples, which is less time-consuming, and provide more objectivity compared to the subjective IFA, as there is no reader-bias due to the use of optical density (OD) by ELISA readers. A diagnostic cut-off OD of ⩾0.5 has been applied previously to the SFG ELISA developed by the US Naval Medical Research Center [74, 75], however, there is no independent validation of the use of this cut-off. However, ELISAs may not recognize all Rickettsia pathogens and may be negative at the acute stage of an infection depending on the isotype (IgM or IgG) (Fig. 1), and results can only be obtained after seroconversion (around 5–14-days post onset of illness) [71,72]. Collecting only a single sample from patients can often make diagnosis difficult [76] and it is recommended that both acute and convalescent samples are collected whereby a significant rise in antibody levels is then considered as indicative of an active infection.
Need to improve diagnosis and clinical awareness of SFG
There is an urgent need to improve diagnosis and awareness of SFG in Asia. Generally, SFG infections are significantly neglected and under recognized in Asia. Spotted fever rickettsiae diseases often have non-specific clinical symptoms and may be overlooked by physicians [61, 77], although awareness is increasing with improved diagnostics [17, 33, 37] and publication of findings. SFG are also under recognized by laboratory staff as SFG are obligate intracellular bacteria whose culture in vitro is complicated, lengthy, expensive and often reserved to specialized laboratories equipped with BSL3 level containment facilities [78].
SFG may be misidentified as TG Rickettsia due to serological cross-reactions in which the rOmpB protein appears to play an important role [79–81]. Moreover, sequencing data indicates that some 50 amino acids in rOmpB of R. japonica (SFG) are identical to that of R. typhi (TG) [82]. Antibodies for R. typhi have demonstrated greater cross-reaction with R. conorii than R. rickettsii and higher cross-reactivity with IgM than IgG antibodies. However, there may be enough difference between end point titers to permit identity between TG and SFG [83]. The cross-reacting antibodies may also depend on the immunogenic responses of each patient [82] and multiple infections and re-infections also make it difficult to distinguish between species due to the broadness of the immune response.
In the case of molecular diagnostic tests, adoption of PCR technologies is limited due to cost, lack of expertise and technical issues. Although often highly sensitive and specific, PCR techniques have limitations as false-negative results may result from low quantities of SFG and the transient nature of these pathogens present in the circulating blood [84]. In reality, the major limiting factor for the use of PCR methods is the need for expensive thermocycler equipment, reagents and specific primers. Despite this, these techniques should be encouraged as PCR assays are highly useful, they allow accurate SFG identification, enable the discovery of novel species, are increasingly affordable, reproducible and less-time consuming with high specificity and sensitivity especially in the early phase of infection [85].
Diagnostic cutoffs in seroprevalence studies
Seroprevalence studies provide important information regarding the distribution of SFG in community- or hospital-based settings. The choice of diagnostic cut off may greatly influence the seroprevalence results, with a low cut off often giving high sensitivity but low specificity, whereas a high cut off will give low sensitivity but high specificity [69]. Seroprevalence studies of SFG in Asia have used IFA, IIP and ELISA techniques (Table 1) with IFA considered to be the diagnostic gold standard for the quantitative detection of rickettsial antibodies [30]. However, there appears to be a lack of consistency in the application of the cut-off titer criteria depending on the geographic location (Table 2). In the case of seroprevalence studies, the application of a consistent regional cut off will more readily reflect true endemicity and enable comparability between countries and regions. In Southeast, South and East Asia, studies have used reasonably consistent cut-off titers of ⩾1:64 in Philippines [86], Sri Lanka [87], Thailand [3] and Bangladesh [88] and in South Korea and Taiwan, a cut-off of 1:40 is considered as positive [89, 90]. However, different cut-off values have been reported for IgM and IgG isotypes, such as ⩾1:128 for IgG and ⩾1:64 for IgM in Thailand [3]. Interestingly, Denmark has used a cutoff as high as 1:512 [79]. There is a need to standardize diagnostic cut-offs for seroprevalence studies to allow easier comparability of results.
