Abstract
Poor cellular uptake, rapid degradation in the presence of serum, and inefficient transfection are some of the major barriers in achieving therapeutic efficacy of naked small interfering RNAs (siRNAs). We investigated the efficacy of the polyplex formulated using our synthesized polymer, polyethylene glycol (PEG)–modified l-arginine oligo(-alkylaminosiloxane) that is grafted with poly(ethyleneimine) (PEI) for siRNA delivery. We hypothesized that the polyplex formulated using the polymer with a balanced composition of PEI for siRNA condensation and its protection, PEG for polyplex stability and to minimize the PEI-associated toxicity, and with arginine facilitating cellular uptake would overcome the aforementioned issues with siRNA delivery. We tested our hypothesis using antiluciferase siRNA in luciferase-expressing metastatic breast cancer cells (MDA-MB-231-Luc-D3H2LN) and anti-ABCB1 siRNA against an efflux membrane protein, ABCB1, in doxorubicin (DOX)-resistant breast cancer cells (MCF-7/Adr). The results demonstrated that the polyplex at an optimal nucleotide/polymer ratio is stable in the presence of excess polyanions, has no cellular toxicity, and protects siRNA from RNase degradation. Transfection of MDA-MB-231-Luc-D3H2LN cells with antiluciferase siRNA polyplex showed almost complete knockdown of luciferase expression. In MCF-7/Adr cells, transfection with anti-ABCB1 siRNA effectively downregulated its target efflux protein, ABCB1; increased cellular uptake of DOX; and enhanced its cytotoxic effect. However, the cotreatment did not completely overcome drug resistance, suggesting that further optimization is needed and/or a mechanism(s) other than the efflux protein ABCB1 may be involved in drug resistance. In conclusion, our polyplex is effective for siRNA delivery and can be explored for different therapeutic applications.
Introduction
Therapeutic promise of small interfering RNAs (siRNA) has gained wide recognition for treating different diseases, such as cancer, viral infections, and neurodegenerative diseases (Dykxhoorn et al., 2003; Burnett et al., 2011; Lan et al., 2015). A major advantage of siRNA lies in its function in the cytoplasm rather than in the nucleus, which prevents nuclear penetration, a major barrier for cDNA delivery for gene transfection (Wang et al., 2010a). In cancer, siRNA can repress cancer cell proliferation, metastasis, and reverse drug resistance (Scamuffa et al., 2008; Yang et al., 2015; Zi et al., 2015). Despite these benefits, translation of siRNA-based therapy has been limited due to several factors, including: 1) poor cellular uptake of siRNAs because of their anionic charge and hydrophilic nature (Ragelle et al., 2013), 2) instability because of their rapid degradation by RNase (Hickerson et al., 2008), and 3) delivery vehicle–associated cytotoxicity (de Fougerolles et al., 2007; Whitehead et al., 2009; Shegokar et al., 2011). Cationic lipids (e.g., lipofectamine) (Rietwyk and Peer, 2017) and polymers can both complex siRNA and facilitate transfection (Zhu and Mahato, 2010). The two commonly used polymers for transfection are poly(ethyleneimine)s (PEIs) and poly(amidoamine)s (Kawakami et al., 2006; Hobel and Aigner, 2010). PEI can spontaneously self-assemble with siRNA to form a polyplex (Wang et al., 2010b; Merkel et al., 2011; Yuan et al., 2011; Wu et al., 2012); however, it faces significant toxicity issues, such as membrane damage and activation of apoptotic pathways by mitochondria (Moghimi et al., 2005; Hunter and Moghimi, 2010; Alameh et al., 2012). Therefore, a general strategy is to minimize the toxic effect of cationic polymers by modifying them with functional groups, such as polyethylene glycol (PEG) or polysaccharides (e.g., dextran, pullulan) (Caroline Diana and Rekha, 2017); however, such modifications tend to reduce transfection efficiency (Brissault et al., 2006). Therefore, a critical balance is required for the polymers with a cationic charge that is effective in condensing nucleic acids but at the same time does not cause cellular toxicity due to cationic charge or aggregation in the presence of negatively charged serum proteins.
Here, we explored the efficacy of the polyplex formulated using our synthesized polymer, l-arginine oligo(alkylaminosiloxane) grafted PEI [P(SiDAAr)5], and modified with PEG, [P(SiDAAr)5PEG3], to deliver siRNA to both metastatic (MDA-MB-231-Luc-D3H2LN) and doxorubicin (DOX)-resistant (MCF-7/Adr) breast cancer cells. Arginine has the ability to facilitate intracellular translocation of macromolecules and nanocarriers, as it has a strong affinity with heparan sulfate expressed on mammalian cell membranes (Morris and Labhasetwar, 2015). Also, the guanidine groups on arginine can form hydrogen bonds with polyhydroxy compounds in cell membranes (Calnan et al., 1991; Richard et al., 2003; Wang et al., 2009). Dendrimers, such as polyamidoamine (PAMAM) and polymer (propylene imine) conjugated to arginine, have shown enhanced transfection efficiency (Choi et al., 2004; Kim et al., 2007). We hypothesized that a well characterized formulation of polyplex would facilitate efficient cellular delivery of siRNA and, hence, its effect. Our data show that the polyplex at an optimized composition is nontoxic to the cells and is effective in transfecting antiluciferase siRNA to metastatic breast cancer cells and therapeutic siRNA against efflux protein ABCB1 in resistant breast cancer cells.
