Abstract
The complete genome sequence of Narcissus yellow stripe potyvirus (NYSV) isolated from Narcissus tazetta cv. Paperwhite exhibiting leaf chlorotic stripe symptoms was determined for the first time from India. The viral genome sequence contained 9650 nucleotides that encode a large polyprotein (372.36 kDa) of 3103 amino acids. The comparison of the NYSV genome sequences with corresponding sequences of other potyviruses revealed 90–97% identities and closest phylogenetic relationships with NYSV-Zhangzhou-1 and -ZZ-2 isolates infecting N. tazetta reported from China. Therefore, the NYSV isolate understudy was considered as a new member of NYSV and designated as NYSV–NAR2.
Keywords: Narcissus tazetta, Potyvirus, Complete genome, Sequence analyses, Narcissus yellow stripe virus
Introduction
Narcissus tazetta L., a member of Amaryllidaceae family, is an ornamental plant grown in garden beds and pots for its attractive blooms. It is used as a cut flower for horticulture industry and in perfumery for its sweet fragrance. Cultivation of N. tazetta under field/polyhouse conditions has been reported to be affected by a number of RNA viruses of the genera Carlavirus, Maculavirus, Nepovirus, Potexvirus and Potyvirus (Brunt 1995; Wylie and Jones 2012; Ohshima et al. 2016; Raj et al. 2018). Amongst them Potyviruses are most prevalent and found to be more infectious to narcissus which resulted in drastic reduction in its quality and quantity of blooms (Brunt 1995; Aminuddin et al. 1999; Wylie and Jones 2012; Raj et al. 2018).
Potyviruses are flexuous filamentous particles of 680–900 nm in length and 11–12 nm in width. They belong to the family Potyviridae that contain a single stranded positive sense RNA of approximately ten thousand base pair long. Various potyviruses such as Narcissus yellow stripe virus (NYSV), Narcissus late season yellow virus (NLSYV), Narcissus degenerate virus (NDV), Cyrtanthus elatus virus A (CyEVA) and Ornithogalum mosaic virus (OrMV) have been reported to infect narcissus (Chen et al. 2006; Ohshima et al. 2016). Previously the full genome sequences of NYSV and NLSYV infecting narcissus were published from China and Australia respectively (Chen et al. 2006; Wylie and Jones 2012).
In India, an uncharacterized potyvirus, Lycoris potyvirus and Cyrtanthus elatus virus-A (CyEVA) associated with leaf yellow stripe disease of N. tazeeta have been investigated (Aminuddin et al. 1999; Yadav and Khan 2008; Kumar et al. 2015). Further, the full length genome sequence of an Indian isolate of CyEVA infecting N. tazeeta has also been published recently (Raj et al. 2018). In present study, the complete genome sequence of NYSV infecting N. tazeeta cv. Paperwhite plants is being reported first time from India.
Materials and methods
The leaf samples of N. tazetta cv. Paperwhite exhibiting chlorotic leaf stripes were collected from the cultivated field at NBRI, Lucknow. Partial purification of virus was done and transmission electron microscopy (TEM) was carried out as described earlier (Kaur et al. 2015). The partially purified virus was inoculated on Chenopodium album, Datura innoxia, and Narcissus tazetta plants at 3–4 leaf stage. After inoculation local and systemic symptoms on inoculated plants were recorded up to 35 days post inoculation (dpi).
For virus detection, the total RNA was extracted from 100 mg leaf tissue of infected narcissus using TRI reagent (Sigma Aldrich, Missouri, USA) and reverse transcription-PCR was performed using potyvirus degenerate primers (Pot-I/Pot-II, Gibbs and Mackenzie 1997). For full length genome amplification, the four set of primers: Pot-I/Pot-II, CI-F/NIb-Pot-3(Ha et al. 2008), HP-F/CI-R (Yakoubi et al. 2008) and 5′RACE/HP-R (Lucinda et al. 2010) yielding the expected size amplicons of ~ 1.5, 3.0, 3.0 and 2.0 kb, respectively were used following the strategy described as earlier (Raj et al. 2018). For amplification of 5′ end, the First Choice RLM-RACE kit was employed. The size of DNA fragment was assessed on 1% agarose gel with comparison of 1 kb DNA ladder. Amplicons were gel eluted using Wizard® SV gel and PCR clean-up system (Promega, USA). The eluted products were ligated into pGEM-T vector and transformed into E. coli (DH5α) cells. The positive transformants were screened by colony PCR and clones were sequenced. The obtained sequence data were edited and assembled using BIOEDIT tool (http://www.mbio.ncsu.edu/bioedit/bioedit.html) to eliminate any sequence ambiguity. The consensus sequence for full length genome was determined and submitted to GenBank.
The NCBI tool open reading frames (ORF) finder (www.ncbi.nlm.nih.gov/projects/gorf/) was used to analyze the ORFs encoded by the genome and their putative proteins were translated using ExPasy tool (http://web.expasy.org/translate/). The complete analyzed sequence of the potyvirus isolate was compared with respective sequences of other potyviruses available in NCBI using BLASTn (http://blast.ncbi.nlm.nih.gov/Blast.cgi) for potyvirus identification. DiAlign tool (http://www.genomatix.de/cgi-bin/dialign/dialign.pl) was used to obtain the nucleotide and amino acid identity of ORFs of selected potyvirus isolates. The phylogenetic analysis of full genome sequence was also performed by Molecular Evolutionary Genetic Analysis (MEGA) v6.1 tool using Maximum Likelihood algorithm at 1000 boostrap value (Tamura et al. 2013).