Conclusions
SFG Rickettsia causes a large number of emerging and re-emerging infectious diseases with a worldwide distribution. Its occurrence in LMICs exacerbates an already problematic diagnostic issue. With a limited range of suitable sensitive and specific tests available, the additional compounding factor of the need for cheap and easy to use diagnostic tests inputs additional burden on the populations exposed to these pathogens. Despite many tests being available, their lack of suitability for use in resource-limited regions is of concern, as many require technical expertise, expensive equipment and reagents. In addition, many existing diagnostic tests still require rigorous validation in the regions and populations where these tests may be used, in particular to establish coherent and worthwhile cut-offs. Possibly the best strategy would be to use a qPCR and IFA in tandem whereby, if the specimen is collected early enough in the infection where there will be no antibodies, there is a greater chance of a PCR-positive result. Conversely, if there are detectable antibodies, it is less likely that there will be a positive PCR result. We are in an age when more and more novel diagnostic tests are coming onto the market, and we need to ensure that these tests are suitable and appropriate for the diagnosis of rickettsial diseases, especially in low-income countries.
Acknowledgements
Stuart D. Blacksell and Matthew T. Robinson are supported by the Wellcome Trust of the United Kingdom.
References
- 1.Parola P et al. (2013) Update on tick-borne rickettsioses around the world: a geographic approach. Clinical Microbiology Reviews 26, 657–702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Acestor N et al. (2012) Mapping the aetiology of non-malarial febrile illness in Southeast Asia through a systematic review – terra incognita impairing treatment policies. PLoS One 7, e44269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bhengsri S et al. (2016) Sennetsu neorickettsiosis, spotted fever group, and typhus group rickettsioses in three provinces in Thailand. American Journal of Tropical Medicine and Hygiene 95, 43–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Aung AK et al. (2014) Rickettsial infections in Southeast Asia: implications for local populace and febrile returned travellers. American Journal of Tropical Medicine and Hygiene 91, 451–460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rodkvamtook W et al. (2013) Scrub typhus outbreak, northern Thailand, 2006–2007. Emerging Infectious Diseases 19, 774–777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Newton PN et al. (2019) A prospective, open-label, randomized trial of doxycycline versus azithromycin for the treatment of uncomplicated murine typhus. Clinical Infectious Diseases 68, 738–747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Vallee J et al. (2010) Contrasting spatial distribution and risk factors for past infection with scrub typhus and murine typhus in Vientiane City, Lao PDR. PLoS Neglected Tropical Diseases 4, e909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chikeka I and Dumler JS (2015) Neglected bacterial zoonoses. Clinical Microbiology and Infection 21, 404–415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Izzard L et al. (2010) Isolation of a novel Orientia species (O. chuto sp. nov.) from a patient infected in Dubai. Journal of Clinical Microbiology 48, 4404–4409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Luce-Fedrow A et al. (2018) A review of scrub typhus (Orientia tsutsugamushi and related organisms): then, now, and tomorrow. Tropical Medicine and Infectious Diseases 3, 8. doi: 10.3390/tropicalmed3010008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kato CY et al. (2013) Assessment of real-time PCR assay for detection of Rickettsia spp. and Rickettsia rickettsii in banked clinical samples. Journal of Clinical Microbiology 51, 314–317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Graves S and Stenos J (2009) Rickettsioses in Australia. Annals of the New York Acadamy of Sciences 1166, 151–155. [DOI] [PubMed] [Google Scholar]
- 13.Niang M et al. (1998) Prevalence of antibodies to Rickettsia conorii, Rickettsia africae, Rickettsia typhi and Coxiella burnetii in Mauritania. European Journal of Epidemiology 14, 817–818. [DOI] [PubMed] [Google Scholar]
