Abstract
The role of starch degradation in non-vascular plants is poorly understood. To expand our knowledge of this area, we have studied this process in Physcomitrella patens. This has been achieved through examination of the step known to initiate starch degradation in angiosperms, glucan phosphorylation, catalysed by glucan, water dikinase (GWD) enzymes. Phylogenetic analysis indicates that GWD isoforms can be divided into two clades, one of which contains GWD1/GWD2 and the other GWD3 isoforms. These clades split at a very early stage within plant evolution, as distinct sequences that cluster within each were identified in all major plant lineages. Of the five genes we identified within the Physcomitrella genome that encode GWD-like enzymes, two group within the GWD1/GWD2 clade and the others within the GWD3 clade. Proteins encoded by both loci in the GWD1/GWD2 clade, named PpGWDa and PpGWDb, are localised in plastids. Mutations of either PpGWDa or PpGWDb reduce starch phosphate abundance, however, a mutation at the PpGWDa locus had a much greater influence than one at PpGWDb. Only mutations affecting PpGWDa inhibited starch degradation. Mutants lacking this enzyme also failed to develop gametophores, a phenotype that could be chemically complemented using glucose supplementation within the growth medium.
Subject terms: Carbohydrates, Plant physiology
Introduction
The bryophyte Physcomitrella patens is a species that has many advantages for use in the study of plant molecular physiology. Its genome has been sequenced1, it is transformable, knockout mutations are easily generated through homologous recombination, and its generally haploid lifestyle means that mutations can be studied directly in the M1 generation2. Physcomitrella and tracheophytes diverged approximately 450 million years ago1, after which vascular plants developed an increased ability to survive without proximate water sources. During this time, their metabolic pathways will have changed in a way that would advantage vascular plants in the new ecological niches they encountered. The use of this plant to study metabolism allows for the analysis of how biochemical pathways have altered in different plant species since this divergence3.
We have decided to examine the pathway of starch metabolism in P. patens. This is because the presence of starch has been demonstrated to be important for plant growth and development in vascular plants4,5, and we wish to examine if the same holds true in this non-vascular species. When grown on artificial media, P. patens spores or explant material develop initially into thread like protonemal tissue comprising two distinct cell types, chloronema and caulonema. Caulonemal cells contain fewer chloroplasts than chloronemal cells, and can also form buds which develop into leafy shoots known as gametophores. Many factors are known to affect gametophore development including alterations in cell wall6, phytohormone synthesis7,8, light perception9 and several regulatory genetic elements10–12. Alterations in carbon metabolism and sensing also alter colony growth and development, alongside influencing starch accumulation13–15. Little is known about the role of starch in Physcomitrella, although it is present14,16 and its degradation has been implicated in freezing tolerance16.
In this study we examine the step that has been shown to initiate starch catabolism in angiosperms, starch phosphorylation17,18. Much, if not all of the pathway of leaf starch degradation has been recently elucidated through studies in a number of species, primarily Arabidopsis, and involves several enzymatic reactions5,19. The initial steps occur within chloroplasts where starch is phosphorylated by glucan, water dikinase (GWD) isoforms20–28, that solubilize the surface of the granule, allowing access to α-amylase, β-amylase, isoamylase and β-limit dextrinase29–32. Degradation by these enzymes releases soluble phosphorylated malto-oligosaccharides into the stroma. The phosphate from these is removed by two phosphoglucan phosphatases33–35, before they can be further degraded to maltose and glucose through the actions of ∝-, β- and isoamylases29–31 alongside disproportionating enzyme 136–38. Maltose and glucose are exported from the plastid into the cytosol by two transporters39–41 where the maltose is further mobilized by disproportionating enzyme 237,38,42,43.
To help understand the role of starch degradation in P. patens we decided to mutate some of the starch phosphorylating enzymes as they catalyse the first step in starch degradation. Their importance is, therefore, likely to be conserved between vascular and non-vascular plants. Three of these have been identified in angiosperms: GWD1 is localized to the plastid and phosphorylates glucose residues within amylopectin at the 6-position20,24. Mutations in this gene lead to starch without covalently bound phosphate and a large decrease in leaf starch degradation25,26,44–47. GWD3 is also plastidial and phosphorylates glucose residues within starch at the 3-position20. It is often named the phosphoglucan, water dikinase (PWD) as it can only phosphorylate starch that has already been acted upon by GWD1. Mutations eliminating GWD3/PWD lead to starch without phosphate bound at the 3-position and a mild repression of leaf starch degradation27,28. GWD2 is present in the cytosol and the gene encoding it is expressed mainly in sieve elements48. Although mutations eliminating it do not affect starch turnover in photosynthetic tissue, they affect plant growth49.
In this study we report on mutations in two GWD1 isoforms, and demonstrate that this leads to the synthesis of starch containing reduced glucose 6-phosphate. Mutations in one of the isoforms lead to increased starch accumulation, and to colonies that do not produce gametophores.
Results
The Physcomitrella patens genome contains multiple GWD isoforms
We examined the presence of sequences encoding GWD-like enzymes within the Physcomitrella genome through a tBLASTn search using the Arabidopsis GWD1 (NCBI accession NM_001331926.1) amino acid sequence at Phytozome (https://phytozome.jgi.doe.gov/pz/portal.html#!search?show=BLAST). This identified five loci on chromosomes 3 (Pp3c3_11200), 8 (Pp3c8_6536), 14 (Pp3c14_19150), 17 (Pp3c17_18900) and 18 (Pp3c18_14870) encoding putative GWD isoforms. BLASTP searches of the A. thaliana genome using the predicted amino acid sequence encoded at each loci indicated that the genes on chromosomes 3 and 8 were most similar to AtGWD1, while the other three were most similar to AtGWD3. We decided to name the two genes that appear to encode GWD1 or GWD2 isoforms as PpGWDa (Pp3c8_6536) and PpGWDb (Pp3c3_11200), while we named the other three loci PpGWDc (Pp3c17_18900), PpGWDd (Pp3c14_19150) and PpGWDe (Pp3c18_14570).
The coding sequences for two of these (PpGWDa & PpGWDb) are predicted to contain 31 introns, one (PpGWDc) contains 6 introns, while the final two (PpGWDd & PpGWDe) are intronless (Fig. 1a). Four of the five loci (PpGWDa, PPGWDb, PpGWDd & PpGWDe) encode proteins containing the histidine known to be involved in transfer of phosphate during the dikinase reaction50, while the PpGWDc locus contains a deletion that eliminates this residue (Fig. 1b). PpGWDa and PpGWDb are approximately 1420 amino acids (aa) in length, PpGWDc is 989 aa and the other two approximately 1160 aa. Amino acid sequences encoded at both PpGWDa and PpGWDb loci contain the CFATC motif thought to be involved in redox regulation51, while in PpGWDd and PpGWDe that motif is altered to VFVTC. The deletion present within PpGWDc, which eliminates the catalytic histidine, also removes the VFVTC containing region (Fig. 1b). All polypeptides encoded by these loci contain C-terminal dikinase domains, similar to those from either pyruvate phosphate dikinase or PEP synthase. PpGWDa and PpGWDb contain N-terminal PLN02784 carbohydrate-binding α-amylase domains, whilst the related CBM20 domain is present at the N-termini of PpGWDc, PpGWDd and PpGWDe (Fig. 1c).
Figure 1.
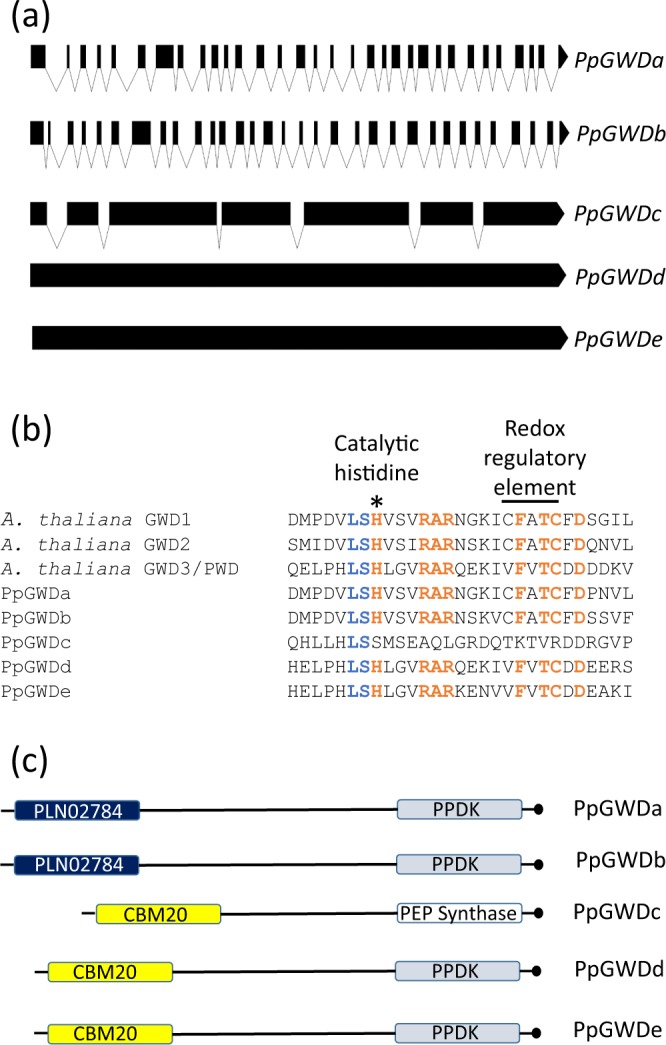
Analysis of putative GWD encoding sequences from Physcomitrella. (a) Predicted exon-intron structure of the five identified loci. (b) Amino acid sequence within the active sites of the predicted proteins in comparison with those from Arabidopsis. (c) Domain structure within the PpGWD proteins. PLN02784, CBM20, Pyruvate phosphate dikinase (PPDK) and PEP synthase domains are shown at the approximate sites that they are found within the polypeptides. The length of the figures represent the relative number of amino acids (aa) present in PpGWDa (1420aa), PpGWDb (1415aa), PpGWDc (989aa), PpGWDd (1170aa) and PpGWDe (1148aa). Black circles denote the C-termini.