Further, the presence of NYSV in infected N. tazetta samples were also confirmed by nucleic acid spot hybridization (NASH) assay performed as described earlier (Raj et al. 2018) using the probe prepared from cloned NYSV (KM066972). The 2 µg RNA of uninfected N. tazetta plant was used as negative control whereas 200 ng cloned DNA of NYSV (KM066972) was used as positive controls. Blotted membranes were hybridized with probe prepared from cloned NYSV (KM066972) by random primer labelling method and pre-hybridization, hybridization and washing steps were performed following the standard method as described by Raj et al. (2018). The hybridization signals were observed after exposure to phosphor imaging screen in Typhoon Imaging Phosphor imager (GE Healthcare Life Sciences, USA).
Results and discussion
During survey in 2014, the N. tazetta plants showing chlorotic stripe disease symptoms with 99% disease incidence were observed in a cultivated field at NBRI, Lucknow. Virus infected plants showed chlorotic stripes on leaves and stunting symptoms (Fig. 1a, b). More or less the similar disease symptoms were described earlier on narcissus plants infected by NYSV, OrMV, NLSYV and CyEVA potyviruses (Chen et al. 2006; Yadav and Khan 2015; Wylie and Jones 2012; Wylie et al. 2014; Raj et al. 2018).
Fig. 1.
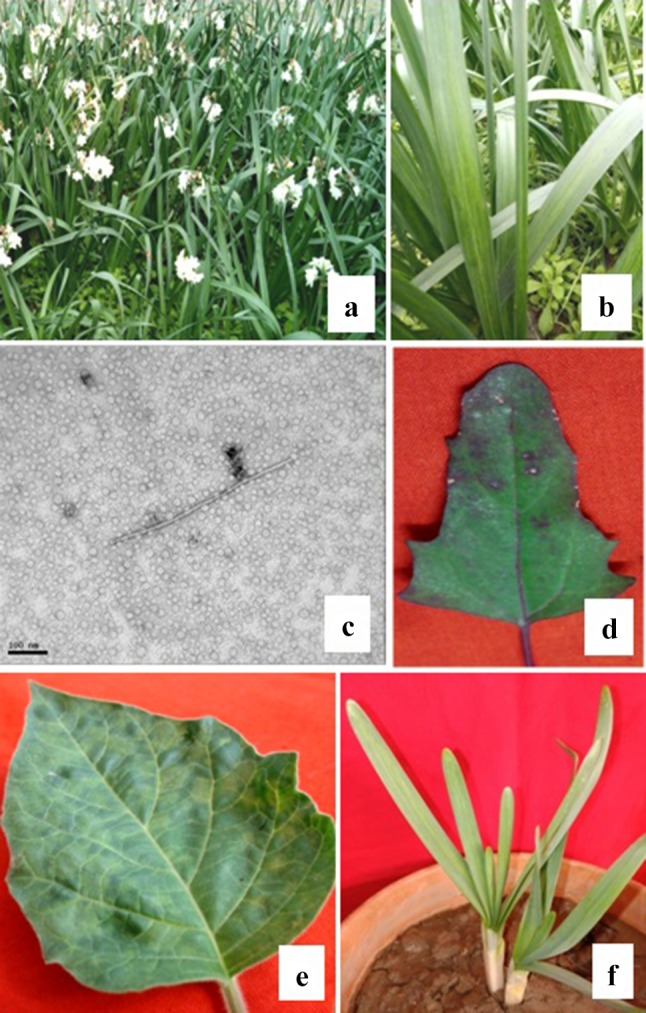
a Field view of N. tazetta plants and a close view of diseased N. tazetta showing yellow stripes on leaves (b). TEM of partially purified preparation of infected N. tazetta showing a typical flexuous rod shaped virus particle of 680 nm × 11 nm in size, bar 100 nm, (c). Partially purified virus inoculated plants showing symptoms: necrotic local lesions on C. album at 10 dpi (d), systemic mosaic on D. innoxia at 30 dpi (e) and systemic yellow stripes on N. tazetta at 35 dpi (f)
The initial virus detection was done by TEM from the partially purified virus preparation, where the typical flexuous rod-shaped virus particles of 680 nm × 11 nm were observed (Fig. 1c) that indicated the presence of potyviruses as reported earlier (Kaur et al. 2015). Further the purified virus was inoculated on some host plants viz., C. album, D. innoxia, and N. tazetta. The virus inoculated C. album plant developed local necrotic lesions at 10 days post inoculation (dpi) while D. innoxia developed systemic mosaic at 30 dpi, and N. tazetta developed yellow stripe at 35 dpi (Fig. 1d–f).