- 14.Parola P (2004) Tick-borne rickettsial diseases: emerging risks in Europe. Comparative Immunology, Microbiology and Infectious Diseases 27, 297–304. [DOI] [PubMed] [Google Scholar]
- 15.Nanayakkara DM et al. (2013) Serological evidence for exposure of dogs to Rickettsia conorii, Rickettsia typhi, and Orientia tsutsugamushi in Sri Lanka. Vector Borne and Zoonotic Diseases 13, 545–549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Brown LD and Macaluso KR (2016) Rickettsia felis, an emerging flea-borne rickettsiosis. Current Tropical Medicine Reports 3, 27–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Luce-Fedrow A et al. (2015) Strategies for detecting rickettsiae and diagnosing rickettsial diseases. Future Microbiology 10, 537–564. [DOI] [PubMed] [Google Scholar]
- 18.Dieme C et al. (2015) Transmission potential of Rickettsia felis infection by Anopheles gambiae mosquitoes. Proceedings of the National Academy of Sciences. 112, 8088–8093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Colonne PM, Eremeeva ME and Sahni SK (2011) Beta interferon-mediated activation of signal transducer and activator of transcription protein 1 interferes with Rickettsia conorii replication in human endothelial cells. Infection and Immunity 79, 3733–3743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rydkina E, Turpin LC and Sahni SK (2010) Rickettsia rickettsii infection of human macrovascular and microvascular endothelial cells reveals activation of both common and cell type-specific host response mechanisms. Infection and Immunity 78, 2599–2606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gaywee J et al. (2007) Human infection with Rickettsia sp. related to R. japonica, Thailand. Emerging Infectious Diseases 13, 671–673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Young RP, Ip M and Bassett DC (1995) Fatal rickettsial meningitis in Hong Kong: a need for rapid laboratory diagnosis. Scandinavian Journal of Infectious Diseases 27, 527–528. [DOI] [PubMed] [Google Scholar]
- 23.Rossio R et al. (2015) Mediterranean spotted fever and hearing impairment: a rare complication. International Journal of Infectious Diseases 35, 34–36. [DOI] [PubMed] [Google Scholar]
- 24.Ben Mansour N et al. (2014) [Acute myocarditis complicating Mediterranean spotted fever. A case report]. Annales de Cardiologie et d'Angéiologie 63, 55–57. [DOI] [PubMed] [Google Scholar]
- 25.Botelho-Nevers E et al. (2005) Cerebral infarction: an unusual complication of Mediterranean spotted fever. European Journal of Internal Medicine 16, 525–527. [DOI] [PubMed] [Google Scholar]
- 26.Kodama K, Noguchi T and Chikahira Y (2000) [Japanese spotted fever complicated by acute respiratory failure]. Kansenshogaku Zasshi 74, 162–165. [DOI] [PubMed] [Google Scholar]
- 27.Takiguchi J et al. (2016) [Severe Japanese spotted fever complicated by acute respiratory failure in Kobe City]. Kansenshogaku Zasshi 90, 120–124. [DOI] [PubMed] [Google Scholar]
- 28.McBride WJ et al. (2007) Severe spotted fever group rickettsiosis, Australia. Emerging Infectious Diseases 13, 1742–1744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Santibanez S et al. (2013) Usefulness of rickettsial PCR assays for the molecular diagnosis of human rickettsioses. Enfermedades Infecciosas y Microbiología Clínica 31, 283–288. [DOI] [PubMed] [Google Scholar]
- 30.La Scola B and Raoult D (1997) Laboratory diagnosis of rickettsioses: current approaches to diagnosis of old and new rickettsial diseases. Journal of Clinical Microbiology 35, 2715–2727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kato C, Chung I and Paddock C (2016) Estimation of Rickettsia rickettsii copy number in the blood of patients with Rocky Mountain spotted fever suggests cyclic diurnal trends in bacteraemia. Clinical Microbiology and Infection 22, 394–396. [DOI] [PubMed] [Google Scholar]
- 32.Sonthayanon P et al. (2009) Association of high Orientia tsutsugamushi DNA loads with disease of greater severity in adults with scrub typhus. Journal of Clinical Microbiology 47, 430–434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kondo M et al. (2015) Improvement in early diagnosis of Japanese spotted fever by using a novel Rick PCR system. Journal of Dermatology 42, 1066–1071. [DOI] [PubMed] [Google Scholar]