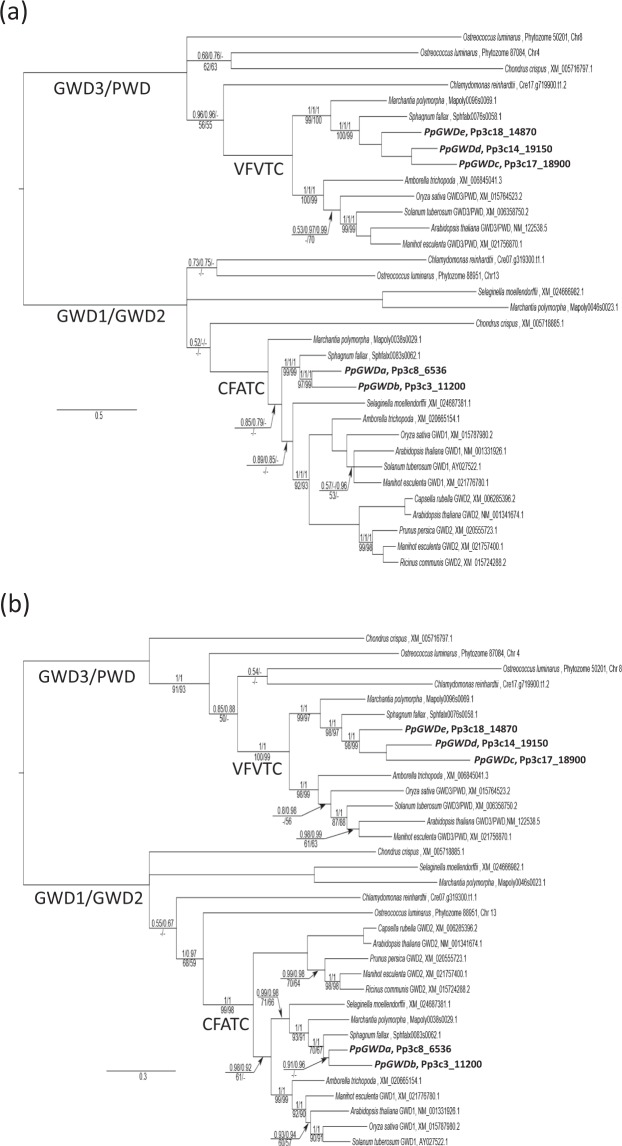
A recent phylogeny of GWD sequences has been published47, but this only included three of the five Physcomitrella sequences, and none from other non-vascular plants. To examine the relationships of the five sequences identified from the Physcomitrella genome with other GWD genes and proteins, we have produced new phylogenetic trees including all Physcomitrella sequences as well as others from a number of red and green algae, non-vascular and vascular plants. Analysis was performed using both nucleotide (DNA; Fig. 2a) and amino acid (AA; Fig. 2b) data in both a maximum likelihood (RAxML) and Bayesian (MrBayes) context.
Figure 2.
Phylogeny of GWD sequences from various plant species. (a) 50% majority rule consensus tree from partitioned MrBayes analysis of the full DNA data set of Archaeplastid GWD/PWD sequences, showing phylogenetic placement of determined P. patens (bold) paralogs. The three numbers listed above branches are Bayesian posterior probabilities (PP) for the full, partitioned analysis, PP for a partitioned analysis with a region of uncertain homology excluded, and PP for the recoded RY analysis. Numbers under each branch are RAxML likelihood bootstrap support (BS) for the full, partitioned analysis and BS for a full, partitioned analysis with a region of uncertain homology excluded. Dashes indicate support for a branch <50%, or where the branch was not present in a particular analysis. Un-numbered branches received maximal support in all analyses. The scale bar is in substitutions per site. (b) 50% majority rule consensus tree from partitioned MrBayes analysis of the full AA data set of Archaeplastid GWD/PWD sequences, showing phylogenetic placement of determined P. patens (bold) paralogs. The two numbers listed above branches are Bayesian posterior probabilities (PP) for the full analysis, and PP for analysis with a region of uncertain homology excluded. Numbers under each branch are RAxML likelihood bootstrap support (BS) for the full analysis and BS for analysis with a region of uncertain homology excluded. Dashes indicate support for a branch <50%, or where the branch was not present in a particular analysis. Unnumbered branches received maximal support in all analyses. The scale bar is in substitutions per site. In (a,b) the branches subtending two land plant clades characterized by specific motifs are labelled with the corresponding amino acid sequence (VFVTC and CFATC). Sequences are identified either by Phytozome AGI codes or NCBI accession numbers.
All MrBayes runs reached stationarity (all Potential Scale Reduction Factors between 0.99 and 1.01) and achieved adequate sample sizes (minimum ESS across combined runs >400). MrBayes AA runs overwhelmingly (posterior probability = 1) chose the WAG model52 as the best-fitting empirical model, which was used for RAxML AA analyses. When rooted on the longest internal branch, the tree consisted of two major clades, one containing GWD3/PWD sequences, and the other containing all GWD1 and GWD2 sequences. The five Physcomitrella sequences grouped in two distantly-positioned portions of the tree. The first position, retrieved in all analyses with near-maximal support, contained the three intronless/intron-poor sequences as each other’s closest relatives, with Sphagnum fallax (Sphfal0076s0058.1) and Marchantia polymorpha (Mapoly0096s0069.1) as successive sisters in all analyses with maximal or near-maximal support. All five bryophyte sequences grouped in a land plant clade with maximal support, and these land plant sequences also uniquely shared the VFVTC sequence. The second position contained the two intron-rich sequences as sister to Sphagnum fallax (Sphfal0083s0062.1), with strong support (DNA) and moderate to strong support (AA) in all analyses. DNA- and AA-based analyses differed on the closest relatives of this moss clade, with DNA analyses equivocal and AA analyses supporting Marchantia polymorpha (Mapoly0038s0029.1) and Selaginella moellendorffii (XM_024687381.1) as successive sisters, with moderate to strong support. These sequences formed part of a strongly supported land plant clade that shared the CFATC redox regulation motif (or minor variants thereof). In addition to the bryophyte/Selaginella sequences, this land plant clade included a clade containing all characterized GWD1 accessions, and a last clade containing all characterized GWD2 accessions, but relationships among these differed.
Both GWDa and GWDb localise to the plastid
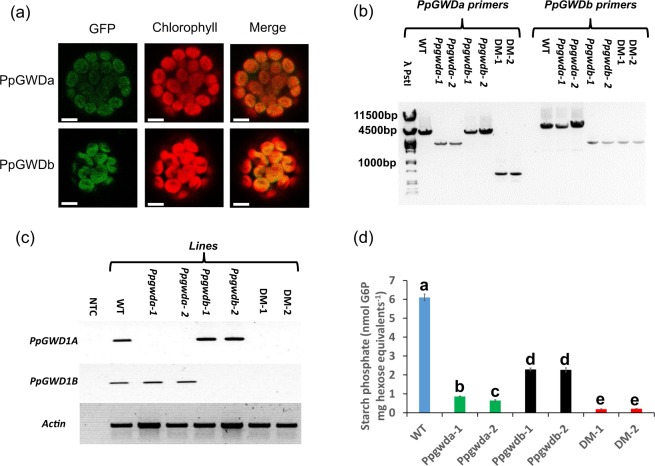
We examined the presence of transit peptides in all five genes using the ChloroP server. PpGWDa, b and e were predicted to target to plastids, while PpGWDc and d were predicted not to contain transit peptides. To confirm the localization of the GWD1 like isoforms we produced constructs where cDNA of either gene was fused in frame with GFP at the C-terminus. These were transformed into P. patens protoplasts and transiently expressing samples imaged by confocal microscopy. In both cases the GFP signal coincided exactly with that of chlorophyll, demonstrating that both proteins are plastidial (Fig. 3a).