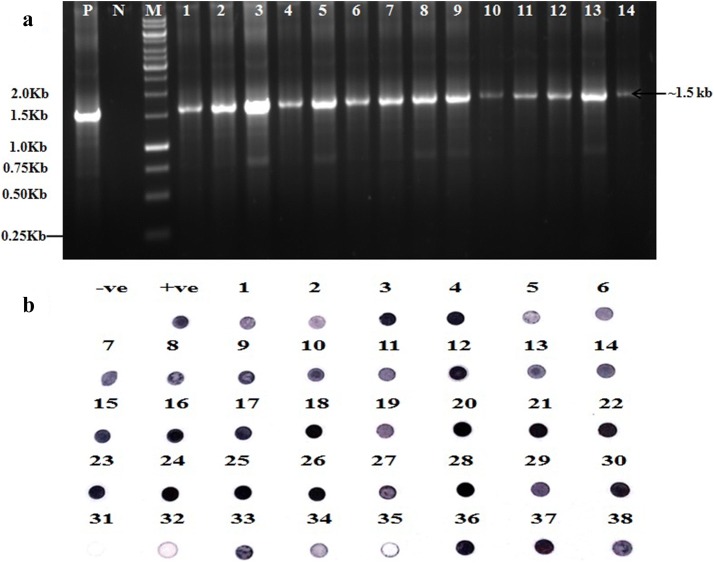
The presence of potyvirus was confirmed by RT-PCR using degenerate primers of potyvirus Pot-I/Pot-II (Gibbs and Mackenzie 1997). The RT-PCR resulted in successful amplification of expected size ~ 1.5 kb band in all 14 symptomatic narcissus samples (Fig. 2a). Sequence analysis of randomly selected two cloned PCR products: accession numbers JQ686724 (NAR-1) and KM066972 (NAR-2) suggested the presence of NYSV potyvirus in N. tazetta. The presence of NYSV in other samples of N. tazetta showing leaf stripe symptoms in field were also confirmed by NASH assay using the probe prepared from cloned NYSV (KM066972). The result revealed presence of NYSV in 37 out of 38 samples that showed positive signals of hybridization with the probe. Among them 35 sample showed the strong signals similar to that of a positive control while two samples showed weak signals (Fig. 2b) confirming ~ 99% disease incidence in field.
Fig. 2.
a RT-PCR using potyvirus degenerate primers (Pot-I/Pot-II, Gibbs and Mackenzie 1997) showing ~ 1.5 kb band in all 14 symptomatic samples of N. tazetta (lanes 1–14) similar as in a positive control (P) but not in healthy N. tazetta plants (N). M = 1.0 kb DNA ladder marker. b Nucleic acid spot hybridization test using a probe developed from cloned coat protein gene of NYSV (KM066972) showing positive signals of hybridization with the probe in 37 N. tazetta leaf samples similar as a positive control (P cloned DNA of NYSV) but not in a negative control (N healthy N. tazetta)
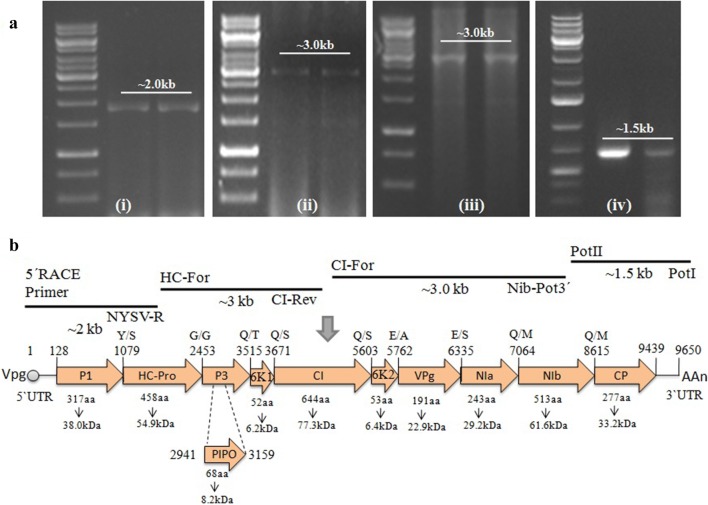
Further, the sample NAR-2 representing partial genome sequence of NYSV was selected for full length genome amplification by RT-PCR and RACE using the available potyvirus degenerate primers targeting the different conserved motifs of genome (Gibbs and Mackenzie 1997; Ha et al. 2008; Yakoubi et al. 2008; Lucinda et al. 2010). The Pot-I/Pot-II primers amplified the ~ 1.5 kb long region of polyadenylated 3′ end of 3′UTR and -GNNS- motif of partial nuclear inclusion B (Nlb) region (Fig. 3a-i) whereas, the other CI-F/NIb-Pot-3 primers amplified the GxVGSGKST motif in cylindrical inclusion (CI) protein and partial Nlb region of ~ 3.0 kb in size (Fig. 3a-ii). The region from partial CI to HC region of ~ 3.0 kb was amplified using HP-F/CI-R primers (Fig. 3a-iii). Finally RACE kit was used for amplification of remaining 5′ end to HC region of ~ 2.0 kb in size (Fig. 3a-iv). The amplified products were cloned, sequenced and assembled sequence data as full length genome was deposited in NCBI GenBank database under the accession number KU516386 (NYSV-NAR2).