- 34.Bechah Y, Socolovschi C and Raoult D (2011) Identification of rickettsial infections by using cutaneous swab specimens and PCR. Emerging Infectious Diseases 17, 83–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Denison AM et al. (2014) Detection of Rickettsia rickettsii, Rickettsia parkeri, and Rickettsia akari in skin biopsy specimens using a multiplex real-time polymerase chain reaction assay. Clinical Infectious Diseases 59, 635–642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ishikura M et al. (2003) Phylogenetic analysis of spotted fever group rickettsiae based on gltA, 17-kDa, and rOmpA genes amplified by nested PCR from ticks in Japan. Microbiology and Immunology 47, 823–832. [DOI] [PubMed] [Google Scholar]
- 37.Paris DH and Dumler JS (2016) State of the art of diagnosis of rickettsial diseases: the use of blood specimens for diagnosis of scrub typhus, spotted fever group rickettsiosis, and murine typhus. Current Opinions in Infectious Diseases 29, 433–439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Prakash JA et al. (2012) Molecular detection and analysis of spotted fever group Rickettsia in patients with fever and rash at a tertiary care centre in Tamil Nadu, India. Pathogens and Global Health 106, 40–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Roux V and Raoult D (2000) Phylogenetic analysis of members of the genus Rickettsia using the gene encoding the outer-membrane protein rOmpB (ompB). International Journal of Systematic Evolution and Microbiology 50, 1449–1455. [DOI] [PubMed] [Google Scholar]
- 40.Choi YJ et al. (2005) Evaluation of PCR-based assay for diagnosis of spotted fever group rickettsiosis in human serum samples. Clinical Diagnostic Laboratory Immunology 12, 759–763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Marquez FJ et al. (1998) Genotypic identification of an undescribed spotted fever group Rickettsia in Ixodes ricinus from southwestern Spain. American Journal of Tropical Medicine and Hygiene 58, 570–577. [DOI] [PubMed] [Google Scholar]
- 42.Jiang J et al. (2013) Molecular detection of Rickettsia felis and Candidatus Rickettsia asemboensis in fleas from human habitats, Asembo, Kenya. Vector Borne and Zoonotic Diseases 13, 550–558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Jiang J et al. (2004) Development of a quantitative real-time polymerase chain reaction assay specific for Orientia tsutsugamushi. American Journal of Tropical Medicine and Hygiene 70, 351–356. [PubMed] [Google Scholar]
- 44.Jiang J, Stromdahl EY and Richards AL (2012) Detection of Rickettsia parkeri and Candidatus Rickettsia andeanae in Amblyomma maculatum Gulf Coast ticks collected from humans in the United States. Vector Borne and Zoonotic Diseases 12, 175–182. [DOI] [PubMed] [Google Scholar]
- 45.Taylor AJ et al. (2016) Large-scale survey for tickborne bacteria, Khammouan Province, Laos. Emerging Infectious Diseases 22, 1635–1639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Maina AN et al. (2017) Molecular characterization of novel mosquito-borne Rickettsia spp. from mosquitoes collected at the Demilitarized Zone of the Republic of Korea. PLoS One 12, e0188327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Massung RF et al. (2001) Epidemic Typhus Meningitis in the Southwestern United States. Clinical Infectious Diseases 32, 979–982. [DOI] [PubMed] [Google Scholar]
- 48.Tzianabos T, Anderson BE and McDade JE (1989) Detection of Rickettsia rickettsii DNA in clinical specimens by using polymerase chain reaction technology. Journal of Clinical Microbiology 27, 2866–2868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Paddock CD et al. (2004) Rickettsia parkeri: a newly recognized cause of spotted fever rickettsiosis in the United States. Clinical Infectious Diseases 38, 805–811. [DOI] [PubMed] [Google Scholar]
- 50.Webb L et al. (1990) Detection of murine typhus infection in fleas by using the polymerase chain reaction. Journal of Clinical Microbiology 28, 530–534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Roux V et al. (1997) Citrate synthase gene comparison, a new tool for phylogenetic analysis, and its application for the rickettsiae. International Journal of Systematic Bacteriology l 47, 252–261. [DOI] [PubMed] [Google Scholar]