Figure 3.
Examination of GWD in P. patens. (a) Protoplasts expressing PpGWDa or PpGWDb fused to GFP. Scale bar is 5 µm. (b) PCR analysis from gDNA demonstrating the presence of mutant alleles in Ppgwda, Ppgwdb or double mutant (DM) lines. Amplicons were separated on a 1% (w/v) agarose gel. λ-PstI represent λ phage DNA digested with PstI. (c) Semi-quantitative RT-PCR analysis of PpGWD1a, PpGWD1b or Actin expression in the wild-type (WT), Ppgwd1a, Ppgwd1b and DM experimental lines. NTC designates the no template control. Original gels are shown in Supplementary Fig. 2. (d) Glucose 6-phosphate amounts in starch from the wild-type and mutant lines. Data represent means of three independent digestions of pooled starch samples ± SEM. Letters represent groups with similar means at the 5% significance level as determined using the Bonferroni-Holm post hoc test following a one-way analysis of variance. Equal variance was determined using Levene’s test.
Mutations in both genes affect starch phosphorylation
We used homologous recombination to manufacture mutants. The constructs were designed to replace DNA encoding the known dikinase domain from either gene (Fig. 2a and Supplementary Fig. 1), which includes the catalytically essential histidine residue50, with a disruption cassette flanked with loxP sites. For each gene we recovered two plants that grew on hygromycin following transformation with gene disruption cassettes, and named these Ppgwda-1 or -2 and Ppgwdb-1 or -2. We also removed the disruption cassettes from both lines containing inserts at the PpGWDa locus using cre recombinase and, in these lines, mutated the PpGWDb locus to produce two double mutant (DM-1 or -2) lines.
To confirm that the lines we created were mutated, PCR analysis was performed using gDNA template and two sets of primers (Fig. 3b). One primer set, which binds either side of the homologous recombination sites, was used to detect the presence of either wild type alleles (amplicon sizes of 4500 bp for PpGWDa and 6000 bp for PpGWDb) or deletions within the PpGWDa locus in the DM (amplicon size approximately 700 bp). This demonstrated that wild type PpGWDa alleles were present in the Ppgwdb mutants and wild type PpGWDb alleles were present in Ppgwda mutants. As expected the PpGWDa amplicon in the DM was approximately 700 bp due to the elimination of the resistance cassette and part of the gene encoding the active site. A second primer set was used where one primer binds upstream of the insertion and another within the disruption cassette. This demonstrated the presence of the cassette within the PpGWDb locus in both single and double mutant lines (Fig. 3b). We attempted to examine protein amounts using a GWD antibody that was raised against the potato protein25, but it did not recognize any Physcomitrella GWD polypeptide. Therefore, we examined gene expression using semi quantitative RT-PCR, and showed that RNA transcribed from either PpGWDa or PpGWDb loci was abolished whenever the appropriate gene disruption cassette was present (Fig. 3c and Supplementary Fig. 2).
As GWD isoforms are known to phosphorylate starch in vascular plants, we examined the amounts of covalently bound starch phosphate in both the single and double mutants (Fig. 3d). There was a significant decrease in all the mutant lines. Starch from Ppgwda mutant lines contained approximately one seventh of the glucose 6-phosphate of the control, while lines containing an insert within the PpGWDb locus contained approximately one third. Glucose 6-phosphate was reduced below that of the Ppgwda lines in the double mutants.
Mutations in PpGWDa affect starch degradation
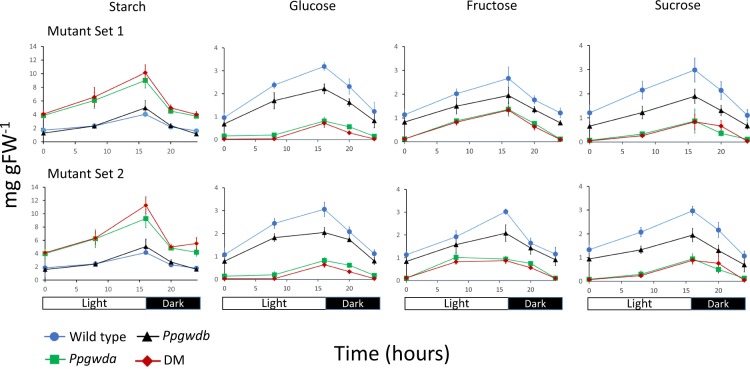
To examine starch degradation in the mutant lines, we grew colonies for 8 weeks on artificial medium before determining starch and soluble sugars over a diurnal cycle from entire colonies (Fig. 4). Starch contents increased during the light period and decreased during the dark. The WT and Ppgwdb mutants accumulated similar amounts of starch at all time points, as did the Ppgwda and DM mutants. Starch was significantly lower (p ≤ 0.05) in the WT and Ppgwdb lines in comparison with Ppgwda and DM mutants at almost all time points. The one exception was at 0 hours in the second set of mutants when the starch content in the WT and the Ppgwda mutant were invariable. Conversely, soluble sugars were reduced in the Ppgwda and DM mutant lines compared with the WT and Ppgwdb mutant. Within both mutant sets glucose was significantly (p ≤ 0.05) reduced in the Ppgwda and DM lines compared with the others at the 0, 8, 16 and 20 hour time points. Similarly fructose was significantly (p ≤ 0.05) reduced at 20 and 24 hours and sucrose at 0, 8, and 20 hours.
Figure 4.
Starch and soluble sugar amounts in colonies from the WT and mutant lines. Plants were grown on BCD medium for 5 weeks under a 16 h/8 h day/night regime. Tissue was sampled at five time points over a 24 hours period and starch, glucose, fructose and sucrose determined. Data represent means of at least 5 colonies. Error bars are SEM and, if not visible, are within the symbol.
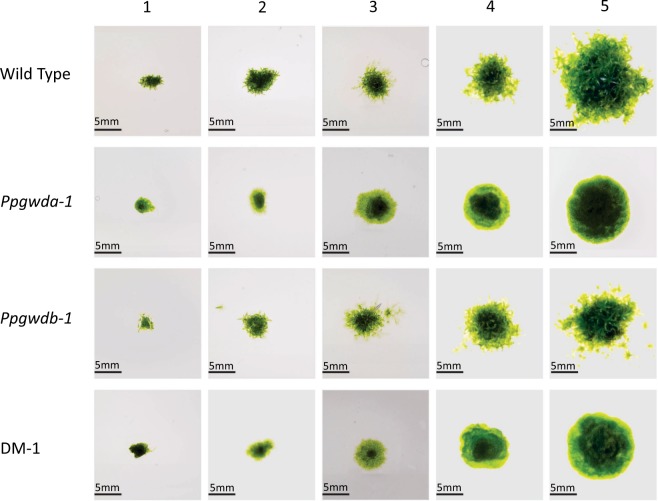
Ppgwda and DM plants demonstrate altered colony morphology
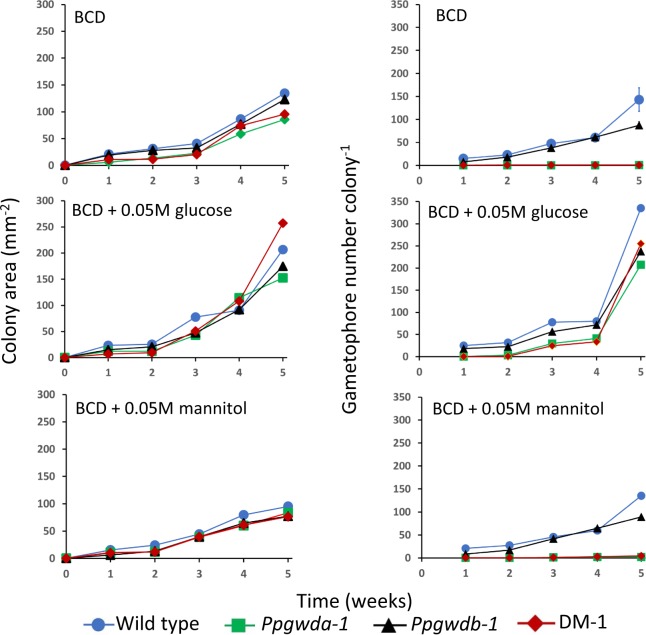
There were no consistent significant differences in growth between the lines when grown on BCD medium, however, we noted alterations in colony morphology with the WT and Ppgwdb lines developing gametophores, while the Ppgwda and DM lines did not. Data from the first mutant set are presented in Figs 4 and 5, while data from the second set are presented in Supplementary Figs 3 and 4. Colonies from the wild type and Ppgwdb mutants contained significantly (p < 0.05) more gametophores compared with Ppgwda and DM lines at every time point, when grown on BCD media or BCD media supplemented with mannitol. Under these growth conditions Ppgwdb mutants also accumulated significantly (p < 0.05) fewer gametophores than the wild-type at week 5. When the plants were grown on media containing glucose, all lines grew faster and all produced significant numbers of gametophores (Fig. 5 and Supplementary Fig. 3). The Ppgwda and DM lines still contained significantly fewer gametophores (p < 0.05) than the WT at weeks 1 and 5, but at weeks 2, 3 and 4 they contained similar numbers (Fig. 5 and Supplementary Fig. 3).
