Abstract
Tree peony (Paeonia suffruticosa Andrew) is a popular ornamental plant due to its large, fragrant and colorful flowers. The floral development is the most important event in its lifecycle. To explore the mechanism that regulate flower development, we sequenced the flower bud transcriptomes of ‘High Noon’, a reblooming cultivar of P. suffruticosa × P. lutea, using both full-length isoform-sequencing (ISO-seq) and RNA-seq were sequenced. A total of 15.94 Gb raw data were generated in full-length transcriptome sequencing of the 3 floral developmental stages, resulting 0.11 M protein-coding transcripts. Over 457.0 million reads were obtained by RNA-seq in the 3 floral buds. Here, we openly released the full-length transcriptome database of ‘High Noon’ and RNA-seq database of floral development. These databases can provide a fundamental genetic information of tree peony to investigate its transcript structure, variants and evolution. Data will facilitate to deep analyses of the transcriptome for flower development.
Subject terms: Shoot apical meristem, Development, Transcriptomics
| Measurement(s) | transcription profiling assay • full-length isoform |
| Technology Type(s) | RNA sequencing • isoform sequencing |
| Factor Type(s) | developmental stage |
| Sample Characteristic - Organism | Paeonia suffruticosa |
Machine-accessible metadata file describing the reported data: 10.6084/m9.figshare.9938360
Background & Summary
Tree peony (Paeonia suffruticosa Andrew), is one of the most important horticultural plants in the world and a culturally important ornamental plants in China, due to its striking ornamental and medicinal values. It is a perennial deciduous shrub with large, fragrant, and colorful flowers. With a long history of cultivation, there are more than 3,000 cultivars all over the world. ‘High Noon’ (P. suffruticosa × P. lutea) is one of the most famous and popular cultivars and is always used for hybrid breeding due to its characteristic of cup shape, semi-double and clear lemon color. On the other hand, High Noon showed an unregular reblooming phenomenon1 which means a twice floral development occurred around a year. These traits made ‘High Noon’ a suitable material for researching the floral development in tree peony.
Floral development is the most important developmental event in the life cycle of higher plants. The flowering timing is determined by processes of flowering transition, floral bud differentiation and floral organ identification2. A complex gene regulatory network was involved in the floral bud differentiation including plant hormone signal pathway and meristem activity regulation. Although there are great progresses in the study of molecular mechanism in floral development of model plants, it remains unclear in perennial plants, especially tree peony, whose genome information was not yet published. A few genes have been identified to be involved in the transition of shoot apical meristem (SAM) to floral bud in tree peony, including SOC13, FT4, and AP15. However, it was still hard to understand the underlying mechanism on floral development of tree peony at transcriptome level.
Next-generation sequencing (NGS) provides precise and comprehensive analysis of RNA transcripts for gene expression. It is applied to explore biological research frequently. Single molecular real time (SMRT) sequencing is a third-generation sequencing technology which offers great improvement than NGS on reads length and avoids the requirement of assembly in NGS6,7. The combination of SMRT and NGS has proceeded the genome assembly and transcriptomic research in several species. The genome assembly of (sunflower) Helianthus annuus and an indica rice Shuhui498 (R498) was completed with PacBio SMRT technology8,9. The combination of these 2 sequencing technologies has also been applied in many ways, such as seeking for the characteristic of transcriptomes and identifying new genes in Sorghum bicolor10, Zea mays11, Phyllostachys edulis12. For species without genome information published, the combination of NGS and SMRT was applied to establish a reliable training set for gene prediction and settle biological questions in Beta vulgaris13, Alternanthera philoxeroides14, and Cassia obtusifolia15. In consideration of the absence of tree peony genome, the information of completed mRNA of transcripts is still unclear, which further limits the exploration of tree peony. Therefore, it is necessary to conduct a combined transcript sequencing for the gene prediction and the floral development research in tree peony.
In this study, we performed both SMRT and NGS to generate large-scale full-length transcripts and collect the gene expression profile for bud development of tree peony. Additionally, the data quality was assessed to verify their reliability. The full-length transcripts will provide gene sequence information for the further study of tree peony, and the gene expression profile will provide comprehensive understanding of the bud development of tree peony.