Fig. 3.
a RT-PCR products of full length genome of NYSV using various potyvirus specific pair of primers: (i) 5′ RACE Inner primer/NYSV-R, (ii) HPFor/CIRev (iii) CIFor/NIbPot-3′ and (iv) Pot I/Pot II showing ~ 2.0 kb, 3.0 kb, 3.0 kb and 1.5 kb bands, respectively in NYSV infected narcissus samples (lanes 1 and 2). M = 1 kb DNA ladder marker. b The genome organization of NYSV-NAR2 isolate (KU516386). Arrow indicates the size, location and orientation of ORFs in the potyvirus genome understudy. Below the arrow, the number of amino acids and predicted molecular weight (in kDa) is shown. The polyadenylated tail is shown by AAn and VPg abbreviates for viral protein genome-linked
The full-length genome of NAR2 isolate (KU516386) was of 9650 nucleotides long excluding the polyA tail. The genome organisation was found similar to a typical member of genus potyvirus (Fig. 3b). The translated product of the ORF contained large polyprotein of 372.36 kDa in size with 3103 amino acids. This polyprotein further yields ten well known proteins (P1, HC-Pro, P3, 6K1, CI, 6K2, NIa-VPg, NIa-Pro, Nib and CP) along with a newly identified PIPO protein (Fig. 3b). These 11 proteins have amino acids/molecular weight of 317/38.0, 458/54.9, 55/6.2, 354/42.4, 644/77.3, 53/6.4, 191/22.9, 243/29.2, 513/61.6, 277/33.2 and 68/8.2, respectively (Fig. 3b). Further, the NYSV-NAR2 isolate contained some putative proteolytic cleavage sites as Y/S, G/G, Q/T, Q/S, Q/S, E/A, E/S, Q/M and Q/M (Fig. 3b) identified from alignments with full length polyprotein sequences of available potyviruses in NCBI database. The proteolytic cleavage sites in NYSV-NAR2 isolate were similar as in case of NYSV isolates (Wylie et al. 2014) with exception of Q/M at the NIb/CP junction instead of Q/S. The NLSYV-Zhangzhou, Marijiniup8 and Marijiniup9 isolates revealed differences at two cleavage sites, one at P1/HC-Pro junction where they possess F/S or F/T instead of Y/S amino acid combination and second at P3/6K1 junction having Q/V or Q/A instead of Q/T (Data not shown). Conserved potyvirus motifs like FRNK and IGN in the central region of the HC-Pro (Shiboleth et al. 2007; Cronin et al. 1995), GDD in the Nlb, and DAG motif in the CP region (Peng et al. 1998) are also present in NYSV-NAR2 isolate.
The full length genome data of the NYSV-NAR2 isolate was also compared with respective sequences of other potyviruses available in NCBI (Table 1) by BLASTn tool for virus identification. During BLASTn analysis, the NYSV-NAR2 isolate (KU516386) shared highest 93% identities with complete nucleotide sequence of NYSV of Chinese narcissus (AM158908, Shuang et al. 2012). It also shared 82% identities with two Narcissus virus-1 isolates of Japan (LC31498, LC31499) and 76% identities with NYSV-ZZ2 isolate of China (JQ911732), two NYSV isolates of Japan (LC31493, LC31495) and 74% identities with two isolates of NLSYV–Marijiniup8 (KC691259, Wylie et al. 2014) of Australia and NLSYV isolate Zhangzhou (JQ326210) of China.
Table 1.
Details of virus isolates of NYSV and other potyvirus isolates used in present and previous studies
| Virus | Isolate | Host | Location | Genome length (nt) | GenBank Accession | Source |
|---|---|---|---|---|---|---|
| NYSV | NAR2 | Narcissus tazetta | India | 9650 | KU516386 | This study |
| NYSV | NY-OI1 | Narcissus tazzeta | Japan | 9626 | LC314391 | Ohshima et al. (2016) |
| NYSV | NY-KM1O | Narcissus tazzeta | Japan | 9637 | LC314392 | Ohshima et al. (2016) |
| NYSV | NY-KM1P | Narcissus tazzeta | Japan | 9639 | LC314393 | Ohshima et al. (2016) |
| NYSV | NY-CB5 | Narcissus tazzeta | Japan | 9630 | LC314394 | Ohshima et al. (2016) |