Figure 5.
Growth and gametophore number in the first set of mutant lines. Colonies were established on BCD media, BCD + 0.05 M glucose and BCD + 0.05 M mannitol and allowed to grow for 5 weeks. Data represent means of at least 3 colonies. Error bars are SEM and, if not visible, are within the symbol.
Discussion
Starch degradation is important for the normal growth and development of some angiosperms5, but knowledge about its role in these processes in non-vascular plants is lacking. Some work has been initiated in Chlamydomonas where a forward genetic approach has identified mutants affecting starch catabolism53, but to date only plants mutated in the plastidial maltose transporter have been characterized54,55. To broaden our knowledge of this process in non-vascular plants, we have examined the role of GWD1 like enzymes in Physcomitrella patens through production of targeted mutants.
Analysis of the Physcomitrella genome identified five genes encoding proteins with significant similarity to GWD1 from Arabidopsis (Fig. 1). Phylogenetic analyses (Fig. 2a,b) demonstrated the presence of two clades, each containing red algal, green algal and land plant representatives in an approximate recapitulation of recognized Archaeplastid phylogeny. This indicates that the duplication leading to GWD3/PWD-type genes and GWD1/GWD2-type genes was present in the last common Archaeplastid ancestor; yet we could find no clear homologs to any GWD-type gene in Gloeomargarita lithophora, the recently identified cyanobacterium thought to be sister to the plastid endosymbiont56. Subsequent duplications led to different isoforms of Ostreococcus-type GWD3/PWD genes, an uncharacterized GWD1/GWD2-type lineage shared by Selaginella and Marchantia, and the separation of GWD1 and GWD2 clades somewhere between the evolution of tracheophytes and angiosperms.
The differing topologies in the DNA/AA phylogenies suggest two potential evolutionary scenarios: the DNA-based trees (Fig. 2a) indicates a duplication event leading to the formation of GWD2 isoforms after the divergence of bryophytes and tracheophytes i.e. acquisition of GWD2 function and localization from a GWD1-type ancestor. Alternatively, the AA trees (Fig. 2b) suggest a much deeper separation of GWD1/GWD2 lineages, at or near the origin of land plants, but with GWD2 genes either deleted from or undetected in all non-angiosperm land plants, and ancestral function uncertain. Our analysis indicates that there are two P. patens genes (PpGWDa and PpGWDb) encoding either GWD1 and/or GWD2 isoforms. Like AtGWD126, but unlike AtGWD248, both encode polypeptides that are targeted to plastids (Fig. 3a). This may indicate that the interpretation from the DNA tree is correct, but similar analysis of GWD1/GWD2 like isoforms in green algae, other non-vascular plants, seedless vascular plants and gymnosperms will be needed to confirm this. The other three genes are most likely GWD3/PWD isoforms as they group in that clade and all contain CBM20 motifs that are found in GWD3/PWD, but not GWD1 isoforms17 (Fig. 1c). Interestingly the two PpGWD1/GWD2 genes contained similar exon intron structures, while PpGWDd and PpGWDe were intron less (Fig. 1a). All four of these genes encode proteins predicted to contain the known active site, and elements identified in vascular plant GWD1’s as redox regulatory, or variants of them (Fig. 1b)50,51. These GWD1/GWD2 and GWD3/PWD gene pairs were most likely formed during the recent genome duplication event that is thought to have occurred in Physcomitrella 30–60 million years ago, and which is known to have led to increased numbers of genes involved in metabolic processes57. PpGWDc contains 7 exons, but also a large deletion. It, therefore, encodes a protein significantly smaller than the others and lacks the catalytically essential histidine58 (Fig. 1), meaning that it is unlikely to be active.
To functionally examine the roles of the PpGWD1/PpGWD2 like enzymes, we manufactured mutants in each gene as well as isolating double mutants (DM) lacking both (Fig. 3b,c). The homologous recombination constructs used to produce the mutants were designed to remove parts of the genes encoding the known active site of the protein (Fig. 2b and Supplementary Fig. 1) meaning that any RNA manufactured from the remaining gDNA would encode an inactive polypeptide. The inserts would, however, potentially allow expression of RNA from gDNA upstream of the insert sites, which includes the PLN02784 glucan binding domains. We cannot, therefore, rule out the possibility that protein produced from this could affect starch metabolism in some way. We believe that this is unlikely as point mutations in the AtGWD1 gene allow production of inactive protein containing the carbohydrate-binding domain, but demonstrate a similar phenotype as knockouts26. For both genes we produced two independent sets of single or double mutants. We examined if the mutations reduced starch bound phosphate and found that insertions in either gene reduced the amount of glucose 6-phosphate in starch (Fig. 3d). The reduction was greater when the insertion was present in the PpGWDa locus than when PpGWDb was mutated. Starch phosphate in the double mutants was, however, lower than either single mutant line. This demonstrates that both genes encode proteins that incorporate phosphate at the 6-position of glucose moieties within the starch polymer in vivo, the known biochemical function of GWD1 isoforms, and that PpGWDa plays a greater role in this process than PpGWDb.
Reduction in starch phosphate caused by mutations in GWD1 has been demonstrated to impair starch catabolism in angiosperms25,26,45,46. This is believed to be due to the phosphate disrupting double helices between amylopectin chains and allowing access to amylases to initiate starch granule degradation17,18. To examine this we grew plants over a diurnal cycle and examined starch and soluble sugar levels (Fig. 4). As in angiosperms starch increased during the light period and decreased when it was dark. Both Ppgwda and DM lines contained significantly more starch than the WT or Ppgwdb line at almost all time points. Conversely, soluble sugar contents were reduced at most time points in the DM and Ppgwda mutants compared with the WT and Ppgwdb lines (Fig. 4). This demonstrates that mutations affecting PpGWDa reduce starch degradation in P. patens as Ppgwda and DM mutant plants develop a starch excess phenotype, but that PpGWDb has little effect on this process. Plants lacking PpGWDa can clearly still degrade starch. Indeed they degraded more starch between time points than either the WT or Ppgwdb (Fig. 4), most likely due to the larger amounts they accumulated prior to the start of the experiment. This is similar to other plants lacking GWD where starch is still degraded in leaves over a diurnal cycle, but where a starch excess phenotype develops over time due to small differences in the amount of starch that is mobilised at night25,26,46.
The influence of the two enzymes on starch catabolism is in line with their observed effects on starch phosphate amounts, where PpGWDa has a greater influence than PpGWDb (Fig. 3d). However, the relationship between starch phosphate reduction and starch catabolism is not linear as mutations in PpGWDb reduce starch phosphate by more than 50%, but do not affect starch degradation. This indicates both that starch phosphate must be reduced below a threshold level before an effect on starch degradation is observed, and that the starch phosphate is above that threshold in the WT and Ppgwdb mutants, but below it in the Ppgwda and DM lines. Interestingly a previous study59 in Arabidopsis demonstrated that GWD amounts had to be reduced below a threshold level before starch degradation was inhibited, although the authors did not examine starch phosphate in that study.
Mutant plants lacking PpGWDa demonstrated an altered appearance when grown on BCD medium, with both Ppgwda and DM plants developing almost no gametophores (Figs 5, 6 and Supplementary Figs 3 and 4). We hypothesized that the decrease in starch degradation would reduce soluble sugars within plant cells, and that this could impact both growth and plant development. To examine this, we grew plants on media containing either 0.05 M glucose or 0.05 M mannitol. When placed on glucose-containing medium, all plants grew more quickly and both Ppgwda and DM lines developed gametophores, although not to the same extent as wild type or Ppgwdb mutants. Neither the increase in growth nor gametophore development occurred when Ppgwda or DM lines were grown on mannitol-containing medium, indicating that the reversion of the mutant phenotype is caused by glucose uptake and not the altered osmotic potential. Interestingly mutations in PpGWDb also reduced gametophore number to about two thirds of the wild-type strain at week 5. It is not clear why this is the case as we could not identify decreased starch degradation in these lines. It is possible that PpGWDb is present at specific growth stages, or in only some cell types, leading to a localized reduction in starch degradation that impacts on gametophore development.