Methods
Design and sample collection
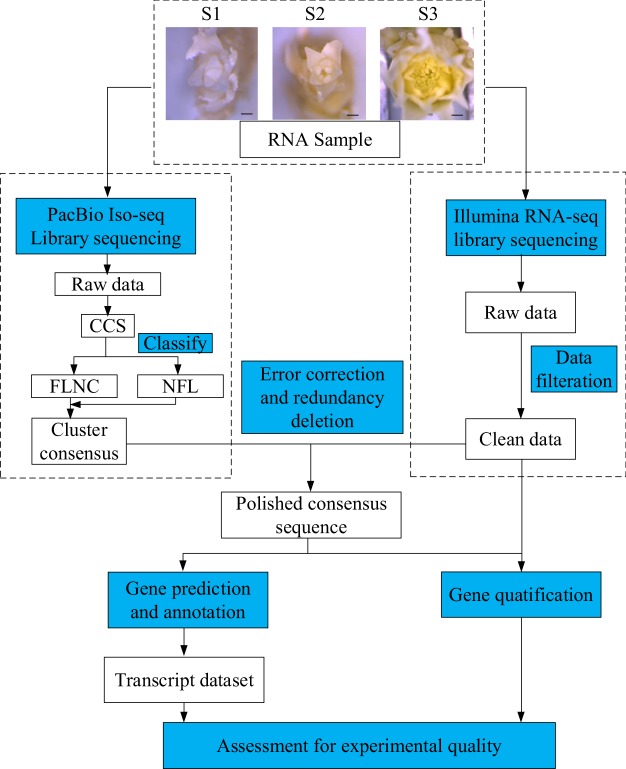
‘High Noon’ is a cultivar of tree peony, which contributed an important genetic resource for extending flowering period. The buds of different developmental stage were obtained from 3–5 years-old plants of a farm in Heze (E, 115°32′30.7818″; N, 35°20′4.794″). After discarding the adjacent scales and leaves, the buds were transferred to liquid nitrogen immediately and stored to −80 °C. The buds were also fixed simultaneously in FAA solution as parallel samples for microscopic observation. And subjected to section in slices and observed under microscope (Zeiss Primo Star, Germany). Through paraffin sections, vegetative meristem (Stage I, S1), floral meristem (Stage II, S2) and floral organ (Stage III, S3) were identified each with at least 3 buds (Fig. 1).
Fig. 1.
Overview of the experimental design and analysis pipeline. The RNA of ‘High Noon’ bud in its three developmental stages of was collected for PacBio Iso-sequencing and Illumina RNA-seq. The raw data of PacBio Iso-seq were classified and corrected to generate the high-quality transcript sequence. Then these sequences were used to predict the gene sequence and annotate the gene function. The raw data of Illumina RNA-seq were filtered using ng_QC package. Then the clean data were mapped to the full-length transcript sequences by RSEM and used to calculate the reads coverage and gene abundance with package DESeq R package and DESeq2, respectively. The sample stage was confirmed through the parallel samples observed through microscope. Stage I represents the vegetative stage. The stage II represents the bud developmental stage during which the floral organ differentiated. Stage III represents the stage during which the floral organ were formed and the floral bud was identified. Bar = 0.5 mm.
RNA extraction, Pacbio cDNA library preparation and sequencing
Total RNA was extracted using RNeasy Plant Mini kit (Qiangen, 74904) and treated with RNase-free DNase I (TAKARA, D2215) according to the manufacture’s protocol. The RNA was used for cDNA synthesis through SMARTer PCR cDNA Synthesis Kit (Clontech). The first strand and second strand were synthesized with SMARTScribe RT, using oligo(dT) primer and PCR Primer, respectively. Then the cDNA was selected with the BluePippin Size Selection System (Sage Science, Beverly, MA) according to the Isoform Sequencing protocol as described by Pacific Biosciences (PN 101-070-200-02). To increase the sequencing yield of >4 kb transcripts, a mixture of unfiltered fractions and fractions with size of >4 kb with a mole ratio of 1:1 was processed with the DNA Template Prep Kit (Pacific Biosciences of California, Inc.). Then the library was ready for sequencing after a binding of primer and DNA polymerase to the mixed transcripts. The final library was sequenced on Pacific Bioscience RS II platform (Pacific Biosciences of California, Inc.) by Novogene technology (Tianjin, China; http://www.novogene.com/).
