Abstract
Background
The role of bacteria on the onset of cardiovascular disease has been suggested. Reciprocally, increased intestinal bacterial translocation and bloodstream infection are common comorbidities associated with heart failure and myocardial infarction (MI). In this context, the aim of this study was to analyze the blood microbiome in patients shortly after acute myocardial infarction.
Methods and Results
We carried out a case control study comparing 103 patients at high cardiovascular risk but free of coronary disease and 99 patients who had an MI. The blood microbiome was analyzed both quantitatively by 16S quantitative polymerase chain reaction and qualitatively by 16S targeted metagenomic sequencing specifically optimized for blood samples. A significant increase in blood bacterial 16S rDNA concentration was observed in patients admitted for MI. This increase in blood bacterial DNA concentration was independent of post‐MI left ventricular function and was more marked in patients with low‐density lipoprotein cholesterol ≥1 g/L. In addition, differences in the proportion of numerous bacterial taxa in blood were significantly modified with the onset of MI, thus defining a blood microbiota signature of MI. Among the bacterial taxa whose proportions are decreased in patients with MI, at least 6 are known to include species able to metabolize cholesterol.
Conclusions
These results could provide the basis for the identification of blood microbiome‐based biomarkers for the stratification of MI patients. Furthermore, these findings should provide insight into the mechanism underlying the negative correlation reported between low‐density lipoprotein cholesterol concentration and the prognosis at the acute onset of MI and mortality.
Clinical Trial Registration
URL: http://www.clinicaltrials.gov. Unique identifier: NCT02405468.
Keywords: bacteria, blood metagenomics, blood microbiota, cholesterol, myocardial infarction
Subject Categories: Biomarkers, Inflammation, Translational Studies, Risk Factors, Myocardial Infarction
Clinical Perspective
What Is New?
Thanks to optimized 16S metagenomic sequencing and 16S quantitative polymerase chain reaction pipelines, we analyzed the blood microbiome in patients admitted for myocardial infarction (MI).
Important differences were found in the blood microbiome of patients admitted for MI.
The increase in blood bacterial DNA concentration in MI patients depends upon blood low‐density lipoprotein cholesterol levels.
What Are the Clinical Implications?
These findings should provide insight into the mechanism of MI and the negative correlation reported between low‐density lipoprotein cholesterol concentration and the prognosis at the acute onset of MI and mortality.
The variation of specific bacterial taxa in the blood could represent potential predictive biomarkers of a cardiovascular disease risk, MI, or associated complications.
Those taxa, if they participate in the disease or have a protective role, can be explored as a potential therapeutic strategy to prevent cardiovascular disease.
Introduction
A systemic role for bacteria and/or bacterial products in the onset of cardiovascular diseases (CVDs) has been suggested in several studies.1, 2, 3, 4, 5 For instance, an association between endotoxemia (elevation of endotoxin from gram‐negative bacteria in the blood) and atherosclerosis in a population‐based study was discovered several decades ago.1 More recently, the deleterious effect of a bacterial metabolite, the trimethyl amine oxide, on the blood vessel wall was shown in an animal model,2, 6 and the relevance of this finding in humans was suggested in a large prospective population‐based study.3 With the recent explosion of the new field of human metagenomics, alteration of gut microbiota has been found in patients with symptomatic atherosclerotic disease,7 and a diversified microbiome within human atherosclerotic plaques has been described,8, 9 suggesting the role of bacterial translocation on atherosclerosis. In this respect, thanks to optimized 16S metagenomic sequencing and 16S quantitative polymerase chain reaction (qPCR) pipelines,10 we recently described the blood microbiome in healthy donors.11 We and others have demonstrated the existence of a specific blood bacterial DNA profile associated with human diseases such as liver fibrosis in obese patients,12 or the onset of cardiovascular events in a large general population.13 In this respect, several myocardial infarction (MI)‐related changes should result in changes in bacterial translocation and consequently in the blood microbiome. Indeed, reciprocally to the role of bacteria in the onset of CVDs, increased intestinal bacterial translocation (“leaky gut”), and bloodstream infection are common comorbidities associated with heart failure and MI.14, 15 Hence, it has been shown that substantial hemodynamic changes associated with post‐MI left ventricular dysfunction, such as intestinal hypoperfusion and congestion, can alter the gut functions, its morphology, and its permeability, which impact the composition of gut microbiota and increase bacterial translocation to the bloodstream.16, 17 Also, MI‐induced systemic inflammation could have an effect both on the intestinal barrier and the transintestinal cholesterol efflux through the crosstalk between the key mediators of these 2 pathways.18 In this respect, a decrease in LDL cholesterol at the acute phase of MI has been reported.19, 20 The intensity of this decrease of LDL cholesterol is associated with significantly higher mortality in patient with acute MI, defining the so‐called lipid paradox.19, 20 In the light of the interplay between bacterial translocation and MI, we undertook here to analyze in a large cohort, both quantitatively (by 16S qPCR) and qualitatively (by 16S metagenomic sequencing), the blood bacterial DNA in patients admitted for MI and to investigate the influence of blood LDL cholesterol concentration on this relationship.
Material and Methods
The data that support the findings of this study are available from the corresponding author upon reasonable request.
Population
We carried out a case control study (NCT02405468) in which control patients (n=103) were at high cardiovascular risk as defined by the presence of at least 2 of the following risk factors: treated dyslipidemia, treated hypertension, treated diabetes mellitus, or current smoking (>1 cigarette per day). All patients were recruited in the cardiology department of Toulouse Teaching Hospital in France. Control patients were free of coronary disease on the basis of a stress test or coronary angiogram. Exclusion criteria for both groups included the following: infectious disease within 1 week before the inclusion, immunocompromised patients, antibiotic treatment within 1 month before the inclusion, chronic viral infection, chronic inflammatory intestinal bowel disease, renal failure (estimated glomerular filtration rate <50 mL/min per 1.73 m2), and pregnancy. The case patients (n=99) had a history of MI. A majority (96/99; 97%) were included within 6 days after the acute ischemic event. Demographic, clinical, and biological characteristics were recorded, and a blood sample was drawn. Coronary data were available for all patients. Left ventricular ejection fraction (LVEF) was available at inclusion for 94 cases. Patients with missing LVEF or LDL concentration were excluded from microbiota analysis regarding these variables but not from the other analyses. Informed consent in writing was obtained from each patient. The study was conducted in accordance with the ethical guidelines of the 1975 Declaration of Helsinki and with INSERM (Institut national de la santé et de la recherche médicale) and Toulouse Hospital guidelines. The protocol and study were approved by the local Ethics Committee.