Figure 6.
Colony morphology of wild-type and the first set of mutant lines grown on BCD medium for 1, 2, 3, 4 and 5 weeks.
These data demonstrate that starch-derived soluble sugars influence the development of gametophores in P. patens. It is possible that decreased soluble sugars inhibit gametophore development directly due to reduction in carbon skeletons, possibly through decreasing substrate supplies to a cellulose synthase which are essential for gametophore development6. It is, however, also possible that downstream processes repressing gametophore initiation are affected by the reduced sugar levels. The development both of gametophore forming caulonemal tissue and gametophores is known to be under hormonal control, being influenced by auxins7 and cytokinins8, as well as a recently identified ancestral gibberrelin60. It is unclear if amounts of any of these hormones have been altered to lead to this phenotype, although it is interesting to note that mutations affecting starch degradation in higher plants affects gibberellin synthesis61. Our future work will examine alterations in these factors within the Ppgwd1a mutant to identify the underlying changes that inhibit gametophore formation.
Methods
Analysis of GWD genes from P. patens and comparison with those from other species
tBLASTn searches of the Physcomitrella patens genome were performed using the GWD1 sequence from Arabidopsis thaliana (UniProtKB# Q9SAC6) at Phytozome 12.1.6. Genomic sequences of putative GWD encoding genes were downloaded and predicted exon-intron boundaries were visualized at www.wormweb.org/exonintron. The presence of predicted chloroplast transit peptides was examined using ChloroP 1.162 (http://www.cbs.dtu.dk/services/ChloroP/) and conserved domains within predicted polypeptides were identified through searches of the CCD database63 at https://www.ncbi.nlm.nih.gov/Structure/cdd/wrpsb.cgi.
DNA sequences encoding GWD polypeptides were obtained from either the NCBI or Phytozome and used to construct a phylogenetic tree. DNA encoding the predicted PPDK domains from GWD were translated to amino acid sequence and aligned using Muscle64 in MegaX65. This was followed by subsequent manual alignment of clearly homoplasious indels, as well as of a region encoding amino acids 214–304 from the start of the alignment, which was highly variable. Phylogenetic analyses were conducted using the CIPRES66 implementation of MrBayes 3.2.667 and RAxML v8.2.468, using both the nucleotide (DNA) and amino acid (AA) alignments. All DNA analyses were partitioned by codon position, which yielded vastly improved log likelihood values over naïve analyses (mean Δln likelihood values MrBayes: >1000; Δln likelihood values RAxML: >540). MrBayes partitions were allowed to average over GTR submodels (lset nst = mixed) and assigned a gamma correction for among-site rate variation. All MrBayes analyses were run twice, with 1 million generations and default burnin fractions. All runs were checked for stationarity and adequate estimated sample sizes using MrBayes’ own diagnostics and the program Tracer v1.6–1.769. RAxML DNA partitions were individually assigned the GTRGAMMA model under otherwise default settings, with the bootstopping criterion to determine adequate bootstrap replicates70. In addition, because of >35% difference in GC content across sampled sequences, which can bias phylogenetic inference71, nucleotide alignments were also transformed to RY-coding (purine-pyrimidine coding) to check for artefacts introduced by base composition heterogeneity and run in MrBayes under the previously mentioned parameters. AA alignments in MrBayes were allowed to average over a set of empirical substitution rate matrices, with a gamma correction applied. Otherwise MrBayes AA analyses were run under the same settings and checked in the same way as the nucleotide results. RAxML AA results used the best-fitting empirical matrix from the MrBayes runs as default, and otherwise were run as for the DNA results. Finally, both DNA and AA matrices were run excluding the highly variable region as defined above, to check whether this region of uncertain homology unduly influenced results. All trees were rooted on the longest internal branch, which was also the branch chosen by midpoint rooting.
Plant material and growth conditions
P. patens ssp. patens strain Gransden was a kind gift of Dr. P. Hills (Institute for Plant Biotechnology, Stellenbosch University, South Africa). Standard growth conditions for cultures were 23 ± 2 °C with a light intensity of 50 μmol photons m−2 s−1 (Osram L 58 V/740, Germany) and a 16/8 hour light/dark regime. Cultures used for starch phosphate analyses were grown on PpNH4 medium (1 mM MgSO4, 1.85 mM KH2PO4, 10 mM KCL, 45 mM FeSO4, 1 mM CaCl2, 9.93 µM H3BO3, 0.103 µM Na2MoO4, 0.266 µM CoCl2, 0.191 µM ZnSO4, 1.97 µM MnCl2, 0.169 µM KI, 0.22 µM CuSO4, 0.57 µM Al2(SO4)3, 5 mM diammonium tartrate) supplemented with 1% (w/v) sucrose. For analysis of growth rates and plant morphology, colonies were divided into approximately 2mm2 pieces and placed on BCD media (1 mM MgSO4, 1.85 mM KH2PO4, 10 mM KNO3, 45 mM FeSO4, 1 mM CaCl2, 9.93 µM H3BO3, 0.103 µM Na2MoO4, 0.266 µM CoCl2, 0.191 µM ZnSO4, 1.97 µM MnCl2, 0.169 µM KI, 0.22 µM CuSO4, 0.57 µM Al2(SO4)3) or the same media supplemented with either 0.05 M glucose or 0.05 M mannitol.
Silencing construct preparation
Genomic DNA was extracted from P. patens by the protocol of Edwards et al.72. Using this as template, amplicons encoding sections of glucan, water dikinase (PpGWD1a or PpGWD1b) genes were amplified by PCR with gene specific primers (GWDa FWD 5′TTCAGCCAGATAGCGTCGTC3′ and GWDa REV 5′GCCATGCACAAGTCGCTATG3′; GWDb FWD 5′TTGCAGGCGGCTTCTGAACTA3′ and GWDb REV 5′GCTCCCACAAGTGTCTCTCC3′). Amplicons were subsequently cloned into the pJET1.2/blunt vector using a CloneJET PCR Cloning Kit, according to manufacturer’s specifications (Thermo Scientific, Waltham, MA, USA). The plasmid containing PpGWDa was digested with SalI and SphI before a loxP-flanked Hygromycin B phosphotransferase (hph) resistance cassette was excised from the pMBLH8a73 plasmid by digesting with the same enzymes and ligating into the genomic fragment. The plasmid containing the GWDb genomic fragment was restricted using StuI and ClaI and the same hph resistance cassette was excised from pMBLH8a using EvoRV and ClaI, before being ligated into the GWDb genomic DNA. The resulting mutations constructs (PpGWDa-HygR-KO and PpGWDb-HygR-KO) were digested with either NotI (PpGWDa-HygR-KO) or XbaI (PpGWDb-HygR-KO) to produce linear plasmids for transformation.
Production of single and double mutants
Protoplasts were transformed using PEG mediated transformation and mutant plants recovered following selection74. For production of double mutants the disruption cassette in the Ppgwda single mutant was removed using Cre recombinase. The DNA sequence encoding Cre-recombinase was amplified by PCR from pMM2375 using the following primers: Cre Forward 5′ATGTCCAATTTACTGACCGTAC3′ and Cre Reverse 5′CTAATCGCCATCTTCCAGC3′. Purified amplicon DNA was cloned into the pCR8/GW/TOPO according to the manufacturers (Life Technologies, USA-CA) specifications. The expression cassette from pBinAR-Hyg76 was excised using the restriction enzymes EcoRI and HindIII, and ligated into the same sites within pCAMBIA2200 (http://www.cambia.org/daisy/cambia/585). A Gateway reading frame cassette (Gateway Vector Conversion Kit, Thermo Fisher Scientific) was ligated in the SmaI site of the expression cassette polylinker in sense orientation with respect to the 35S promoter. The cre-sequence was transferred into this vector using LR clonase (Life Technologies) as described in the manufacturer’s instructions to obtain pCAMBIA2200-Cre. Protoplasts isolated from Ppgwda mutant lines were transformed with pCAMBIA2200-Cre using PEG74. Protoplasts were regenerated on BCDAT (BCD medium containing 5 Mm ammonium tartrate) medium lacking antibiotics for two weeks, after which they were transferred to BCDAT containing 50 µg/mL geneticin. After one 1 week, colonies that had survived the selection stage were divided into two, plated on both selective (BCDAT plus Hygromycin-B) and non-selective (BCDAT) media and allowed to grow for another week. Replicates growing only on non-selection medium were screened for loss of hph resistance cassette. Plants confirmed as lacking the disruption cassette and containing the deletion within PpGWD1a were further mutated using PpGWD1b-HygR-KO using the same method as for the production of single mutants.