Illumina cDNA library construction and sequencing
After total RNA was extracted as above, mRNA was enriched by Oligo dT beads and broke into short fragment in fragmentation buffer. Then the first-strand cDNA and second-strand cDNA was synthesized using random hexamers and dNTPs, respectively. The cDNA was subjected to purification and size fractioned by AMPure XP beads, with end pairing, “A” base and Illumina adapter ligation. Then the cDNA libraries were generated by a PCR amplification. After quality control with an Agilent2100 Bioanalyzer, the cDNA libraries were sequenced with a PE mode of 150 bp on an Illumina HiSeq 2000 platform by Novogene technology (Tianjin, China; http://www.novogene.com/).
Data filtering and error correction
Sequence data were processed using the SMRTlink 5.1 software. Circular consensus sequence (CCS) was generated from the raw subreads with a parameter of minimum length > 200 and minimum predicted accuracy > 0.8. The generated CCS sequences were then classified into Full-length non-chimeric reads (FLNC) and non-full length non-chimeric reads (NFL) according to the containment of 5′ primer, 3′ primer and poly A. FLNC were then fed into the cluster step, which underwent an isoform-level clustering (ICE), followed by a final Arrow polishing with NFL, with a minimum accuracy of 0.99. The resulting consensus reads were subjected to a correction using the Illumina RNA-seq data with the software LoRDEC. Then, after a redundancy deletion by CD-HIT software (−c 0.95, −aS 0.99), the final high quality, full-length, polished consensus sequences were generated after a redundancy deletion by CD-HIT software.
Gene quantification
The raw reads of Illumina RNA-seq were filtered by software ng_QC (−t 4, −L 20, −N 0.001). The clean data was mapped to the Polished consensus sequence by bowtie2 using end-to-end and sensitive mode. The readcounts of each transcript were calculated using RSEM and transformed into FPKM value. The expressional differential analysis was conducted by DESeq R package with a criterion of fold change > 2 and qvalue < 0.001.
Data Records
The sequencing raw data and files of gene abundance analysis in this study were deposited in NCBI Gene Expression Omnibus (GEO) and NCBI Sequence Read Archive (SRA) with accessions GSE133476 and SRP21225416,17. The annotation information of full-length transcripts in this study was deposited in figshare18. The Supplementary material including quality assessment data of raw reads was deposited in figshare18. The differentially expressed gene list relative to plant hormone biosynthesis and signaling pathways was deposited in figshare18. The flow cytometry analysis of ‘High Noon’ was deposited in figshare18.
Technical Validation
RNA qualities
The purity and integrity of the total RNA was assessed with Nanodrop 2000 and Agilent 2100. The RNA samples with RIN > 8.0 were used for sequencing library. Qubit 2.0 was used to measure the quantity of RNA sample and cDNA library. The RNA quality values in this study are listed in Table 1.
Table 1.
Summary of sequencing strategies in this study.