16S rDNA Quantitation by Real‐Time qPCR and Targeted Metagenomic Sequencing of Blood Samples
For each patient, an EDTA blood tube (4 mL per patient) was collected in the morning in fasting condition and transported at 4°C. The sample was then aliquoted and immediately frozen in liquid nitrogen before storage at −80°C until DNA extraction. Total DNA was extracted from 200 μL of whole blood (in EDTA) using a protocol carefully designed to minimize any risk of contamination between samples or from the experimenters’ environment as described previously.11 This protocol consists of a mechanical lysis step performed twice for 30 seconds at 20 Hz in a bead beater (TissueLyser, Qiagen, Venlo, Netherlands) with 0.1‐mm glass beads (MoBio, Qiagen, Venlo, Netherlands). Then the NucleoSpin blood kit (Macherey‐Nagel, Düren, Germany) was used following the manufacturer protocol (this protocol includes an enzymatic lysis with proteinase K at 70°C for 15 minutes followed by classical silica‐based DNA extraction steps). The quality and quantity of extracted nucleic acids were controlled by gel electrophoresis (1% w/w agarose in Tris‐borate‐EDTA 0.5x) and absorbance spectroscopy using a NanoDrop 2000 UV spectrophotometer (Thermo Fisher Scientific, Waltham, MA). The V3‐V4 hypervariable regions of the 16S ribosomal DNA (16S rDNA) were quantified by qPCR, sequenced using the Illumina‐MiSeq technology and clustered into operational taxonomic unit before taxonomic assignment, as described previously.10, 11, 12 We performed numerous controls both in vitro and in silico to ensure the absence of artifacts such as bacterial DNA contaminants from reagents or nonspecific amplification of eukaryotic DNA. Figure S1 illustrates the low overall background signal (from reagents, consumables, and potential contamination by the experimenter) obtained in qPCR (Figure S1A and S1B) and in the 16S sequencing pipeline (Figure S1C).
Statistical Analysis
The output matrix containing the relative abundance of operational taxonomic units per sample was processed with the linear discriminant analysis effect size (LEfSe) algorithm using an alpha cut‐off of 0.05 and an effect size cut‐off of 2.0. LEfSe is an algorithm for high‐dimensional biomarker discovery and explanation that can identify taxonomic groups characterizing the differences between 2 or more biological conditions. It emphasizes both statistical significance and biological relevance to identify differentially abundant features.21 Then bacterial taxa were studied at the phylum, class, order, family, and genus levels.
Further statistical analyses on clinical data, bacterial concentration, and bacterial taxa proportions were conducted using either SAS statistical software version 9.4 (SAS Institute Inc., Cary, NC), the R environment version 3.3.1, or Prism version 7.04 (GraphPad Software Inc., La Jolla, CA). The main features of cases and controls were first compared using the chi‐square test (or the Fisher exact test if necessary) for categorical variables and Students t test for continuous variables. When the distribution of the continuous variables departed from normality (triglycerides, fasting blood glucose, blood bacterial concentration, alpha diversity, and bacterial taxa relative proportions), the Mann–Whitney (2 biological groups) or Kruskal–Wallis (more than 2 biological groups) tests were used instead. Linear regression models were built to test the association between case/control status and 16S rDNA concentration, alpha diversity, or taxa relative proportion, with adjustment for age, sex, body mass index, smoking status, serum high‐density lipoprotein and LDL cholesterol, fasting blood glucose, and history of treated cardiovascular risk factors (hypertension, hypercholesterolemia, and diabetes mellitus).
Results
The characteristics of the study population are presented in Table 1 using statistical tests displayed in Table S1. Compared with cases, control patients were on average significantly older, had a higher body mass index, and a higher prevalence of treated hypertension and dyslipidemia. Proportions of male, current smoker, high‐density lipoprotein cholesterol average, and fasting glycemia were higher in cases compared with controls. No significant difference was observed in terms of total cholesterol, LDL cholesterol, triglycerides, and measured blood pressure. All patients were recruited in the cardiology department of Toulouse Teaching Hospital in France, which specializes in the treatment of patients with cardiovascular risk factors. This explains the high percentages of patients with hypertension, dyslipidemia, and diabetes mellitus in the control group. Most of these patients were treated for their cardiovascular risk factors, thus explaining why, despite a higher prevalence of hypertension in the control group, measurements of systolic and diastolic blood pressures were not so different between cases and controls.
Table 1.