Semi-quantitative reverse-transcriptase PCR
Total RNA was extracted from 2 week old P. patens protonemal tissue using Qiagen RNeasy® Plant Mini Kit (Whitehead Scientific, Cape Town, South Africa), according to the manufacturer’s instructions. Synthesis of cDNA was performed with the purified RNA by using the RevertAid™ H Minus First Strand cDNA Synthesis Kit (Thermo Scientific, Waltham, MA, USA). The following primers were used in the PCR: GWD1AsqFwd 5′ACTTGTGCATGGCAGTGTT3′ GWD1AsqRev 5′AACGACGTACAGTTCACCATC3′, GWD1BsqFwd 5′GTGGATCCGTCTTCCAACAT3′ GWD1BsqRev 5′AATATGCTCCCACAAGTGTCTC3′ to examine expression of PpGWD genes. The linear amplification range was determined using P. patens actin primers77 (ActsqFwd 5′AAGGCGAACAGGGAGAAGAT3′; ActsqRev 5′TCCACGAGACGACGTACAAC3′), and 20 cycles was chosen as the optimum number of cycles to use for all semi-quantitative RT-PCR reactions.
GFP fusions and confocal microscopy
GWDa and GWDb cDNA were amplified by PCR using the following primers: PpGWD1a-GFP forward (5′ATGCAGAGACACGGAGTTCT3′) and reverse (5′TTCAAACGAGACCCCAGATG3′); PpGWD1b-GFP forward (5′ATGAAGAGCTTCAGAGCTCA3′) and reverse (5′TTCAAACAAGACCGCAAATG3′) and cloned into pENTR/D-TOPO (ThermoFischer Scientific). The inserts were transferred to the pMPL1382 GFP vector (http://labs.biology.ucsd.edu/estelle/moss2.html) using LR clonase. The GFP vectors were linearized using SphI and protoplasts were transformed with this DNA as described above. Protoplasts transiently expressing GFP were imaged using a Carl Zeiss LSM780 confocal microscope with an ELRYA S.1 super-resolution platform. GFP signal was detected using a laser excitation of 488 nm and GFP filter detection range of 490–579 nm. Chloroplast auto-fluorescence was detected using a laser excitation of 405 nm and chlorophyll filter detection range of 625–738 nm. Images were analysed with the Zen (black edition, version 2.3) imaging software (Carl Zeiss, Germany).
Growth and gametophore analysis
Plants grown on BCD medium or BCD supplemented with sugars were imaged and surface area determine by image analysis using ImageJ 1.52a78. Gametophore numbers were counted manually.
Starch, sugar and starch phosphate determination
Starch was measure enzymatically following digestion to glucose. Briefly, 20–50 mg plant material was placed in a microcentrifuge tube and sugars were removed by adding 1 mL of 80% (v/v) ethanol and heating at 95 °C for 1 hour before the ethanol was decanted. This step was repeated twice more. After the final wash, 0.4 mL of 0.2 M KOH was added and the sample heated at 95 °C for one hour before the KOH was neutralized through addition of 70 μL of 1 M acetic acid. At this point 1.5 mL of 100 mM NaAC (pH 5.5) was added alongside 65 U of amyloglucosidase (Aspergillus niger, Megazyme) and 60 U of thermostable α-amylase (Bacillus sp., Megazyme). This was incubated at 50 °C for 1 hour before 100 μL was mixed with 200 μL of 100 mM Tris-HCl (pH 7.0), 5 mM MgCl2, 1 mM ATP and 1 mM NAD. This was assayed at 340 nm in a microtitreplate reader for increase in absorbance after addition of 1 U of hexokinase (Yeast, Megazyme) and 0.5 U of glucose 6-phosphate dehydrogenase (Leuconostoc mesenteroides, Megazyme). Glucose, fructose and sucrose were determined in the ethanol extracts by a previously described method79. Starch phosphate was determined by a previously published method80 using starch purified81 from protonemal tissue.
Statistical analysis
Data were analysed by one way analysis of variance followed by a Bonferroni-Holm post hoc test using Daniel’s XL Toolbox add-in for Excel82, version 7.3.2 (www.xltoolbox.net). Equal variance was demonstrated using Levene’s test83.
Supplementary information
Acknowledgements
We would like to thank Dr David Ow (Plant Gene Engineering Center, South China Botanical Garden) for a gift of the Cre recombinase plasmid and Prof Mark Estelle and Dr Michael Prigge (University of California, San Diego) for the gift of pMP1382. This project was funded by the by the Swiss/South African Joint Research Program grant 87391 and the NRF SARCHI chair “Genetic Tailoring of Biopolymers”. Bursary funding was obtained from the South African National Research Foundation for Ms Mdodana (SFH13091440165) and Mr Jewell (SFH14072881440). We would especially like to thank Mr Michael Rosmarin for assistance with the photography and Mr Adegbaju Muyiwa Seyi for help with preparing some of the figures.
Author contributions
The project was conceptualized by J.R.L. and J.K. Plants were initially transformed by N.T.M., J.F.J. and S.M. and all subsequent experimental work was performed by N.T.M. M.J.S. helped produce the Cre recombinease expressing vector used to make the double mutants. The initial phylogeny was established by E.E.P. and expanded by J.R.L. and K.O. The manuscript was written by N.T.M. and J.R.L. All authors proofread the paper and provided feedback.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
is available for this paper at 10.1038/s41598-019-51632-9.
References
- 1.Rensing SA, et al. The Physcomitrella genome reveals evolutionary insights into the conquest of land by plants. Science. 2008;319:64–69. doi: 10.1126/science.1150646. [DOI] [PubMed] [Google Scholar]
- 2.Cove, D. J. & Cuming, A. C. Genetics and Genomics of Moss Models: Physiology Enters the Twenty-first Century. In Photosynthesis in Bryophytes and Early Land Plants (eds Hanson, D. T. & Rice, S. K.) 187–199, 10.1007/978-94-007-6988-5_11 (Springer, 2014).
- 3.Cove D, Bezanilla M, Harries P, Quatrano R. Mosses as model systems for the study of metabolism and development. Annu. Rev. Plant Biol. 2006;57:497–520. doi: 10.1146/annurev.arplant.57.032905.105338. [DOI] [PubMed] [Google Scholar]
- 4.Lloyd JR, Kossmann J. Starch trek: The search for yield. Front. Plant Sci. 2019;9:1930. doi: 10.3389/fpls.2018.01930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Stitt M, Zeeman SC. Starch turnover: Pathways, regulation and role in growth. Curr. Opin. Plant Biol. 2012;15:282–292. doi: 10.1016/j.pbi.2012.03.016. [DOI] [PubMed] [Google Scholar]
- 6.Goss CA, Brockmann DJ, Bushoven JT, Roberts AW. A CELLULOSE SYNTHASE (CESA) gene essential for gametophore morphogenesis in the moss Physcomitrella patens. Planta. 2012;235:1355–1367. doi: 10.1007/s00425-011-1579-5. [DOI] [PubMed] [Google Scholar]
- 7.Thelander M, Landberg K, Sundberg E. Auxin-mediated developmental control in the moss Physcomitrella patens. J. Exp. Bot. 2018;69:277–290. doi: 10.1093/jxb/erx255. [DOI] [PubMed] [Google Scholar]
- 8.von Schwartzenberg K, et al. Cytokinins in the bryophyte Physcomitrella patens: Analyses of activity, distribution, and cytokinin oxidase/dehydrogenase overexpression reveal the role of extracellular cytokinins. Plant Physiol. 2007;145:786–800. doi: 10.1104/pp.107.103176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lopez-Obando M, et al. Physcomitrella patens MAX2 characterization suggests an ancient role for this F-box protein in photomorphogenesis rather than strigolactone signalling. New Phytol. 2018;219:743–756. doi: 10.1111/nph.15214. [DOI] [PubMed] [Google Scholar]
- 10.Cho SH, Coruh C, Axtell MJ. miR156 and miR390 regulate tasiRNA accumulation and developmental timing in Physcomitrella patens. Plant Cell. 2012;24:4837–4849. doi: 10.1105/tpc.112.103176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Moody LA, Kelly S, Rabbinowitsch E, Langdale JA. Genetic regulation of the 2D to 3D growth transition in the moss Physcomitrella patens. Curr. Biol. 2018;28:473–478. doi: 10.1016/j.cub.2017.12.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Moody LA. The 2D to 3D growth transition in the moss Physcomitrella patens. Curr. Opin. Plant Biol. 2019;47:88–95. doi: 10.1016/j.pbi.2018.10.001. [DOI] [PubMed] [Google Scholar]
- 13.Olsson T, Thelander M, Ronne H. A novel type of chloroplast stromal hexokinase is the major glucose-phosphorylating enzyme in the moss Physcomitrella patens. J. Biol. Chem. 2003;278:44439–44447. doi: 10.1074/jbc.M306265200. [DOI] [PubMed] [Google Scholar]