| Samples | Sequencing Strategy | Library Layout | Platform | Instrument model | Tissue | OD260/280 | OD260/230 | 25S/18S | RIN |
|---|---|---|---|---|---|---|---|---|---|
| S1-1 | RNA-seq | paired | Illumina | Illumina HiSeq 2000 | floral bud | 2.194 | 1.388 | 2.1 | 8.8 |
| S1-2 | RNA-seq | paired | Illumina | Illumina HiSeq 2000 | floral bud | 2.326 | 0.877 | 1.4 | 8.3 |
| S1-3 | RNA-seq | paired | Illumina | Illumina HiSeq 2000 | floral bud | 2 | 1.053 | 1.6 | 9.1 |
| S2-1 | RNA-seq | paired | Illumina | Illumina HiSeq 2000 | floral bud | 2.115 | 1.743 | 1.8 | 10 |
| S2-2 | RNA-seq | paired | Illumina | Illumina HiSeq 2000 | floral bud | 2.069 | 2.011 | 1.8 | 9.3 |
| S2-3 | RNA-seq | paired | Illumina | Illumina HiSeq 2000 | floral bud | 2.152 | 2.073 | 1.9 | 9.8 |
| S3-1 | RNA-seq | paired | Illumina | Illumina HiSeq 2000 | floral bud | 2.161 | 1.463 | 1.8 | 9.6 |
| S3-2 | RNA-seq | paired | Illumina | Illumina HiSeq 2000 | floral bud | 2.153 | 1.859 | 2 | 10 |
| S3-3 | RNA-seq | paired | Illumina | Illumina HiSeq 2000 | floral bud | 2.092 | 0.823 | 1.8 | 9.3 |
| HN | RNA-seq | single | PACBIO_SMART | PacBio RS II | floral bud | mixed | mixed | mixed | mixed |
Pacbio ISO-seq quality validation
A total of 15.94 Gb raw data was generated by 15,654,254 subreads in the Pacbio ISO-seq. After a single molecular self-correction, circular consensus sequences (CCSs) of 714,643 reads was obtained, which was subsequently classified to full-length non-chimeric (FLNC) with 5′ primer, 3′ primer and poly A and non-full length (NFL) with a proportion of 61.78% and 38.22%, respectively. Consequently, a total of 441,507 high-quality FLNC reads was obtained through the cluster of FLNC and correction by NFL.
As SMRT sequencing generates a high error rate, it is necessary to perform error correction, which includes self-correction by iterative clustering of circular-consensus reads and correction with high-quality NGS short reads. To this end, the NGS sequence data in this study was used to correct the SMRT sequences using LoRDEC software. After that, redundant transcripts were removed by CD-HIT, and a total of 115,439 non-redundant transcripts (Polished consensus sequences) with an average length of 2,060 bp were obtained (see Table 2).
Table 2.
Statistic of ISO-sequencing in this study.
| Type | Total_nucleotides (bp) | Total_reads_number | Average length (bp) |
|---|---|---|---|
| subreads | 15,936,030,572 | 15,654,254 | 1,018 |
| CCS | 1,236,332,390 | 714,643 | 1,730 |
| FLNC | 760,275,054 | 441,507 | 1,722 |
| Consensus | 397,594,800 | 214,800 | 1,851 |
| polished consensus | 387,499,200 | 214,800 | 1,804 |
| Before_correct | 387,469,755 | 214,800 | 1,804 |
| After_correct | 388,005,257 | 214,800 | 1,807 |
| CD-HIT | 237,754,844 | 115,439 | 2,060 |
Predictions of coding sequence (CDS) and function annotation
To obtain comprehensive information of gene function in tree peony, the 115,439 transcripts were mapped to 7 databases, including NR, NT, Pfam, KOG, Swiss-Prot, KDGG, GO for the gene annotation. As a result, at least 32,416 transcripts could be mapped to all these seven databases (Fig. 2a). The length distribution of successfully annotated genes was showed in Fig. 2b. The completeness of transcripts generated by CD-HIT was assessed by BUSCO 2.3. The results showed that 83.68% transcripts were complete of which single copy BUSCOs and duplicated BUSCOs account for 25.97% and 57.71%, respectively. Of the total 1,440 BUSCO groups searched, only 52 fragmented BUSCOs and 183 missing BUSCOs were found in our database (Fig. 2c). All these results showed that our database was complete and available for subsequent research.