Characteristics of the Study Population
| Categorical Variables | Controls (N=102) | Cases (N=99) | P Value |
|---|---|---|---|
| Fisher Exact Test | |||
| Male sex, n=201 (%) | 61 (59.8%) | 81 (81.8%) | <0.001 |
| Hypertension, n=201 (%) | 92 (90.2%) | 42 (42.4%) | <0.0001 |
| Dyslipidemia, n=201 (%) | 80 (78.4%) | 60 (60.6%) | 0.006 |
| Type 2 diabetes mellitus, n=201 (%) | 30 (29.4%) | 19 (19.2%) | 0.092 |
| Current smoking, n=201 (%) | 20 (19.6%) | 40 (40.4%) | 0.001 |
| Quantitative Variables | Median | 25% | 75% | Median | 25% | 75% | t Test or Mann–Whitney |
|---|---|---|---|---|---|---|---|
| Age, y (n=201) | 61.6 | 54.9 | 67.2 | 58.5 | 49.9 | 64.2 | 0.007 |
| BMI kg/m2 (n=201) | 28.7 | 24.7 | 31.4 | 26.3 | 23.5 | 28.4 | 0.001 |
| Systolic blood pressure, mm Hg (n=197) | 140 | 130 | 150 | 130 | 120 | 149 | 0.044 |
| Diastolic blood pressure, mm Hg (n=197) | 80 | 72 | 85 | 80 | 70 | 90 | 0.325 |
| Total cholesterol, mmol/L (n=200) | 4.91 | 4.14 | 5.69 | 4.66 | 4.27 | 5.69 | 0.703 |
| Triglycerides, mmol/L (n=199) | 1.32 | 0.95 | 2.00 | 1.36 | 1.01 | 1.90 | 0.393 |
| LDL cholesterol, mmol/L (n=197)* | 2.85 | 2.13 | 3.62 | 2.85 | 2.33 | 3.62 | 0.877 |
| HDL cholesterol, mmol/L (n=197) | 1.32 | 1.04 | 1.66 | 1.19 | 1.01 | 1.40 | 0.023 |
| Fasting blood glucose, mmol/L (n=197) | 5.23 | 4.92 | 6.17 | 5.67 | 5.17 | 6.71 | 0.012 |
Fisher exact tests were conducted on categorical variables. Student t test or nonparametric Mann–Whitney tests were conducted on quantitative variables depending on the results of the tests of normality performed (displayed in Table S1). BMI indicates body mass index; HDL, high‐density lipoprotein; LDL, low‐density lipoprotein.
Not estimated in 2 patients with triglycerides >3.5 mmol/L.
Among the 99 cases of MI, 43.4% had an anterior MI, and the LVEF was decreased to <50% in 24.5% of the case patients.
Blood Bacterial 16S rDNA Concentration Is Increased in MI Patients
To assess whether blood bacterial DNA was quantitatively altered in patients who underwent MI, we measured the blood 16S rDNA concentration by qPCR in control and case patients. A 1.31‐fold significant increase (P<0.0001) in 16S rDNA concentration was observed in the blood of cases compared with controls (Table 2 and Figure 1A). To take into account the difference between the 2 populations (summarized in Table 1), we also applied the analysis on data after adjustment for age, sex, body mass index, cardiovascular risk factors, and smoking status. After this adjustment, blood bacterial 16S rDNA concentration remained significantly increased (Table 2 and Table S2). No correlation was observed in case patients between blood bacterial 16S rDNA and left ventricular dysfunction at admission (data not shown).
Table 2.
Nonadjusted and Adjusted Comparisons of Blood Bacterial 16S rDNA Concentration According to the Onset of Acute MI
| Controls (N=102) | Cases (N=99) | P Value* | Adjusted P Value† | Adjusted P Value‡ | |
|---|---|---|---|---|---|
| 16S copies/mL of blood±SD | 14.7e+6±6.2e+6 | 20.1e+6±7.8e+6 | <0.0001 | 0.001 | 0.010 |
Results of multiple linear regression models (beta coefficients standard errors and P values) are displayed in Table S2. HDL indicates high‐density lipoprotein; MI, myocardial infarction.
Nonadjusted (non‐parametric Mann–Whitney tests).
Adjustment for age, sex, smoking status, body mass index, hypertension, dyslipidemia, and diabetes mellitus (P value obtained with linear regression).
Additional adjustment for HDL cholesterol and fasting blood glucose, both significantly different between cases and controls.
Figure 1.
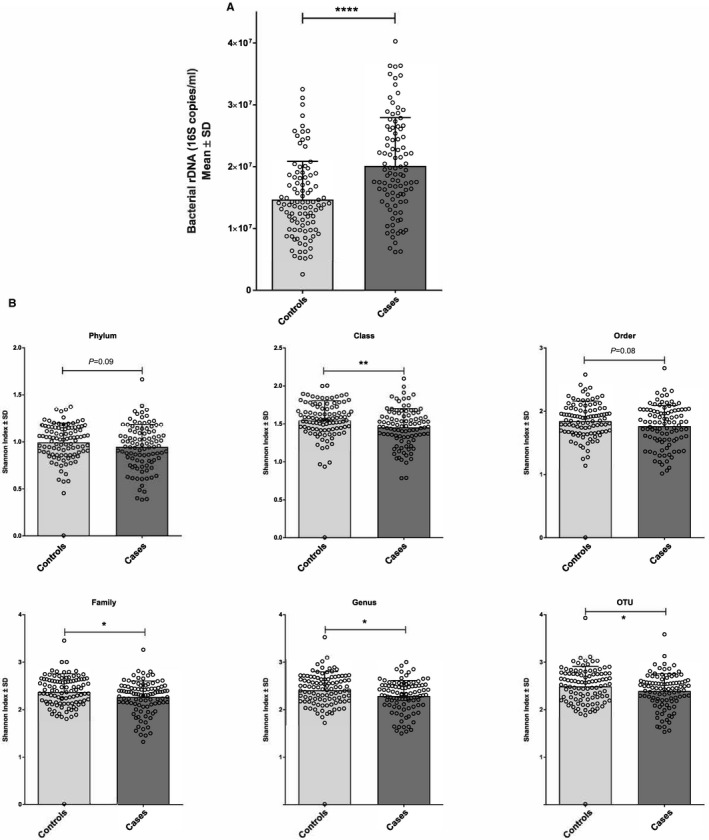
Modifications of blood bacterial 16S rDNA concentration and diversity differentiate patients who had an MI. A, Mean concentrations (bar plot) and individual values (dot plot) of bacterial 16S rDNA assessed by qPCR in patients after MI or control patients. B, Mean concentrations (bar plots) and individual values (dot plots) of bacterial diversity (Shannon index) assessed by 16S metagenomic sequencing in patients after MI or control patients (*P<0.05; **P<0.01; ****P<0.0001). MI indicates myocardial infarction; qPCR, quantitative polymerase chain reaction.
Patients With MI Have Lower Blood Bacterial Diversity
We then investigated whether the blood bacterial dysbiosis also involved a modification of the taxonomic diversity. The 16S rDNA was analyzed by 16S targeted metagenomic sequencing, and the Shannon index, which is a measure of the bacterial alpha diversity (evenness), was calculated at different taxonomic levels. The mean Shannon index of patients with MI was significantly lower (0.0018≤P≤0.0949 depending on the taxonomic level) than control patients (Figure 1B).