- 14.Thelander M, Olsson T, Ronne H. Snf1-related protein kinase 1 is needed for growth in a normal day–night light cycle. EMBO J. 2004;23:1900–1910. doi: 10.1038/sj.emboj.7600182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Thelander M. Effect of the energy supply on filamentous growth and development in Physcomitrella patens. J. Exp. Bot. 2005;56:653–662. doi: 10.1093/jxb/eri040. [DOI] [PubMed] [Google Scholar]
- 16.Nagao M, Minami A, Arakawa K, Fujikawa S, Takezawa D. Rapid degradation of starch in chloroplasts and concomitant accumulation of soluble sugars associated with ABA-induced freezing tolerance in the moss Physcomitrella patens. J. Plant Physiol. 2005;162:169–180. doi: 10.1016/j.jplph.2004.06.012. [DOI] [PubMed] [Google Scholar]
- 17.Mahlow S, Orzechowski S, Fettke J. Starch phosphorylation: Insights and perspectives. Cell. Mol. Life Sci. 2016;73:2753–2764. doi: 10.1007/s00018-016-2248-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Blennow A, Engelsen SB. Helix-breaking news: Fighting crystalline starch energy deposits in the cell. Trends Plant Sci. 2010;15:236–240. doi: 10.1016/j.tplants.2010.01.009. [DOI] [PubMed] [Google Scholar]
- 19.MacNeill GJ, et al. Starch as a source, starch as a sink: The bifunctional role of starch in carbon allocation. J. Exp. Bot. 2017;68:4433–4453. doi: 10.1093/jxb/erx291. [DOI] [PubMed] [Google Scholar]
- 20.Ritte G, et al. Phosphorylation of C6- and C3-positions of glucosyl residues in starch is catalysed by distinct dikinases. FEBS Lett. 2006;580:4872–4876. doi: 10.1016/j.febslet.2006.07.085. [DOI] [PubMed] [Google Scholar]
- 21.Hejazi M, et al. Glucan, water dikinase phosphorylates crystalline maltodextrins and thereby initiates solubilization. Plant J. 2008;55:323–334. doi: 10.1111/j.1365-313X.2008.03513.x. [DOI] [PubMed] [Google Scholar]
- 22.Edner C, et al. Glucan, water dikinase activity stimulates breakdown of starch granules by plastidial β-amylases. Plant Physiol. 2007;145:17–28. doi: 10.1104/pp.107.104224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ritte G, Scharf A, Eckermann N, Haebel S, Steup M. Phosphorylation of transitory starch is increased during degradation. Plant Physiol. 2004;135:2068–2077. doi: 10.1104/pp.104.041301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ritte G, et al. The starch-related R1 protein is an alpha-glucan, water dikinase. Proc. Natl. Acad. Sci. USA. 2002;99:7166–7171. doi: 10.1073/pnas.062053099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lorberth R, Ritte G, Willmitzer L, Kossmann J. Inhibition of a starch-granule-bound protein leads to modified starch and repression of cold sweetening. Nat. Biotechnol. 1998;16:473–477. doi: 10.1038/nbt0598-473. [DOI] [PubMed] [Google Scholar]
- 26.Yu, T.-S. et al. The Arabidopsis sex1 mutant is defective in the R1 protein, a general regulator of starch degradation in plants, and not in the chloroplast hexose transporter. Plant Cell13 (2001). [DOI] [PMC free article] [PubMed]
- 27.Baunsgaard L, et al. A novel isoform of glucan, water dikinase phosphorylates pre-phosphorylated α-glucans and is involved in starch degradation in Arabidopsis. Plant J. 2005;41:595–605. doi: 10.1111/j.1365-313X.2004.02322.x. [DOI] [PubMed] [Google Scholar]
- 28.Kötting O, et al. Identification of a novel enzyme required for starch metabolism in Arabidopsis leaves. The phosphoglucan, water dikinase. Plant Physiol. 2005;137:242–252. doi: 10.1104/pp.104.055954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Streb S, Eicke S, Zeeman SC. The simultaneous abolition of three starch hydrolases blocks transient starch breakdown in Arabidopsis. J. Biol. Chem. 2012;287:41745–41756. doi: 10.1074/jbc.M112.395244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Scheidig A, Frohlich A, Schulze S, Lloyd JR, Kossmann J. Downregulation of a chloroplast-targeted β-amylase leads to a starch-excess phenotype in leaves. Plant J. 2002;30:581–591. doi: 10.1046/j.1365-313X.2002.01317.x. [DOI] [PubMed] [Google Scholar]
- 31.Fulton DC, et al. β-AMYLASE4, a noncatalytic protein required for starch breakdown, acts upstream of three active β-amylases in Arabidopsis chloroplasts. Plant Cell. 2008;20:1040–1058. doi: 10.1105/tpc.107.056507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kaplan F, Guy CL. RNA interference of Arabidopsis β-amylase8 prevents maltose accumulation upon cold shock and increases sensitivity of PSII photochemical efficiency to freezing stress. Plant J. 2005;44:730–743. doi: 10.1111/j.1365-313X.2005.02565.x. [DOI] [PubMed] [Google Scholar]
- 33.Kötting O, et al. STARCH-EXCESS4 is a Laforin-like phosphoglucan phosphatase required for starch degradation in Arabidopsis thaliana. Plant Cell. 2009;21:334–346. doi: 10.1105/tpc.108.064360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Santelia D, et al. The phosphoglucan phosphatase Like Sex Four2 dephosphorylates starch at the C3-position in Arabidopsis. Plant Cell. 2011;23:4096–4111. doi: 10.1105/tpc.111.092155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Samodien E, et al. Repression of Sex4 and Like Sex Four2 orthologs in potato increases tuber starch bound phosphate with concomitant alterations in starch physical properties. Front. Plant Sci. 2018;9:1044. doi: 10.3389/fpls.2018.01044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Critchley JH, Zeeman SC, Takaha T, Smith AM, Smith SM. A critical role for disproportionating enzyme in starch breakdown is revealed by a knock-out mutation in Arabidopsis. Plant J. 2001;26:89–100. doi: 10.1046/j.1365-313x.2001.01012.x. [DOI] [PubMed] [Google Scholar]
- 37.Lütken, H. et al. Repression of both isoforms of disproportionating enzyme leads to higher malto-oligosaccharide content and reduced growth in potato. Planta232 (2010). [DOI] [PubMed]
- 38.George GM, Bauer R, Blennow A, Kossmann J, Lloyd JR. Virus-induced multiple gene silencing to study redundant metabolic pathways in plants: Silencing the starch degradation pathway in Nicotiana benthamiana. Biotechnol. J. 2012;7:884–890. doi: 10.1002/biot.201100469. [DOI] [PubMed] [Google Scholar]
- 39.Niittylä T, et al. A previously unknown maltose transporter essential for starch degradation in leaves. Science. 2004;303:87–89. doi: 10.1126/science.1091811. [DOI] [PubMed] [Google Scholar]
- 40.Weber A. Identification, purification, and molecular cloning of a putative plastidic glucose translocator. Plant Cell. 2000;12:787–802. doi: 10.1105/tpc.12.5.787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cho M-H, et al. Role of the plastidic glucose translocator in the export of starch degradation products from the chloroplasts in Arabidopsis thaliana. New Phytol. 2011;190:101–112. doi: 10.1111/j.1469-8137.2010.03580.x. [DOI] [PubMed] [Google Scholar]
- 42.Chia T, et al. A cytosolic glucosyltransferase is required for conversion of starch to sucrose in Arabidopsis leaves at night. Plant J. 2004;37:853–863. doi: 10.1111/j.1365-313X.2003.02012.x. [DOI] [PubMed] [Google Scholar]
- 43.Lloyd JR, Blennow A, Burhenne K, Kossmann J. Repression of a novel isoform of disproportionating enzyme (stDPE2) in potato leads to inhibition of starch degradation in leaves but not tubers stored at low temperature. Plant Physiol. 2004;134:1347–1354. doi: 10.1104/pp.103.038026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nashilevitz S, et al. The legwd mutant uncovers the role of starch phosphorylation in pollen development and germination in tomato. Plant J. 2009;57:1–13. doi: 10.1111/j.1365-313X.2008.03664.x. [DOI] [PubMed] [Google Scholar]
- 45.Vriet C, et al. A suite of Lotus japonicus starch mutants reveals both conserved and novel features of starch metabolism. Plant Physiol. 2010;154:643–655. doi: 10.1104/pp.110.161844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hirose T, et al. Disruption of a rice gene for α-glucan water dikinase, OsGWD1, leads to hyperaccumulation of starch in leaves but exhibits limited effects on growth. Front. Plant Sci. 2013;4:147. doi: 10.3389/fpls.2013.00147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhou W, et al. Alpha-Glucan, Water Dikinase 1 affects starch metabolism and storage root growth in Cassava (Manihot esculenta Crantz) Sci. Rep. 2017;7:9863. doi: 10.1038/s41598-017-10594-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Glaring MA, et al. An extra-plastidial α-glucan, water dikinase from Arabidopsis phosphorylates amylopectin in vitro and is not necessary for transient starch degradation. J. Exp. Bot. 2007;58:3949–3960. doi: 10.1093/jxb/erm249. [DOI] [PubMed] [Google Scholar]