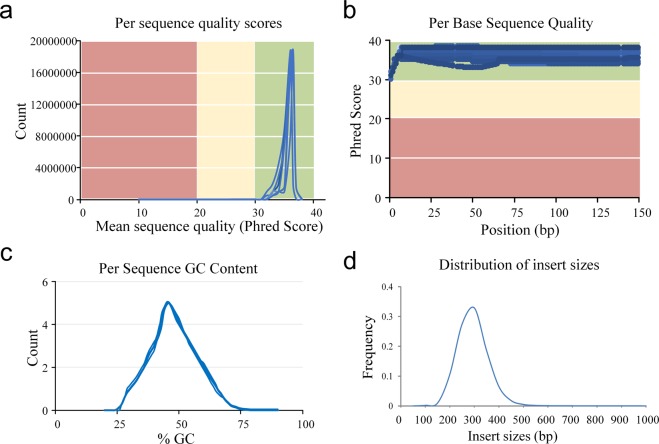
Fig. 2.
The reads and alignment quality in RNA-seq. (a) Per sequence quality scores. The x-axis represents the mean sequence quality of RNA-seq reads. The y-axis represents the count of reads with specified Phred score. (b) Per base sequence quality. The x-axis represents the position of reads in RNA-seq. The y-axis represents the Phred scores of each base. (c) Per sequence GC content. The x-axis represents the percentage of GC of each read. The y-axis represents the count of the reads with. (d) Distribution of insert size after alignment. The x-axis represents the insert size of library after alignment. The y-axis represents the frequency of each size.
Illumina RNA-seq quality validation and floral development gene identification
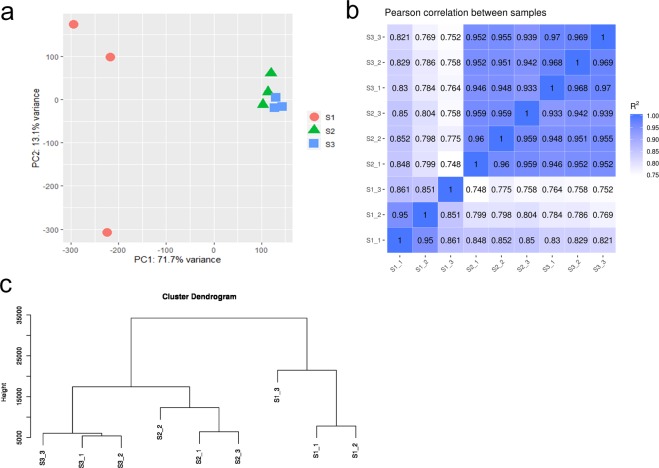
The reads quality of clean reads in Illumina RNA-seq was assessed using FastQC, including the mean per sequence quality scores, per base quality scores, and GC contents. The per base quality scores were higher than phred quality 30, and most sequences had a quality over 20 (Fig. 3a,b). The GC contents of the samples showed a similar normal distribution, which indicated a sequencing data free of contamination (Fig. 3c). The reads quality of the samples showed that the RNA-seq reads in this study have a high quality. The clean reads of 9 samples were aligned to the 115,439 non-redundant transcripts (reference) using bowtie2 with end-to-end and sensitive mode. The distribution of library insert length after alignment was measured which showed a 270–320 bp distribution (Fig. 3d). The mapping rate of Illumina RNA-seq reads to the high-quality polished sequence ranged from 83.85–88.27% (Table 3). The reliability of the RNA-seq data between the 9 samples was measured with PCA analysis, Pearson correlation and clustering analysis (Fig. 4a–c). The results all showed a reliable biological duplication, indicated that the data obtained in this study could be used for subsequent research.
Fig. 3.
The statistic results of gene annotation and predicted CDS length distribution. (a) The transcripts were annotated in 7 databases. The y-axis represents the annotated gene number. (b) The CDS length distribution of transcripts in ISO-seq. The x-axis represents CDS length. The y-axis in left represent the frequency of different CDS length. The right y-axis represents the percentage of different CDS length. (c) The completeness of transcripts in ISO-seq. The completeness of transcripts was assessed by benchmarking universal single-copy ortholog (BUSCO). The x-axis represents the percentage of detected BUSCOs. The light blue diamond represents the complete (C) and single-copy (S) genes; the dark blue represent complete and duplicated (D) genes; the yellow diamond represents fragmented (F) genes; the red diamond represents the missing (M) genes. Total number of core genes queried was 1440.