| NYSV | NY-EH173 | Narcissus tazzeta | Japan | 9630 | LU314395 | Ohshima et al. (2016) |
| NYSV | NY-HG19 | Narcissus tazzeta | Japan | 9629 | LU314396 | Ohshima et al. (2016) |
| NYSV | NY-HG27 | Narcissus tazzeta | Japan | 9629 | LU314397 | Ohshima et al. (2016) |
| NYSV | Zhangzhou-1 | Narcissus tazetta | China | 9650 | AM158908 | Chen et al. (2006) |
| NYSV | Zhangzhou-2 | Narcissus tazetta | China | 9650 | NC_011541 | Chen et al. (2006) |
| NYSV | ZZ2 | Narcissus tazetta | China | 9654 | JQ911732 | Shuang et al. (2012) |
| NYSV | Marijiniup3 | Narcissus spp. | Australia | 9647 | JQ395042 | Wylie and Jones (2012) |
| NLSYV | Zhangzhou | Narcissus tazetta | China | 9651 | JQ326210 | Jenner et al. (2000) |
| NLSYV | Marijiniup8 | Narcissus spp. | Australia | 9687 | KC691259 | Wylie et al. (2014) |
| NLSYV | Marijiniup9 | Narcissus spp. | Australia | 9577 | JX156421 | Wylie et al. (2014) |
| CyEVA | NBRI16 | Narcissus tazetta | India | 9942 | KX575832 | Raj et al. (2018) |
| CyEVA | Marijiniup7-1 | Cyrtanthus elatus | Australia | 9908 | NC_017977 | Wylie and Jones (2012) |
| ScaMV | -1 | Allium chinense | China | 9324 | NC_003399 | Brunt (1995) |
| ScaMV | -2 | Allium chinense | China | 9324 | AJ316084 | Chen et al. (2006) |
| TuMV | TIGD | Tigridia spp. | Germany | 9798 | AB701735 | Nguyen et al. (2013) |
| TuMV | NZ403B | Lepidium oleraceum | New Zealand | 9796 | AB989658 | Yasaka et al. (2015) |
| JYMV | M | Dioscorea japonica | Japan | 9760 | NC_000947 | Chen et al. (2006) |
| PWV | MU2 | Passiflora caerulea | Australia | 9682 | NC_014790 | Wylie et al. (2014) |
Virus acronyms: NYSV = Narcissus yellow stripe virus, JYMV = Japanese yam mosaic virus, NLSYV = Narcissus late season yellows virus, CyEVA = Cyrtanthus elatus virus-A, ScaMV = Scallion mosaic virus, TuMV = Turnip mosaic virus, PWV = Passion fruit woodiness virus
The Genomatix tool was used to identify ORFs from nucleotide and amino acid sequence of the selected potyvirus isolates (Table 2). The pairwise sequence comparisons of NYSV-NAR2 isolate (KU516386) with other potyvirus isolates considered for study at complete genome nucleotides and deduced amino acids of polyprotein gene revealed highest 90% and 97% identities, respectively with NYSV-Zhangzhou-1 and ZZ-2 isolates reported from China infecting N. tazetta cv. Chinensis (Table 2). While the identities were 58–74% and 77–85% at nt of complete genome and at aa of polyprotein gene, respectively of other NYSV isolates considered for the study. It also showed 54-55% (nt) and 75% (aa) identities with Zhangzhou, Marijiniup8 and Marijiniup9 isolates of NLSYV reported from China and Australia, respectively (Table 2).
Table 2.
Percent (%) identities of full length genome of NAR-2 isolate (KU516386) with respective sequences of other potyviruses at nucleotides and amino acid levels obtained using DiAlign tool
| GenBank accession | Virus | Isolate | Location | Percent identity | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Complete genome (polypeptide) | Open reading frames | |||||||||||||
| P1 | HC-Pro | P3 | 6K1 | CI | 6K2 | VPg | NIa | NIb | CP | |||||
| AM158908 | NYSV | Zhangzhou-1 | China | 90 (97) | 100 (100) | 86 (93) | 100 (99) | 100 (100) | 83 (96) | 77 (94) | 87 (97) | 100 (100) | 92 (96) | 95 (96) |
| NC_011541 | NYSV | Zhangzhou-2 | China | 90 (97) | 100 (100) | 86 (93) | 100 (99) | 100 (100) | 83 (96) | 77 (94) | 87 (97) | 100 (100) | 92 (96) | 95 (96) |
| LU314396 | NYSV | NY-HG19 | Japan | 74 (85) | 36 (42) | 77 (88) | 57 (66) | 64 (87) | 88 (96) | 96 (100) | 92 (99) | 82 (98) | 86 (93) | 90 (97) |