- 49.Pirone C, et al. The analysis of the different functions of starch-phosphorylating enzymes during the development of Arabidopsis thaliana plants discloses an unexpected role for the cytosolic isoform GWD2. Physiol. Plant. 2017;160:447–457. doi: 10.1111/ppl.12564. [DOI] [PubMed] [Google Scholar]
- 50.Mikkelsen R, Baunsgaard L, Blennow A. Functional characterization of alpha-glucan,water dikinase, the starch phosphorylating enzyme. Biochem. J. 2004;377:525–532. doi: 10.1042/bj20030999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Mikkelsen R, Mutenda KE, Mant A, Schurmann P, Blennow A. α-glucan, water dikinase (GWD): A plastidic enzyme with redox-regulated and coordinated catalytic activity and binding affinity. Proc. Natl. Acad. Sci. 2005;102:1785–1790. doi: 10.1073/pnas.0406674102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Whelan S, Goldman N. A general empirical model of protein evolution derived from multiple protein families using a maximum-likelihood approach. Mol. Biol. Evol. 2001;18:691–699. doi: 10.1093/oxfordjournals.molbev.a003851. [DOI] [PubMed] [Google Scholar]
- 53.Tunçay H, et al. A forward genetic approach in Chlamydomonas reinhardtii as a strategy for exploring starch catabolism. PLoS One. 2013;8:e74763. doi: 10.1371/journal.pone.0074763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Jang S, et al. Characterization of a Chlamydomonas reinhardtii mutant defective in a maltose transporter. J. Plant Biol. 2015;58:344–351. doi: 10.1007/s12374-015-0377-1. [DOI] [Google Scholar]
- 55.Findinier, J. et al. The Chlamydomonas mex1 mutant shows impaired starch mobilization without maltose accumulation. J. Exp. Bot. 68 (2017). [DOI] [PubMed]
- 56.Ponce-Toledo RI, et al. An early-branching freshwater Cyanobacterium at the origin of plastids. Curr. Biol. 2017;27:386–391. doi: 10.1016/j.cub.2016.11.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Rensing SA, et al. An ancient genome duplication contributed to the abundance of metabolic genes in the moss Physcomitrella patens. BMC Evol. Biol. 2007;7:130. doi: 10.1186/1471-2148-7-130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Mikkelsen R, Baunsgaard L, Blennow A. Functional domain organization of the potato alpha-glucan, water dikinase (GWD): evidence for separate site catalysis as revealed by limited proteolysis and deletion mutants. Biochem. J. 2005;385:355–361. doi: 10.1042/BJ20041119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Skeffington AW, Graf A, Duxbury Z, Gruissem W, Smith AM. Glucan, water dikinase exerts little control over starch degradation in Arabidopsis leaves at night. Plant Physiol. 2014;165:866–879. doi: 10.1104/pp.114.237016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Miyazaki S, et al. An ancestral gibberellin in a moss Physcomitrella patens. Mol. Plant. 2018;11:1097–1100. doi: 10.1016/j.molp.2018.03.010. [DOI] [PubMed] [Google Scholar]
- 61.Paparelli E, et al. Nighttime sugar starvation orchestrates gibberellin biosynthesis and plant growth in Arabidopsis. Plant Cell. 2013;25:3760–3769. doi: 10.1105/tpc.113.115519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Emanuelsson O, Nielsen H, Heijne G. Von. ChloroP, a neural network-based method for predicting chloroplast transit peptides and their cleavage sites. Protein Sci. 1999;8:978–984. doi: 10.1110/ps.8.5.978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Marchler-Bauer A, et al. CDD: NCBI’s conserved domain database. Nucleic Acids Res. 2015;43:D222–D226. doi: 10.1093/nar/gku1221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kumar S, Stecher G, Li M, Knyaz C, Tamura K. MEGA X: Molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 2018;35:1547–1549. doi: 10.1093/molbev/msy096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Miller, M., Pfeiffer, W. & Schwartz, T. Creating the CIPRES science gateway for inference of large phylogenetic trees. in 2010 Gateway Computing Environments Workshop (GCE), 10.1109/GCE.2010.5676129 (2010).
- 67.Ronquist F, et al. MrBayes 3.2: Efficient Bayesian phylogenetic inference and model choice across a large model space. Syst. Biol. 2012;61:539–542. doi: 10.1093/sysbio/sys029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Stamatakis A. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 2014;30:1312–1313. doi: 10.1093/bioinformatics/btu033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Rambaut A, Drummond AJ, Xie D, Baele G, Suchard MA. Posterior summarization in Bayesian phylogenetics using Tracer 1.7. Syst. Biol. 2018;67:901–904. doi: 10.1093/sysbio/syy032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Pattengale ND, Alipour M, Bininda-Emonds ORP, Moret BME, Stamatakis A. How many bootstrap replicates are necessary? J. Comput. Biol. 2010;17:337–354. doi: 10.1089/cmb.2009.0179. [DOI] [PubMed] [Google Scholar]
- 71.Hasegawa M, Hashimoto T. Ribosomal RNA trees misleading? Nature. 1993;361:23–23. doi: 10.1038/361023b0. [DOI] [PubMed] [Google Scholar]
- 72.Edwards K, Johnstone C, Thompson C. A simple and rapid method for the preparation of plant genomic DNA for PCR analysis. Nucleic Acids Res. 1991;19:1349–1349. doi: 10.1093/nar/19.6.1349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Knight, C., Cove, D., Cumming, A. & Quatrano, R. Moss gene technology. In Molecular Plant Biology: A Practical Approach, vol 2 (eds Gilmartin, P. & Bowler, C.) 285–301 (Oxford University Press, 2002).
- 74.Liu, Y.-C. & Vidali, L. Efficient polyethylene glycol (PEG) mediated transformation of the moss Physcomitrella patens. J. Vis. Exp. e2560, 10.3791/2560 (2011). [DOI] [PMC free article] [PubMed]
- 75.Qin M, Bayley C, Stockton T, Ow DW. Cre recombinase-mediated site-specific recombination between plant chromosomes. Proc. Natl. Acad. Sci. 1994;91:1706–1710. doi: 10.1073/pnas.91.5.1706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Abel GJW, Springer F, Willmitzer L, Kossmann J. Cloning and functional analysis of a cDNA encoding a novel 139 kDa starch synthase from potato (Solanum tuberosum L) Plant J. 1996;10:981–991. doi: 10.1046/j.1365-313X.1996.10060981.x. [DOI] [PubMed] [Google Scholar]
- 77.Aoki S, Kato S, Ichikawa K, Shimizu M. Circadian expression of the PpLhcb2 gene encoding a major light-harvesting chlorophyll a/b-binding protein in the moss Physcomitrella patens. Plant Cell Physiol. 2004;45:68–76. doi: 10.1093/pcp/pch006. [DOI] [PubMed] [Google Scholar]
- 78.Schneider CA, Rasband WS, Eliceiri KW. NIH Image to ImageJ: 25 years of image analysis. Nat. Methods. 2012;9:671–675. doi: 10.1038/nmeth.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Müller-Röber B, Sonnewald U, Willmitzer L. Inhibition of the ADP-glucose pyrophosphorylase in transgenic potatoes leads to sugar-storing tubers and influences tuber formation and expression of tuber storage protein genes. Embo J. 1992;11:1229–1238. doi: 10.1002/j.1460-2075.1992.tb05167.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Nielsen TH, Wischmann B, Enevoldsen K, Möller BL. Starch phosphorylation in potato-tubers proceeds concurrently with de-novo biosynthesis of starch. Plant Physiol. 1994;105:111–117. doi: 10.1104/pp.105.1.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Tomlinson, K. L., Lloyd, J. R. & Smith, A. M. Importance of isoforms of starch-branching enzyme in determining the structure of starch in pea leaves. Plant J. 11 (1997).
- 82.Kraus D. Consolidated data analysis and presentation using an open-source add-in for the Microsoft Excel spreadsheet software. Med. Writ. 2014;23:25–28. doi: 10.1179/2047480613Z.000000000181. [DOI] [Google Scholar]
- 83.Glantz, S. A. & Slinker, B. K. One-way analysis of variance. In Primer of Applied Regression and Analysis of Variance. 2nd Edition. 274–336 (McGraw-Hill, 2000).
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.