A Specific Blood Taxonomic Signature Characterizes Patients With MI
Next, we performed a taxonomic assignment of the operational taxonomic unit representative sequences present in the blood for patients with or without MI. As shown in Figure 2A and previously reported in other studies,11, 12 the bacterial DNA sequences found in blood belonged mainly to the Proteobacteria (57.7%) and Actinobacteria (25.7%) phyla and to a lesser extent to the Firmicutes (10.4%) and Bacteroidetes (5.6%) phyla. Beta diversity analysis such as principal coordinate analysis usually does not show separation between biological groups in a blood microbiome study because of a high variability of blood microbiome composition between individuals. However, a slight separation between cases and controls could be observed on the principal coordinate analysis (Figure S2).
Figure 2.
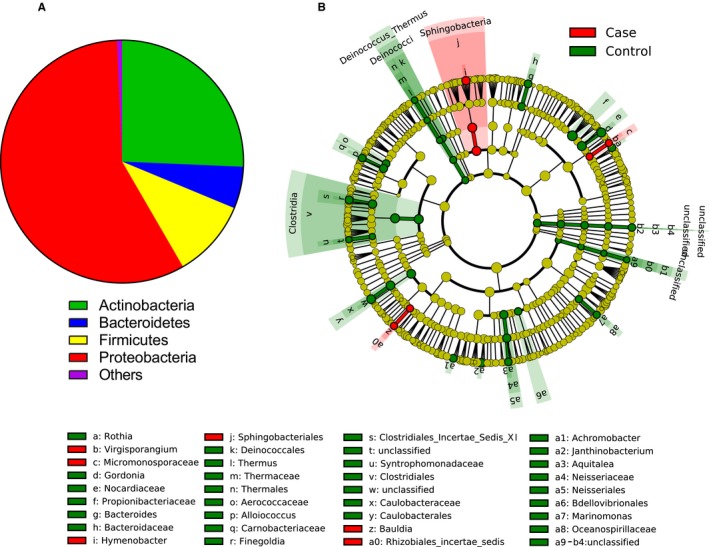
Specific blood bacterial profiles assessed by 16S metagenomic sequencing characterize patients who had an MI. A, Mean relative proportions of bacterial phyla in blood of the overall study population. B, LEfSe analysis of the blood bacterial taxa correlated (red) or inversely correlated (green) with MI status. The cladograms represented here display all bacterial taxa sorter by phylogenic levels (from phyla at the center to genus at the outside rim). The bacterial taxa that are significantly different (P<0.05 with Mann–Whitney test) between the 2 groups being compared are displayed in green when more present in the control patients and in red when more present in the case patients. LEfSe, linear discriminant analysis effect size; MI, myocardial infarction.
Analysis using the LEfSe algorithm (Figure 2B) or using the Mann–Whitney test (Figure 3) showed specific differences in the proportion of blood taxa depending on the presence or absence of MI, therefore defining a specific signature of MI. To take into account the fact that proportions of some taxa are null in a significant part of the study population, a situation for which the Mann–Whitney test is not ideal, we also analyzed the results using a threshold strategy with the chi‐square/Fisher exact test with (adjusted P value) or without adjustments (P value) for risk factors (Table 3). Among the bacterial taxa that were significantly different, the Caulobacterales order and the Caulobacteraceae family were significantly decreased in the MI group with both statistical strategies (Figure 3 and Table 3), and their presence in the blood of patients with MI tended to be negatively correlated with LVEF at inclusion (P=0.072 with chi‐square test; Figure S3).
Figure 3.
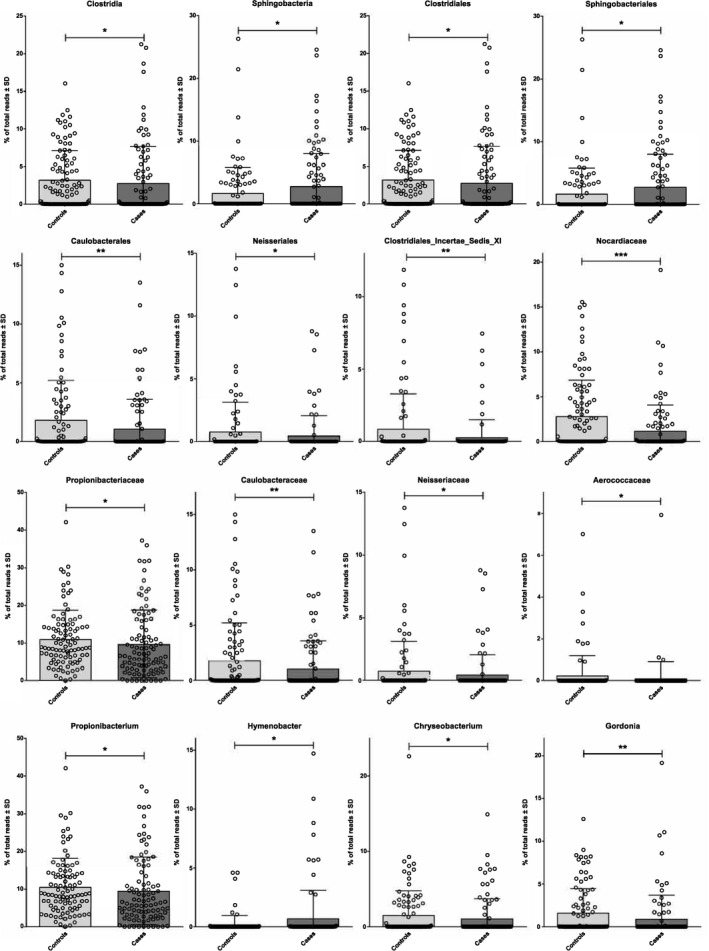
Modification of proportions of blood bacterial taxa assessed by 16S metagenomic sequencing characterizes patients who had an MI. Mean (bar plots) and individual values (dot plots) of relative proportions of relevant blood bacterial taxa in patients after MI or control patients (*P<0.05; **P<0.01; ***P<0.001). MI indicates myocardial infarction.