Table 3.
Summary of Illumina RNA-sequencing.
| Sample | Raw Reads | Clean Reads | Clean Bases (G) | Total mapped reads | Mapping rate (%) |
|---|---|---|---|---|---|
| S1_1 | 49,022,606 | 48,474,096 | 7.27 | 41,483,460 | 85.58 |
| S1_2 | 48,375,304 | 47,857,452 | 7.18 | 40,130,002 | 83.85 |
| S1_3 | 50,839,914 | 49,433,588 | 7.42 | 41,993,190 | 84.95 |
| S2_1 | 58,645,596 | 58,018,926 | 8.7 | 50,391,040 | 86.85 |
| S2_2 | 55,158,816 | 54,515,864 | 8.18 | 47,983,332 | 88.02 |
| S2_3 | 46,277,122 | 45,839,116 | 6.88 | 40,461,148 | 88.27 |
| S3_1 | 47,068,706 | 45,955,414 | 6.89 | 40,193,166 | 87.46 |
| S3_2 | 48,714,736 | 47,383,326 | 7.11 | 41,088,574 | 86.72 |
| S3_3 | 52,899,086 | 51,759,904 | 7.76 | 45,315,526 | 87.55 |
Fig. 4.
Summary of the sample clustering. (a) Principal component analysis of the 9 samples based on expression levels. (b) The person correlation between the 9 samples based on expression levels. (c) The cluster analysis of the 9 samples based on expression levels.
After mapping to the non-redundant transcript, the gene expressional level was analyzed and the differential expressional genes (DEGs) were screened with a parameter of fold change > 2 and q value < 0.001. According to the annotation, DEGs relative to floral development and regulation were analyzed. A total of 143 genes in plant hormone biosynthesis and signaling pathways including auxin and cytokinin which were believed to regulate the floral initiation and bud development were identified19. In addition, a total of 26 floral-developmental-relative-DEGs were identified, which might play important roles in floral development process, including the establishment of floral meristem, the specification of flower organ identity and the regulation of floral organogenesis in this study20,21. These DEGs were listed in Table 4 and citation 5. These results indicate that our data were valuable for understanding the floral development in tree peony.
Table 4.
DEGs relative to floral development.
| Gene ID | Gene ID in model plant | Gene Symbol | Iidentity (%) | Alignment length (bp) | Pathway |
|---|---|---|---|---|---|
| i1_HQ_HNbud_c49771/f2p0/1159 | AT1G24260.1 | SEP3 | 54.47 | 246 | floral developmental processes |
| i1_LQ_HNbud_c231904/f1p0/1102 | AT1G24260.1 | SEP3 | 74.49 | 247 | floral developmental processes |
| i2_HQ_HNbud_c9324/f2p0/2669 | AT4G32980.1 | ATH1 | 56.04 | 323 | floral developmental processes |
| i2_LQ_HNbud_c47156/f1p3/2045 | AT3G03090.1 | ATVGT1 | 58.49 | 477 | floral developmental processes |
| i1_HQ_HNbud_c27914/f7p7/1955 | AT3G03090.1 | ATVGT1 | 58.07 | 477 | floral developmental processes |
| i1_LQ_HNbud_c91035/f1p1/1218 | AT5G60910.1 | FUL | 68.45 | 206 | floral developmental processes |
| i4_HQ_HNbud_c4367/f6p1/4832 | AT5G67100.1 | ICU2 | 60.67 | 1556 | floral developmental processes |
| i1_LQ_HNbud_c105452/f1p0/1482 | AT5G61850.1 | LFY | 65.64 | 390 | floral developmental processes |
| i1_LQ_HNbud_c42466/f1p1/1326 | AT1G69490.1 | NAP | 63.58 | 162 | floral developmental processes |
| i3_LQ_HNbud_c3353/f1p4/3804 | AT4G04885.1 | PCFS4 | 60.67 | 239 | floral developmental processes |