- 52.Jiang J et al. (2005) Human Infection with Rickettsia honei, Thailand. Emerging Infectious Diseases 11, 1473–1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Regnery RL, Spruill CL and Plikaytis BD (1991) Genotypic identification of rickettsiae and estimation of intraspecies sequence divergence for portions of two rickettsial genes. Journal of Bacteriology 173, 1576–1589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Labruna MB et al. (2004) Rickettsia species infecting Amblyomma cooperi ticks from an area in the state of Saõ Paulo, Brazil, where Brazilian spotted fever is endemic. Journal of Clinical Microbiology 42, 90–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Stenos J, Graves SR and Unsworth NB (2005) A highly sensitive and specific real-time PCR assay for the detection of spotted fever and typhus group Rickettsiae. American Journal of Tropical Medicine and Hygiene 73, 1083–1085. [PubMed] [Google Scholar]
- 56.Fournier PE and Raoult D (2009) Current knowledge on phylogeny and taxonomy of Rickettsia spp. Annals of the New York Acadamy of Sciences 1166, 1–11. [DOI] [PubMed] [Google Scholar]
- 57.Eremeeva ME, Dasch GA and Silverman DJ (2003) Evaluation of a PCR assay for quantitation of Rickettsia rickettsii and closely related spotted fever group rickettsiae. Journal of Clinical Microbiology 41, 5466–5472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wikswo ME et al. (2008) Detection and identification of spotted fever group rickettsiae in Dermacentor species from southern California. Journal of Medical Entomology 45, 509–516. [DOI] [PubMed] [Google Scholar]
- 59.Jiang J et al. (2005) Phylogenetic analysis of a novel molecular isolate of spotted fever group Rickettsiae from northern Peru: Candidatus Rickettsia andeanae. Annals of the New York Acadamy of Sciences 1063, 337–342. [DOI] [PubMed] [Google Scholar]
- 60.Pan L et al. (2012) Rapid, simple, and sensitive detection of the ompB gene of spotted fever group rickettsiae by loop-mediated isothermal amplification. BMC Infectious Diseases 12, 254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Raoult D and Dasch GA (1995) Immunoblot cross-reactions among Rickettsia, Proteus spp. and Legionella spp. in patients with Mediterranean spotted fever. FEMS Immunology and Medical Microbiology 11, 13–18. [DOI] [PubMed] [Google Scholar]
- 62.McQuiston JH et al. (2014) Inadequacy of IgM antibody tests for diagnosis of Rocky Mountain Spotted Fever. American Journal of Tropical Medicine and Hygiene 91, 767–770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Cruickshank R (1927) The Weil-Felix reaction in typhus fever. Journal of Hygiene (Lond) 27, 64–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Amano K et al. (1992) Serological studies of antigenic similarity between Japanese spotted fever rickettsiae and Weil-Felix test antigens. Journal of Clinical Microbiology 30, 2441–2446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kaplan JE and Schonberger LB (1986) The sensitivity of various serologic tests in the diagnosis of Rocky Mountain spotted fever. American Journal of Tropical Medicine and Hygiene 35, 840–844. [DOI] [PubMed] [Google Scholar]
- 66.Kularatne SA and Gawarammana IB (2009) Validity of the Weil-Felix test in the diagnosis of acute rickettsial infections in Sri Lanka. Transactions of the Royal Society of Tropical Medicine and Hygiene 103, 423–424. [DOI] [PubMed] [Google Scholar]
- 67.Tay ST and Rohani MY (2002) The use of the indirect immunoperoxidase test for the serodiagnosis of rickettsial diseases in Malaysia. Southeast Asian Journal of Tropical Medicine and Public Health 33, 314–320. [PubMed] [Google Scholar]
- 68.Premaratna R et al. (2008) Rickettsial infections and their clinical presentations in the Western Province of Sri Lanka: a hospital-based study. International Journal of Infectious Diseases s 12, 198–202. [DOI] [PubMed] [Google Scholar]
- 69.Newhouse VF et al. (1979) A comparison of the complement fixation, indirect fluorescent antibody, and microagglutination tests for the serological diagnosis of rickettsial diseases. American Journal of Tropical Medicine and Hygiene 28, 387–395. [DOI] [PubMed] [Google Scholar]
- 70.Shirai A, Dietel JW and Osterman JV (1975) Indirect hemagglutination test for human antibody to typhus and spotted fever group rickettsiae. Journal of Clinical Microbiology 2, 430–437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Clements ML et al. (1983) Serodiagnosis of Rocky Mountain spotted fever: comparison of IgM and IgG enzyme-linked immunosorbent assays and indirect fluorescent antibody test. Journal of Infectious Diseases 148, 876–880. [DOI] [PubMed] [Google Scholar]