Table 3.
Bacterial Taxa Found to Be Differentially Present in Blood According to the Onset of MI After Adjustment on Cardiovascular Risk Factors (Chi‐Square/Fisher Exact Test for Presence/Absence)
| Controls, N (%) | Cases, N (%) | P Value | Adjusted P Value* | Adjusted Odds Ratio (95% CI) | |
|---|---|---|---|---|---|
| Order level | |||||
| Caulobacterales | 54 (54.5) | 30 (30.9) | 0.001 | <0.001 | 0.24 (0.11–0.54) |
| Neisseriales | 27 (27.3) | 14 (14.4) | 0.027 | 0.019 | 0.32 (0.12–0.83) |
| Family level | |||||
| Caulobacteraceae | 53 (53.5) | 30 (30.9) | 0.001 | 0.001 | 0.27 (0.12–0.59) |
| Clostridiales‐incertae‐sedis‐XI | 29 (29.3) | 13 (13.4) | 0.007 | 0.026 | 0.34 (0.13–0.88) |
| Microbacteriaceae | 82 (82.8) | 56 (57.7) | <0.001 | 0.001 | 0.27 (0.12–0.59) |
| Neisseriaceae | 27 (27.3) | 14 (14.4) | 0.027 | 0.020 | 0.32 (0.12–0.84) |
| Genus level | |||||
| Brevundimonas | 26 (26.3) | 14 (14.4) | 0.040 | 0.002 | 0.15 (0.04–0.49) |
| Chryseobacterium | 46 (46.5) | 29 (29.9) | 0.017 | 0.026 | 0.42 (0.19–0.90) |
| Gordonia | 50 (50.5) | 24 (24.7) | <0.001 | 0.012 | 0.39 (0.18–0.81) |
| Hymenobacter | 10 (10.1) | 23 (23.7) | 0.011 | 0.036 | 2.79 (1.07–7.30) |
| Microbacterium | 68 (68.7) | 45 (46.4) | 0.002 | 0.005 | 0.35 (0.17–0.73) |
MI indicates myocardial infarction.
Adjustment for age, sex, treated hypertension, treated dyslipidemia, treated diabetes mellitus, and current smoking.
Cholesterol‐Degrading Bacteria Are Decreased in Patients With MI
Among the relatively few known cholesterol‐degrading bacterial families and genera,22, 23 several (Norcardiaceae, Aerococcaceae, Gordonia, Propionibacterium, Chryseobacterium, Rhodococcus) are present in the blood of our control and case patients, and remarkably the relative proportions of all of them are decreased (0.0002≤P≤0.0624) in patients with MI (Figure 4A). Moreover, in a subgroup analysis with separation of the case and control patients between patients with low LDL levels (<1 g/L) and high LDL levels (≥1 g/L), we showed that the increase in blood bacterial 16S rDNA concentration in cases as compared with controls is significant (P<0.0001) only in patients with LDL cholesterol ≥1 g/L (Figure 4B). This difference remained significant after adjustment for age, sex, smoking status, body mass index, treated dyslipidemia, and treated hypertension (Table 4). Interestingly, this finding did not depend on either the prescription of statin (Figure S4A) or the time between the onset of MI and blood sample collection (2‐day threshold; Figure S4B). Spearman analysis between lipid parameters and microbiome did not show strong individual correlation (Table S3).
Figure 4.
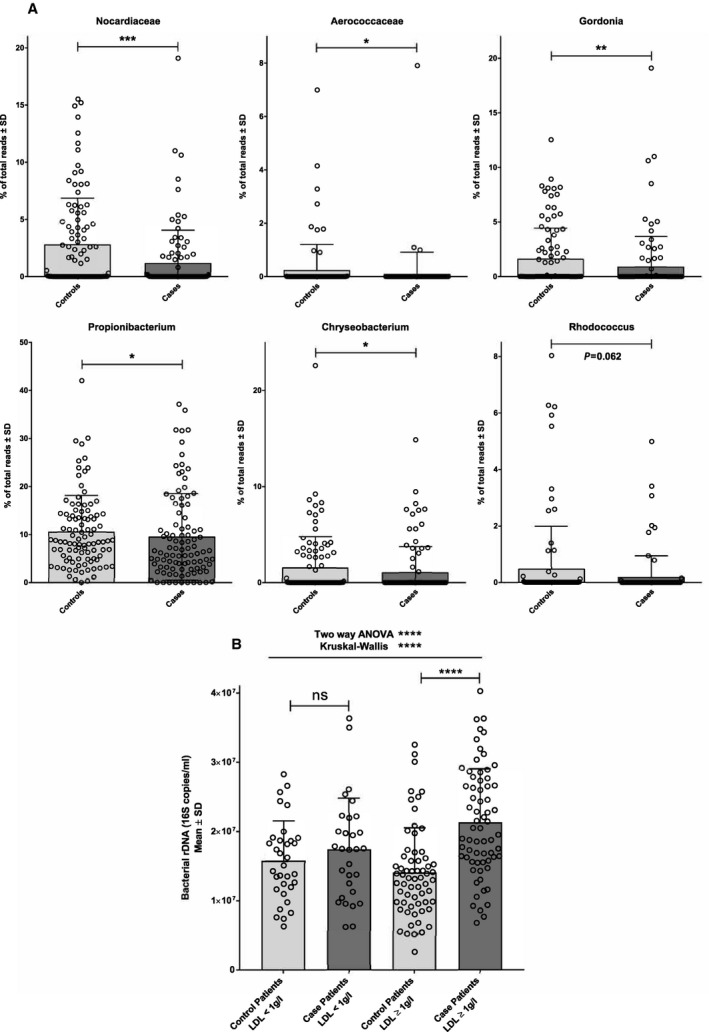
Cholesterol‐degrading bacteria are decreased in patients with MI. A, Mean (bar plots) and individual values (dot plots) of relative proportions of blood bacterial taxa known to degrade cholesterol in patients after myocardial infarction or control patients. B, Modification of blood bacterial 16S rDNA concentration in patients who had an MI depends on the blood LDL cholesterol concentration. Mean (bar plots) and individual values (dot plots) concentrations of bacterial 16S rDNA assessed by qPCR in patients after myocardial infarction or control patients in subgroup of patients depending of LDL cholesterol concentration (*P<0.05; **P<0.01; ***P<0.001; ****P<0.0001; ns: not significant). LDL indicates low‐density lipoprotein; MI, myocardial infarction; qPCR, quantitative polymerase chain reaction.