| i3_LQ_HNbud_c2041/f1p5/3526 | AT4G04890.1 | PDF2 | 74.9 | 741 | floral developmental processes |
| i0_HQ_HNbud_c9787/f2p4/869 | AT5G20240.1 | PI | 56.13 | 212 | floral developmental processes |
| i0_LQ_HNbud_c42964/f1p0/955 | AT5G20240.1 | PI | 57.35 | 211 | floral developmental processes |
| i0_LQ_HNbud_c22315/f1p0/746 | AT5G37055.1 | SEF | 70.91 | 165 | floral developmental processes |
| i0_LQ_HNbud_c30233/f1p0/911 | AT3G02310.1 | SEPALLATA2 | 68.8 | 250 | floral developmental processes |
| i1_LQ_HNbud_c213752/f1p14/1085 | AT1G14400.1 | UBC1 | 94.74 | 152 | floral developmental processes |
| i1_LQ_HNbud_c10259/f1p0/1559 | AT1G30950.1 | UFO | 62.14 | 420 | floral developmental processes |
| i1_LQ_HNbud_c105095/f1p0/1458 | AT1G30950.1 | UFO | 56.9 | 420 | floral developmental processes |
| i0_LQ_HNbud_c79621/f1p2/890 | AT1G65480.1 | FT | 79.29 | 169 | floral integrator |
| i1_LQ_HNbud_c105115/f1p1/1458 | AT5G13790.1 | AGL15 | 59.32 | 236 | floral repressor |
| i1_LQ_HNbud_c133723/f1p1/1875 | AT5G13790.1 | AGL15 | 60.17 | 236 | floral repressor |
| i1_LQ_HNbud_c49765/f1p3/1453 | AT1G68840.1 | TEM2 | 63.32 | 368 | floral repressor |
| i1_HQ_HNbud_c33701/f2p5/1226 | AT2G01290.1 | RPI2 | 75.31 | 243 | flower meristem identity |
| i1_LQ_HNbud_c51781/f2p3/1522 | AT2G01290.1 | RPI2 | 73.68 | 266 | flower meristem identity |
| i1_LQ_HNbud_c128329/f1p0/1130 | AT1G69120.1 | AP1 | 69.14 | 256 | flower meristem identity |
| i1_LQ_HNbud_c51214/f1p0/1256 | AT1G69120.1 | AP1 | 69.66 | 178 | flower meristem identity |
Acknowledgements
This work was funded by Fundamental Research Funds of ICBR (Nos 1632018023 and 1632019009).
Author contributions
Y.-T.C., T.H., designed the experiments, collected samples, extracted RNA, processed the data, and wrote the manuscript. W.-B.Z. collected samples, extracted RNA, processed the data, and reviewed the manuscript. L.Z. designed the experiments, collected samples and reviewed the manuscript. Y.W. and Z.-H.J. designed the experiments, reviewed the manuscript and supervised the study.
Code availability
CD-HIT: http://www.bioinformatics.org/cd-hit/ (version 4.6.6).
BUSCO: https://gvolante.riken.jp/index.html (version 2.3).
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Yanting Chang, Tao Hu and Wenbo Zhang.
Contributor Information
Yan Wang, Email: chwy8915@sina.com.
Zehui Jiang, Email: zhjiang2015@163.com.
References
- 1.Kessenich, G. A. P. Saunders hybrid peonies (Lutea hybrid tree peonies). In: Peonies: the history of the peonies and their originations1, 146–152 (1976).
- 2.Irish VF. The flowering of Arabidopsis flower development. Plant J. 2010;61:1014–1028. doi: 10.1111/j.1365-313X.2009.04065.x. [DOI] [PubMed] [Google Scholar]
- 3.Wang S, et al. Molecular cloning and potential function prediction of homologous SOC1 genes in tree peony. Plant Cell Reports. 2015;34:1459–1471. doi: 10.1007/s00299-015-1800-2. [DOI] [PubMed] [Google Scholar]
- 4.Zhu, F. Y. et al. Isolation of Florigen Gene PdFT and Its Effects on Flowering of Tree Peony (Paeonia delavayi Franch.). Scientia Agricultura Sinica 47, 2613–2624 (2014).