| LU314397 | NYSV | NY-HG27 | Japan | 72 (84) | 37 (42) | 77 (88) | 54 (67) | 65 (87) | 86 (95) | 96 (94) | 90 (97) | 83 (98) | 85 (92) | 89 (97) |
| LC314391 | NYSV | NY-OI1 | Japan | 73 (84) | 41 (42) | 70 (87) | 59 (68) | 66 (84) | 75 (94) | 77 (94) | 86 (94) | 98 (99) | 90 (96) | 94 (96) |
| LC314393 | NYSV | NY-KM1P | Japan | 60 (78) | 36 (42) | 67 (84) | 56 (63) | 70 (81) | 69 (90) | 62 (64) | 67 (82) | 72 (90) | 71 (85) | 73 (85) |
| LC314394 | NYSV | NY-CB5 | Japan | 59 (78) | 30 (41) | 66 (89) | 60 (65) | 67 (78) | 68 (92) | 64 (70) | 72 (81) | 69 (86) | 67 (84) | 72 (88) |
| JQ911732 | NYSV | ZZ-2 | China | 59 (78) | 29 (38) | 63 (85) | 60 (65) | 52 (71) | 70 (91) | 61 (61) | 68 (79) | 69 (85) | 67 (84) | 71 (88) |
| LU314395 | NYSV | NY-EH173 | Japan | 58 (78) | 33 (40) | 67 (90) | 65 (65) | 59 (75) | 69 (91) | 66 (70) | 70 (81) | 71 (86) | 68 (84) | 73 (88) |
| LC314392 | NYSV | NY-KM1O | Japan | 58 (77) | 39 (41) | 66 (85) | 53 (63) | 66 (81) | 69 (90) | 58 (58) | 65 (82) | 73 (91) | 69 (84) | 71 (85) |
| JQ395042 | NYSV | Marijiniup 3 | Australia | 58 (78) | 35 (42) | 67 (84) | 54 (59) | 69 (84) | 70 (88) | 60 (70) | 65 (82) | 72 (90) | 69 (85) | 71 (81) |
| JQ326210 | NLSYV | Zhangzhou | China | 55 (75) | 33 (38) | 65 (83) | 55 (64) | 71 (84) | 65 (88) | 64 (64) | 71 (83) | 71 (87) | 63 (80) | 68 (81) |
| KC691259 | NLSYV | Marijiniup8 | Australia | 54 (75) | 32 (36) | 64 (83) | 56 (64) | 63 (78) | 67 (84) | 68 (70) | 67 (82) | 66 (88) | 64 (79) | 68 (81) |
| JX156421 | NLSYV | Marijiniup9 | Australia | 54 (75) | 35 (39) | 67 (83) | 56 (65) | 73 (84) | 65 (73) | 66 (64) | 69 (84) | 68 (87) | 65 (79) | 68 (80) |
| AJ316084 | ScaMV | – | China | 43 (63) | 25 (22) | 56 (66) | 41 (47) | 60 (65) | 53 (73) | 45 (47) | 61 (70) | 57 (70) | 58 (71) | 58 (78) |
| NC_003399 | ScaMV | – | China | 43 (63) | 25 (22) | 56 (66) | 41 (47) | 60 (65) | 53 (73) | 45 (47) | 61 (70) | 57 (70) | 58 (71) | 58 (78) |
| AB701735 | TuMV | TIGD | – | 42 (59) | 15 (15) | 58 (68) | 25 (36) | 56 (75) | 55 (74) | 48 (29) | 53 (65) | 60 (72) | 62 (73) | 64 (76) |
| NC_000947 | JYMV | – | – | 40 (57) | 19 (19) | 53 (72) | 40 (36) | 58 (65) | 55 (70) | 52 (35) | 55 (66) | 50 (54) | 57 (69) | 56 (72) |
| AB989658 | TuMV | NZ403B | New Zealand | 39 (60) | 21 (23) | 55 (67) | 30 (37) | 64 (78) | 55 (74) | 54 (29) | 54 (64) | 59 (72) | 59 (74) | 60 (75) |
| NC_014790 | PWV | PWV-MU2 | Australia | 25 (42) | 13 (13) | 35 (47) | 19 (22) | 23 (40) | 44 (55) | 40 (29) | 37 (44) | 37 (45) | 44 (57) | 50 (65) |
| NC_017977 | CyEVA | Marijiniup7-1 | Australia | 21 (38) | 16 (15) | 33 (41) | 16 (17) | 42 (43) | 34 (52) | 35 (23) | 34 (37) | 43 (39) | 43 (54) | 48 (58) |
| KX575832 | CyEVA | NBRI16 | India | 20 (36) | 10 (15) | 32 (39) | 15 (14) | 37 (40) | 32 (50) | 18 (23) | 33 (43) | 41 (39) | 46 (55) | 44 (59) |
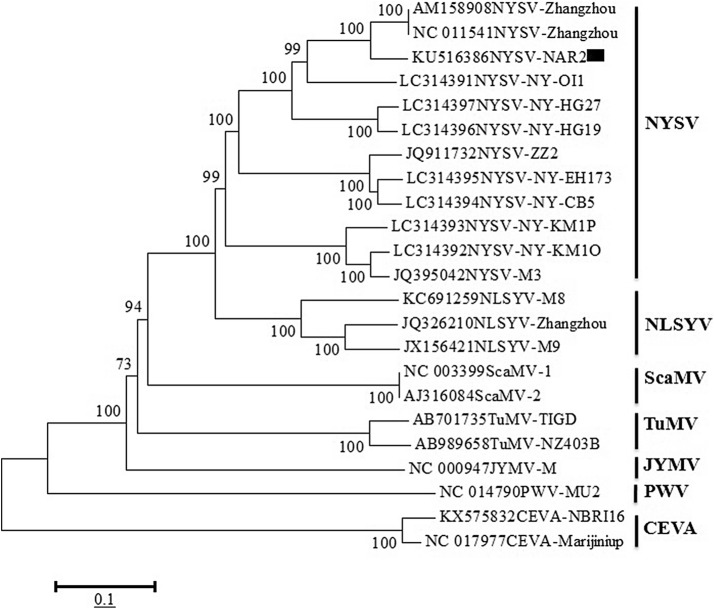
The phylogeny analysis of NYSV-NAR2 isolate with eleven full genome sequences of NYSV isolates (Zhangzhou-1, Zhangzhou-2, ZZ2, NY-OI1, NY-KM1O, NY-KM1P, NY-CB5, NY-EH173, NY-HG19, NY-HG27 and M3) along with other potyvirus sequences (Table 1) was performed. During the analysis, NYSV-NAR2 isolate clearly grouped with NYSV members and showed close phylogenetic relationships with them (Fig. 4). The clustering also revealed a close homology of NYSV members with NLSYV reported on N. tazetta (Wylie et al. 2010), whereas distant relationships with other potyviruses. The results of BLASTn analysis (90% identities) and close phylogenetic relationship with NYSV members was at par with recommendations of International Committee on Taxonomy of Viruses (ICTV, Adams et al. 2005), hence the NYSV-NAR2 isolate (KU516386) was considered as a new member of NYSV.