Table 4.
Interaction Between Blood LDL Cholesterol Concentration and the Onset of MI on Blood Bacterial Burden
| LDL‐C <1 g/L | LDL‐C ≥1 g/L | |||||||
|---|---|---|---|---|---|---|---|---|
| Controls (N=33) | Cases (N=29) | P Value* | P Value† | Controls (N=64) | Cases (N=66) | P Value* | Adj. P Value† | |
| 16S copies/mL of blood±SD | 15.7e+6±5.7 e+6 | 17.3e+6±7.5e+6 | 0.494 | 0.477 | 14.0e+6±6.5e+6 | 21.2e+6±7.8e+6 | <0.0001 | <0.001 |
LDL‐C indicates low‐density lipoprotein cholesterol; MI, myocardial infarction.
Nonadjusted (nonparametric Mann–Whitney tests).
Adjustment for age, sex, body mass index, hypertension, dyslipidemia, and diabetes mellitus (P value obtained with linear regression).
Discussion
The study revealed differences in the blood microbiome in patients admitted for acute MI compared with patients at high cardiovascular risk, free of coronary disease. First, we showed an increase in blood bacterial 16S rDNA concentrations in patients admitted for MI. Then, we observed qualitative differences in blood microbiota in MI patients as compared with controls. The blood microbiome derives at least partially from the gut microbiome as a result of several mechanisms of bacterial translocation.11, 12, 24 Furthermore, as we previously demonstrated,12 the dramatic difference in the composition of the blood and the gut microbiomes suggests that the intestinal barrier, the immune cells, and the liver play a role of filter during bacterial translocation.24 The lack of significant correlation between LVEF and blood bacterial 16S rDNA concentration in case patients suggests that MI influences this filter function not only by intestinal hypoperfusion or intestinal congestion,16, 17 but also through neurohormonal activation via the so‐called gut‐brain axis.25, 26 Indeed, a large body of evidence demonstrates that ischemic event–induced changes in the gut‐brain axis contributes to substantial changes in intestinal microbial composition, which in turn is associated with intestinal barrier dysfunction, bacterial translocation, and even the onset of post–ischemic event infection.26, 27 In addition to the potential bacterial contamination of the patient with MI resulting from invasive clinical procedures, the bacterial translocation, because of different mechanisms, induces a systemic inflammation that contributes to MI comorbidities and cardiac failure and could also trigger bloodstream infection.14, 15 In this respect, in the present cohort, we detected in a patient admitted for right ventricular infection an unusual bloodstream infection involving Staphylococcus, Microbacterium, and Janibacter, which we published as a case report.28 This patient was consequently excluded from the current study. The observed increase in blood 16S rDNA concentration was more pronounced in patients with MI with LDL cholesterol >1 g/L. Interestingly, this finding is independent of statin therapy, as a more pronounced increased in blood 16S rDNA concentration can be observed in patients with MI with LDL cholesterol >1 g/L whether they received statin therapy or not. This interaction could be explained by several mechanisms. Indeed, MI‐induced inflammation should have an effect on transintestinal cholesterol efflux since peroxisome proliferator–activated receptor signaling and liver X receptor signaling, which are the key pathways for transintestinal cholesterol excretion, are also directly connected to the inflammation (via Toll‐like receptors and nuclear factor kappa light‐chain enhancer of activated B cells signaling pathways).18 Furthermore, it has been demonstrated that the brain can detect a rise in lipids and then orchestrate a biochemical, molecular, neuronal, and physiological network of responses through the gut‐brain axis.29 Thus, it can be speculated that blood LDL cholesterol level, via this feedback loop, influences bacterial translocation from the intestine to the bloodstream at the acute phase of MI. Also, a decrease in LDL cholesterol levels has been shown soon after the acute MI, and a lipid paradox has been repeatedly observed that low LDL cholesterol is associated with significantly higher mortality in patients presenting an acute MI.19 Whether the LDL‐related change in blood microbiota for patients admitted for acute MI contributes to explain this paradox should be explored. Interestingly, the observation that the LDL level is correlated to both an increase in bacterial concentration and a better prognosis at the onset of MI raises the possibility that there is a bacterial translocation at the acute phase of MI that could be beneficial on cardiovascular prognosis. In line with this hypothesis, a deleterious effect of depletion of gut microbiota by broad‐spectrum antibiotics in a murine model of stroke has been reported.19 In addition to a quantitative variation, we also observed qualitative differences in the blood microbiome in patients admitted for acute MI compared with control patients with high cardiovascular risk, free of coronary disease. It is difficult to determine which parts of the significant modifications of the bacterial taxa were preexisting or a result of the MI and whether the striking decrease of all the cholesterol‐degrading taxa is related to the LDL interaction. Examples such as the variation of the Caulobacterales order and the Caulobacteraceae family are nevertheless interesting. Those taxa were significantly decreased in the patients with MI compared with the control patients; in addition, among the patients with MI, Caulobacterales and Caulobacteraceae tended to be increased in patients with preserved LVEF. A decrease of Caulobacterales and Caulobacteraceae in the gut has been reported to be associated with a risk factor of CVD such as diabetes mellitus type 1 in human30 or lack of exercises and high fat diet in mice models.31 Those taxa could represent either biomarkers of a CVD risk, or if they have a protective role, be explored as potential therapeutic strategy to prevent CVD.