- 5.Ren, L., Wang, Y., Zhou, L. & Peng, Z. H. Cloning and Expression of PsAP1 Gene Related to Flowering in Tree Peony. Acta Botanica Boreali-Occidentalia Sinica 9, 1719–1725 (2011).
- 6.Eid J, et al. Real-time DNA sequencing from single polymerase molecules. Science. 2009;323:133–138. doi: 10.1126/science.1162986. [DOI] [PubMed] [Google Scholar]
- 7.Sharon D, Tilgner H, Grubert F, Snyder M. A single-molecule long-read survey of the human transcriptome. Nat Biotechnol. 2013;31:1009–1014. doi: 10.1038/nbt.2705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Badouin H, et al. The sunflower genome provides insights into oil metabolism, flowering and Asterid evolution. Nature. 2017;546:148–152. doi: 10.1038/nature22380. [DOI] [PubMed] [Google Scholar]
- 9.Du H, et al. Sequencing and de novo assembly of a near complete indica rice genome. Nat Commun. 2017;8:15324. doi: 10.1038/ncomms15324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Abdel-Ghany SE, et al. A survey of the sorghum transcriptome using single-molecule long reads. Nat Commun. 2016;7:11706. doi: 10.1038/ncomms11706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang B, et al. Unveiling the complexity of the maize transcriptome by single-molecule long-read sequencing. Nat Commun. 2016;7:11708. doi: 10.1038/ncomms11708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang T, et al. Comprehensive profiling of rhizome-associated alternative splicing and alternative polyadenylation in moso bamboo (Phyllostachys edulis) Plant J. 2017;91:684–699. doi: 10.1111/tpj.13597. [DOI] [PubMed] [Google Scholar]
- 13.Minoche AE, et al. Exploiting single-molecule transcript sequencing for eukaryotic gene prediction. Genome Biol. 2015;16:184. doi: 10.1186/s13059-015-0729-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jia D, et al. SMRT sequencing of full-length transcriptome of flea beetle Agasicles hygrophila (Selman and Vogt) Sci Rep. 2018;8:2197. doi: 10.1038/s41598-018-20181-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Deng, Y. et al. Full-Length Transcriptome Survey and Expression Analysis of Cassia obtusifolia to Discover Putative Genes Related to Aurantio-Obtusin Biosynthesis, Seed Formation and Development, and Stress Response. Int J Mol Sci19, 10.3390/ijms19092476 (2018). [DOI] [PMC free article] [PubMed]
- 16.2019. Gene Expression Omnibus. GSE133476
- 17.2019. NCBI Sequence Read Archive. SRP212254
- 18.Chang Y, 2019. Transcriptome profiling for floral development in reblooming cultivar ‘High Noon’ of Paeonia suffruticosa. figshare. [DOI] [PMC free article] [PubMed]
- 19.Shan, H., Cheng, J., Zhang, R., Yao, X. & Kong, H. Developmental mechanisms involved in the diversification of flowers. Nat Plants, 10.1038/s41477-019-0498-5 (2019). [DOI] [PubMed]
- 20.Okamuro JK, Caster B, Villarroel R, Van Montagu M, Jofuku KD. The AP2 domain of APETALA2 defines a large new family of DNA binding proteins in Arabidopsis. Proc Natl Acad Sci USA. 1997;94:7076–7081. doi: 10.1073/pnas.94.13.7076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Preston JC, Hileman LC. Developmental genetics of floral symmetry evolution. Trends Plant Sci. 2009;14:147–154. doi: 10.1016/j.tplants.2008.12.005. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Citations
- 2019. Gene Expression Omnibus. GSE133476
- 2019. NCBI Sequence Read Archive. SRP212254
- Chang Y, 2019. Transcriptome profiling for floral development in reblooming cultivar ‘High Noon’ of Paeonia suffruticosa. figshare. [DOI] [PMC free article] [PubMed]
Data Availability Statement
CD-HIT: http://www.bioinformatics.org/cd-hit/ (version 4.6.6).
BUSCO: https://gvolante.riken.jp/index.html (version 2.3).