Fig. 4.
Phylogenetic tree representing the relationships of NYSV-NAR2 isolate (KU516386) with other potyviruses. The phylogenetic tree was constructed using MEGA v6.1 tool (Tamura et al. 2013) and using maximum likelihood algorithm at 1000 bootstrap value
It is suggested by Wylie and Jones (2012) that the vegetative propagation of narcissus through bulbs and its international trading favours accumulation and spreading of potyviruses from one place to other, therefore, the nucleic acid hybridization test and RT-PCR utilizing NYSV probe and potyvirus specific primers, respectively may help in diagnosis of NYSV and other potyviruses in narcissus bulbs to check their dissemination from one to other field/country.
Conclusion
The present study reports the complete genome sequence of 9650 nucleotides of Narcissus yellow stripe virus (NYSV-NAR2, KU516386) for the first time from India as per best of our knowledge. The viral genome sequence of NYSV-NAR2 isolate shared highest 90 and 97% identities at nucleotides and deduced amino acid levels, respectively with NYSV-Zhangzhou-1 and ZZ-2 isolates reported from China infecting N. tazetta. It also showed closest phylogenetic relationships with full-length genome sequences of two Zhangzhou isolates of NYSV while distant relationships with other potyvirus isolates, therefore, the virus isolated form N. tazetta has been considered as a new member of NYSV and designated as NYSV-NAR2. The information on complete genome of NYSV-NAR2 available from present study will also help in designing the efficient management strategy through genetic engineering.
Acknowledgements
Authors are thankful to the Director, CSIR-NBRI, Lucknow, India for facilities. RR is thankful to University Grant Commission for Rajiv Gandhi National fellowships and AcSIR for Ph.D. registration. This study was funded by the In-house project OLP105.
Compliance with ethical standards
Conflict of interest
Authors declare that they have no conflict of Interest.
References
- Adams MJ, Antoniw JF, Fauquet CM. Molecular criteria for genus and species discrimination within the family Potyviridae. Arch Virol. 2005;150:459–479. doi: 10.1007/s00705-004-0440-6. [DOI] [PubMed] [Google Scholar]
- Aminuddin, Khan JA, Raj SK. Association of an unknown potyvirus isolate with severe mosaic of Narcissus tazetta L. Indian J Exp Biol. 1999;137:1034–1036. [Google Scholar]
- Brunt AA. Narcissus. In: Loebenstein G, Lawson RH, Brunt AA, editors. Virus and virus-like diseases of bulb and flower crops. Chichester: Wiley; 1995. pp. 322–334. [Google Scholar]
- Chen J, Lu YW, Shi YH, Adams MJ, Chen JP. Complete nucleotide sequence of the genomic RNA of Narcissus yellow stripe virus from Chinese narcissus in Zhangzhou city, China. Arch Virol. 2006;15:1673–1677. doi: 10.1007/s00705-006-0788-x. [DOI] [PubMed] [Google Scholar]
- Cronin S, Verchot J, Haldeman-Cahill R, Schaad MC, Carrington JC. Long distance movement factor: a transport functions of the potyvirus helper component proteinase. Plant Cell. 1995;7:549–559. doi: 10.1105/tpc.7.5.549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibbs A, Mackenzie A. A primer pair for amplifying part of the genome of all potyvirids by RT-PCR. J Virol Methods. 1997;63:9–16. doi: 10.1016/S0166-0934(96)02103-9. [DOI] [PubMed] [Google Scholar]
- Ha C, Coombs S, Revill PA, Harding RM, Vu M, Dale JL. Design and application of two novel degenerate primer pairs for the detection and complete genomic characterization of potyviruses. Arch Virol. 2008;153:25–36. doi: 10.1007/s00705-007-1053-7. [DOI] [PubMed] [Google Scholar]
- Jenner CE, Sanchez F, Nettleship SB, Foster GD, Ponz F, Walsh JA. The cylindrical inclusion gene of Turnip mosaic virus encodes a pathogenic determinant to the Brassica resistance gene TuRB01. Mol Plant Microbe Interact. 2000;13(10):1102–1108. doi: 10.1094/MPMI.2000.13.10.1102. [DOI] [PubMed] [Google Scholar]
- Kaur C, Kumar S, Raj SK, Chauhan PS, Sharma N. Characterization of a new isolate of Bean yellow mosaic virus group-IV associated with mosaic disease of gladiolus in India. J Plant Pathol Microbiol. 2015;6:309. doi: 10.4172/2157-7471.1000309. [DOI] [Google Scholar]