Despite interesting findings, our study has several limitations. First, only blood sampled after the MI was available, making impossible to distinguish which part of blood microbiome differences observed in case patients precede the MI or is a consequence of the MI. This information would be very helpful to develop predictive biomarkers of the MI and/or therapeutic targets to prevent the MI or its complications such as cardiac failure. Second, the molecular approach based on the quantitation and identification of the bacterial DNA, despite many advantages, such as high sensibility and exhaustivity, cannot distinguish DNA from living bacteria and DNA resulting from bacterial degradation. Even if this is not an issue for biomarker discovery, it is more problematic when trying to understand the mechanisms associated with the variation of the blood microbiome in patients with MI. Finally, except for the blood bacterial DNA itself, other markers of intestinal leakage of inflammatory bacterial products in the circulation such as endotoxin, could not be measured, while they could have helped to understand the interplay between MI and systemic bacterial inflammation.
However, the primary goal of our pilot study was to demonstrate that a variation of the blood microbiome exists in patients shortly after acute MI. In this context, our study is a promising proof of concept and opens a novel pathway for using the blood microbiome as a potential diagnostic and therapeutic tool in MI. The above limitations should be addressed in future studies.
In conclusion, we described here important differences in the blood microbiome of patients admitted for MI. The increase in blood bacterial DNA concentration in MI patients depends upon blood LDL cholesterol levels. Whether these changes have an impact on the cardiovascular outcomes remains to be explored, but the variation of specific bacterial taxa in the blood could represent potential predictive biomarkers of the MI or associated complications.
Disclosures
None.
Supporting information
Table S1. Tests of Normality
Table S2. Results of Multiple Linear Regression Models Predicting 16S in Cases as Compared With Controls
Table S3. Correlations Between Lipid Parameters and Blood Microbiota (16S copies/mL of Blood and Relative Proportion of Taxa Displayed in Figures 3 and 4)
Figure S1. Assessment of the background of the 16S qPCR and sequencing assays.
Figure S2. Principal coordinate analysis of the 16S sequencing of the blood samples (controls and cases) using distance calculated with Generalized UniFrac with α=0.2.
Figure S3. Presence of Caulobacterales in blood of patient with MI tended to correlate with LVEF.
Figure S4. Interaction between LDL concentration and modification of blood bacterial 16S rDNA concentration in patients who underwent MI does not depend on statin treatment or time of sampling.
(J Am Heart Assoc. 2019;8:e011797 DOI: 10.1161/JAHA.118.011797.)
References
- 1. Wiedermann CJ, Kiechl S, Dunzendorfer S, Schratzberger P, Egger G, Oberhollenzer F, Willeit J. Association of endotoxemia with carotid atherosclerosis and cardiovascular disease: prospective results from the Bruneck Study. J Am Coll Cardiol. 1999;34:1975–1981. [DOI] [PubMed] [Google Scholar]
- 2. Wang Z, Klipfell E, Bennett BJ, Koeth R, Levison BS, Dugar B, Feldstein AE, Britt EB, Fu X, Chung YM, Wu Y, Schauer P, Smith JD, Allayee H, Tang WH, DiDonato JA, Lusis AJ, Hazen SL. Gut flora metabolism of phosphatidylcholine promotes cardiovascular disease. Nature. 2011;472:57–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Tang WHW, Wang Z, Levison BS, Koeth RA, Britt EB, Fu X, Wu Y, Hazen SL. Intestinal microbial metabolism of phosphatidylcholine and cardiovascular risk. N Engl J Med. 2013;368:1575–1584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Rogler G, Rosano G. The heart and the gut. Eur Heart J. 2014;35:426–430. [DOI] [PubMed] [Google Scholar]
- 5. Ahmadmehrabi S, Tang WHW. Gut microbiome and its role in cardiovascular diseases. Curr Opin Cardiol. 2017;32:761–766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Chen K, Zheng X, Feng M, Li D, Zhang H. Gut microbiota‐dependent metabolite trimethylamine N‐oxide contributes to cardiac dysfunction in Western diet‐induced obese mice. Front Physiol. 2017;8:139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Karlsson FH, Fåk F, Nookaew I, Tremaroli V, Fagerberg B, Petranovic D, Bäckhed F, Nielsen J. Symptomatic atherosclerosis is associated with an altered gut metagenome. Nat Commun. 2012;3:1245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Mitra S, Drautz‐Moses DI, Alhede M, Maw MT, Liu Y, Purbojati RW, Yap ZH, Kushwaha KK, Gheorghe AG, Bjarnsholt T, Hansen GM, Sillesen HH, Hougen HP, Hansen PR, Yang L, Tolker‐Nielsen T, Schuster SC, Givskov M. In silico analyses of metagenomes from human atherosclerotic plaque samples. Microbiome. 2015;3:38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Lindskog Jonsson A, Hållenius FF, Akrami R, Johansson E, Wester P, Arnerlöv C, Bäckhed F, Bergström G. Bacterial profile in human atherosclerotic plaques. Atherosclerosis. 2017;263:177–183. [DOI] [PubMed] [Google Scholar]
- 10. Lluch J, Servant F, Païssé S, Valle C, Valière S, Kuchly C, Vilchez G, Donnadieu C, Courtney M, Burcelin R, Amar J, Bouchez O, Lelouvier B. The characterization of novel tissue microbiota using an optimized 16S metagenomic sequencing pipeline. PLoS One. 2015;10:e0142334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Païssé S, Valle C, Servant F, Courtney M, Burcelin R, Amar J, Lelouvier B. Comprehensive description of blood microbiome from healthy donors assessed by 16S targeted metagenomic sequencing. Transfusion. 2016;56:1138–1147. [DOI] [PubMed] [Google Scholar]