- 72.Rathi NB et al. (2011) Rickettsial diseases in central India: proposed clinical scoring system for early detection of spotted fever. Indian Pediatrics 48, 867–872. [DOI] [PubMed] [Google Scholar]
- 73.Kalal BS et al. (2016) Scrub typhus and spotted fever among hospitalised children in South India: Clinical profile and serological epidemiology. Indian Journal of Medical Microbiology 34, 293–298. [DOI] [PubMed] [Google Scholar]
- 74.Trung NV et al. (2017) Seroprevalence of scrub typhus, typhus, and spotted fever among rural and urban populations of Northern Vietnam. American Journal of Tropical Medicine and Hygiene 96, 1084–1087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Khan SA et al. (2016) Seroepidemiology of rickettsial infections in Northeast India. Transactions of the Royal Society of Tropical Medicine and Hygiene 110, 487–494. [DOI] [PubMed] [Google Scholar]
- 76.Tsai KH et al. (2009) African tick bite Fever in a Taiwanese traveller returning from South Africa: molecular and serologic studies. American Journal of Tropical Medicine and Hygiene 81, 735–739. [DOI] [PubMed] [Google Scholar]
- 77.Salgado Lynn M et al. (2018) Spotted fever rickettsiosis in a wildlife researcher in Sabah, Malaysia: a case study. Tropical Medicine and Infectious Diseases 3, 29–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Ammerman NC, Beier-Sexton M and Azad AF (2008) Laboratory maintenance of Rickettsia rickettsii. Current Protocols in Microbiology 11, 1:3A.5.1–3A.5.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Kantso B et al. (2009) Evaluation of serological tests for the diagnosis of rickettsiosis in Denmark. Journal of Microbiological Methods 76, 285–288. [DOI] [PubMed] [Google Scholar]
- 80.Keysary A and Strenger C (1997) Use of enzyme-linked immunosorbent assay techniques with cross-reacting human sera in diagnosis of murine typhus and spotted fever. Journal of Clinical Microbiology 35, 1034–1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Ormsbee R et al. (1978) Antigenic relationships between the typhus and spotted fever groups of rickettsiae. American Journal of Epidemiology 108, 53–59. [PubMed] [Google Scholar]
- 82.Uchiyama T et al. (1995) Cross-reactivity of Rickettsia japonica and Rickettsia typhi demonstrated by immunofluorescence and Western immunoblotting. Microbiology and Immunology 39, 951–957. [DOI] [PubMed] [Google Scholar]
- 83.Hechemy KE et al. (1989) Cross-reaction of immune sera from patients with rickettsial diseases. Journal of Medical Microbiology 29, 199–202. [DOI] [PubMed] [Google Scholar]
- 84.Kidd L et al. (2008) Evaluation of conventional and real-time PCR assays for detection and differentiation of Spotted Fever Group Rickettsia in dog blood. Veterinary Microbiology 129, 294–303. [DOI] [PubMed] [Google Scholar]
- 85.Znazen A et al. (2015) Comparison of two quantitative real time PCR assays for Rickettsia detection in patients from Tunisia. PLoS Neglected Tropical Diseases 9, e0003487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Camer GA et al. (2003) Detection of antibodies against spotted fever group Rickettsia (SFG), typhus group Rickettsia (TGR), and Coxiella burnetii in human febrile patients in the Philippines. Jpn Journal of Infectious Diseases 56, 26–28. [PubMed] [Google Scholar]
- 87.Premaratna R et al. (2014) Rickettsial infection among military personnel deployed in Northern Sri Lanka. BMC Infectious Diseases 14, 3864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Faruque LI et al. (2017) Prevalence and clinical presentation of Rickettsia, Coxiella, Leptospira, Bartonella and chikungunya virus infections among hospital-based febrile patients from December 2008 to November 2009 in Bangladesh. BMC Infectious Diseases 17, 141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Jang WJ et al. (2005) Seroepidemiology of spotted fever group and typhus group rickettsioses in humans, South Korea. Microbiology and Immunology 49, 17–24. [DOI] [PubMed] [Google Scholar]
- 90.Lai CH et al. (2014) Human spotted fever group rickettsioses are underappreciated in southern Taiwan, particularly for the species closely-related to Rickettsia felis. PLoS One 9, e95810. [DOI] [PMC free article] [PubMed] [Google Scholar]