- Kumar S, Raj R, Kaur C, Raj SK, Roy RK. First report of Cyrtanthus elatus virusA in Narcissus tazetta in India. Plant Dis. 2015;99:1658. doi: 10.1094/PDIS-04-15-0492-PDN. [DOI] [Google Scholar]
- Lucinda N, Inoue-Nagata AK, Kitajima EW, Nagata T. Complete genome sequence of Brugmansia suaveolens mottle virus, a potyvirus from an ornamental shrub. Arch Virol. 2010;155:1729–1732. doi: 10.1007/s00705-010-0798-6. [DOI] [PubMed] [Google Scholar]
- Nguyen HD, Tomitaka Y, Ho SYW, Duchene S, Vetten HJ, Lesemann D, Walsh JA, Gibbs AJ, Ohshima K. Turnip mosaic potyvirus probably first spread to eurasian brassica crops from wild orchids about 1000 years ago. PLoS One. 2013;8:e55336. doi: 10.1371/journal.pone.0055336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohshima K, Nomiyama R, Mitoma S, Honda Y, Yasaka R, Tomimura K. Evolutionary rates and genetic diversities of mixed potyviruses in Narcissus infection. Genet Evol. 2016;45:213–223. doi: 10.1016/j.meegid.2016.08.036. [DOI] [PubMed] [Google Scholar]
- Peng YH, Kadoury D, Gal-On A, Huet H, Wang Y, Raccah B. Mutations in HC-Pro gene of Zucchini yellow mosaic potyvirus: effects on aphid transmission and binding to purified virions. J Gen Virol. 1998;79:897–904. doi: 10.1099/0022-1317-79-4-897. [DOI] [PubMed] [Google Scholar]
- Raj R, Kaur C, Agrawal L, Chauhan PS, Kumar S, Raj SK. Full-length genome sequence of Cyrtanthus elatus virus-A isolated from Narcissus tazetta in India. 3Biotech. 2018;8:168. doi: 10.1007/s13205-018-1189-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiboleth YM, Haronsky E, Leibman D, Arazi T, Wassenegger M, Whitham SA, Gaba V, Gal-On A. The conserved FRNK box in HC-Pro, a plant viral suppressor of gene silencing, is required for small RNA binding and mediates symptom development. J Virol. 2007;81:13135–13148. doi: 10.1128/JVI.01031-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shuang QL, Jian GS, Fang LG, Wei C, Zhen H, Li YX, Zu JW. Complete genome sequence of Narcissus late season yellows virus infecting Chinese narcissus in China. Arch Virol. 2012;157:1821–1824. doi: 10.1007/s00705-012-1328-5. [DOI] [PubMed] [Google Scholar]
- Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol. 2013;30:2725–2729. doi: 10.1093/molbev/mst197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wylie SJ, Jones MGK. Complete genome sequences of seven carlavirus and potyvirus isolates from Narcissus and Hippeastrum plants in Australia, and proposals to clarify their naming. Arch Virol. 2012;157:1471–1480. doi: 10.1007/s00705-012-1319-6. [DOI] [PubMed] [Google Scholar]
- Wylie SJ, Nouri S, Coutts BA, Jones MGK. Narcissus late season yellows virus and Vallota speciosa virus found infecting domestic and wild populations of Narcissus species in Australia. Arch Virol. 2010;155:1171–1174. doi: 10.1007/s00705-010-0682-4. [DOI] [PubMed] [Google Scholar]
- Wylie SJ, Li H, Sivasithamparam K, Jones MGK. Complete genome analysis of three isolates of Narcissus late season yellows virus and two of Narcissus yellow stripe virus: three species or one? Arch Virol. 2014;159:1521–1525. doi: 10.1007/s00705-013-1969-z. [DOI] [PubMed] [Google Scholar]
- Yadav N, Khan JA. Identification of a potyvirus associated with mosaic disease of Narcissus sp. in India. Plant Pathol. 2008;57:394. doi: 10.1111/j.1365-3059.2007.01655.x. [DOI] [Google Scholar]
- Yadav N, Khan JA. Molecular identification of Narcissus yellow stripe virus strain associated with severe mosaic disease of narcissus from India. Indian Phytopathol. 2015;68:444–448. [Google Scholar]
- Yakoubi S, Lecoq H, Desbiez C. Algerian watermelon mosaic virus (AWMV): a new potyvirus species in the PRSV cluster. Virus Genes. 2008;37:103–109. doi: 10.1007/s11262-008-0237-x. [DOI] [PubMed] [Google Scholar]
- Yasaka R, Ohba K, Schwinghamer M, Fletcher J, Ochoa-Corona F, Thomas J, Ho S, Gibbs A, Ohshima KJ. Phylodynamic evidence of the migration of turnip mosaic potyvirus from Europe to Australia and New Zealand. J Gen Virol. 2015;96(3):701–713. doi: 10.1099/jgv.0.000007. [DOI] [PubMed] [Google Scholar]