- 12. Lelouvier B, Servant F, Païssé S, Brunet A‐C, Benyahya S, Serino M, Valle C, Ortiz MR, Puig J, Courtney M, Federici M, Fernández‐Real J‐M, Burcelin R, Amar J. Changes in blood microbiota profiles associated with liver fibrosis in obese patients: a pilot analysis. Hepatology. 2016;64:2015–2027. [DOI] [PubMed] [Google Scholar]
- 13. Amar J, Lange C, Payros G, Garret C, Chabo C, Lantieri O, Courtney M, Marre M, Charles MA, Balkau B, Burcelin R. Blood microbiota dysbiosis is associated with the onset of cardiovascular events in a large general population: the D.E.S.I.R. study. PLoS One. 2013;8:e54461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Krack A, Sharma R, Figulla HR, Anker SD. The importance of the gastrointestinal system in the pathogenesis of heart failure. Eur Heart J. 2005;26:2368–2374. [DOI] [PubMed] [Google Scholar]
- 15. Zabell A, Tang WHW. Targeting the microbiome in heart failure. Curr Treat Options Cardiovasc Med. 2017;19:27. [DOI] [PubMed] [Google Scholar]
- 16. Sandek A, Bauditz J, Swidsinski A, Buhner S, Weber‐Eibel J, von Haehling S, Schroedl W, Karhausen T, Doehner W, Rauchhaus M, Poole‐Wilson P, Volk H‐D, Lochs H, Anker SD. Altered intestinal function in patients with chronic heart failure. J Am Coll Cardiol. 2007;50:1561–1569. [DOI] [PubMed] [Google Scholar]
- 17. Sandek A, Swidsinski A, Schroedl W, Watson A, Valentova M, Herrmann R, Scherbakov N, Cramer L, Rauchhaus M, Grosse‐Herrenthey A, Krueger M, von Haehling S, Doehner W, Anker SD, Bauditz J. Intestinal blood flow in patients with chronic heart failure: a link with bacterial growth, gastrointestinal symptoms, and cachexia. J Am Coll Cardiol. 2014;64:1092–1102. [DOI] [PubMed] [Google Scholar]
- 18. Herbert KE, Erridge C. Regulation of low‐density lipoprotein cholesterol by intestinal inflammation and the acute phase response. Cardiovasc Res. 2018;114:226–232. [DOI] [PubMed] [Google Scholar]
- 19. Cheng KH, Chu CS, Lin TH, Lee KT, Sheu SH, Lai WT. Lipid paradox in acute myocardial infarction‐the association with 30‐day in‐hospital mortality. Crit Care Med. 2015;43:1255–1264. [DOI] [PubMed] [Google Scholar]
- 20. Reddy VS, Bui QT, Jacobs JR, Begelman SM, Miller DP, French WJ. Relationship between serum low‐density lipoprotein cholesterol and in‐hospital mortality following acute myocardial infarction (the lipid paradox). Am J Cardiol. 2015;115:557–562. [DOI] [PubMed] [Google Scholar]
- 21. Segata N, Izard J, Waldron L, Gevers D, Miropolsky L, Garrett WS, Huttenhower C. Metagenomic biomarker discovery and explanation. Genome Biol. 2011;12:R60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Kreit J, Sampson NS. Cholesterol oxidase: physiological functions. FEBS J. 2009;276:6844–6856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. García JL, Uhía I, Galán B. Catabolism and biotechnological applications of cholesterol degrading bacteria. Microb Biotechnol. 2012;5:679–699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Balmer ML, Slack E, de Gottardi A, Lawson MA, Hapfelmeier S, Miele L, Grieco A, Van Vlierberghe H, Fahrner R, Patuto N, Bernsmeier C, Ronchi F, Wyss M, Stroka D, Dickgreber N, Heim MH, McCoy KD, Macpherson AJ. The liver may act as a firewall mediating mutualism between the host and its gut commensal microbiota. Sci Transl Med. 2014;6:237ra66. [DOI] [PubMed] [Google Scholar]
- 25. Husic M, Nørager B, Egstrup K, Lang RM, Møller JE. Diastolic wall motion abnormality after myocardial infarction: relation to neurohormonal activation and prognostic implications. Am Heart J. 2005;150:767–774. [DOI] [PubMed] [Google Scholar]
- 26. Wen SW, Wong CHY. An unexplored brain‐gut microbiota axis in stroke. Gut Microbes. 2017;8:601–606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Stanley D, Mason LJ, MacKin KE, Srikhanta YN, Lyras D, Prakash MD, Nurgali K, Venegas A, Hill MD, Moore RJ, Wong CHY. Translocation and dissemination of commensal bacteria in post‐stroke infection. Nat Med. 2016;22:1277–1284. [DOI] [PubMed] [Google Scholar]
- 28. Lelouvier B, Servant F, Delobel P, Courtney M, Elbaz M, Amar J. Identification by highly sensitive 16S metagenomic sequencing of an unusual case of polymicrobial bacteremia. J Infect. 2017;75:278–280. [DOI] [PubMed] [Google Scholar]
- 29. Rasmussen BA, Breen DM, Lam TKT. Lipid sensing in the gut, brain and liver. Trends Endocrinol Metab. 2012;23:49–55. [DOI] [PubMed] [Google Scholar]
- 30. Qi C‐J, Zhang Q, Yu M, Xu J‐P, Zheng J, Wang T, Xiao X‐H. Imbalance of fecal microbiota at newly diagnosed type 1 diabetes in Chinese children. Chin Med J (Engl). 2016;129:1298–1304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Kang SS, Jeraldo PR, Kurti A, Miller ME, Cook MD, Whitlock K, Goldenfeld N, Woods JA, White BA, Chia N, Fryer JD. Diet and exercise orthogonally alter the gut microbiome and reveal independent associations with anxiety and cognition. Mol Neurodegener. 2014;9:36. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. Tests of Normality
Table S2. Results of Multiple Linear Regression Models Predicting 16S in Cases as Compared With Controls
Table S3. Correlations Between Lipid Parameters and Blood Microbiota (16S copies/mL of Blood and Relative Proportion of Taxa Displayed in Figures 3 and 4)
Figure S1. Assessment of the background of the 16S qPCR and sequencing assays.
Figure S2. Principal coordinate analysis of the 16S sequencing of the blood samples (controls and cases) using distance calculated with Generalized UniFrac with α=0.2.
Figure S3. Presence of Caulobacterales in blood of patient with MI tended to correlate with LVEF.
Figure S4. Interaction between LDL concentration and modification of blood bacterial 16S rDNA concentration in patients who underwent MI does not depend on statin treatment or time of sampling.