Materials and Methods
Materials.
3-(2-Aminoethylamino)propyl-methyl-dimethoxysilane, 25-kDa branched PEI, aspartic acid, l-arginine, O-(2-aminoethyl)-O′-(2-carboxyethyl)polyethylene glycol-3K hydrochloride, 1-ethyl-3-(3-dimethylaminopropyl)carbodiimide hydrochloride (EDC), heparin sodium, N-hydroxysuccinimide (NHS), 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT), sodium bicarbonate powder, hydroxylamine hydrochloride, uranyl acetate, and Tris acetate-EDTA buffer were purchased from Sigma-Aldrich (St. Louis, MO). Fetal bovine serum (FBS) and trypsin/EDTA were obtained from Gibco (Grand Island, NY). RNase I and Silencer Negative Control No. 1 siRNA were purchased from Life Technologies (Carlsbad, CA). Dulbecco’s modified Eagle’s medium (DMEM), Dulbecco’s phosphate-buffered saline, penicillin, and streptomycin were purchased from the Cell Services’ Media Core at the Cleveland Clinic.
Synthesis of P(SiDAAr)5PEG3.
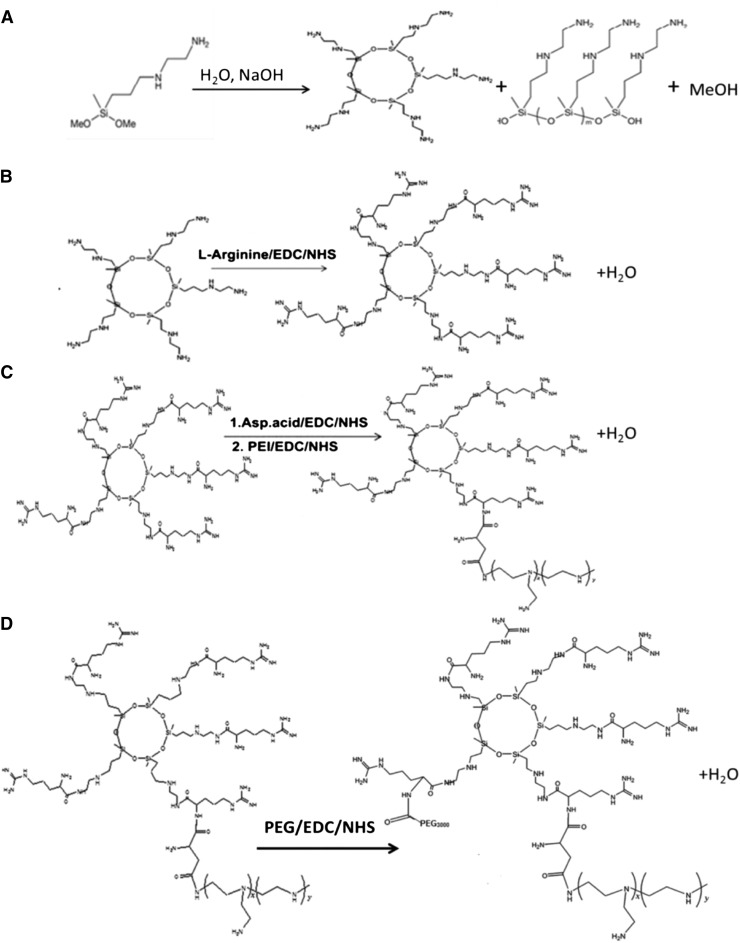
The polymer that consisted of 3 M PEG and 5 M P(SiDAAr)5PEG3 (where numbers in subscript indicate relative molar ratio per mole of polymer) was synthesized as per the Scheme. In brief, following the synthesis of oligo(-alkylaminosiloxane) (step A) and modification with arginine [step B, l-arginine oligo(alkylaminosiloxane) (SiDAAr)], PEI conjugation to SiDAAr was carried out using aspartic acid as a linker. Specifically, acid groups of aspartic acid were activated using EDC and NHS at room temperature in 2-(N-morpholino)ethanesulfonic acid (MES) buffer at pH 6. Thereafter, 5 M SiDAAr and 1 M PEI were added to the reaction mixture with stirring for 18 hours at room temperature, followed by dialysis using 12K molecular weight cutoff (MWCO) dialysis membrane (Spectrum Laboratories, Rancho Dominguez, CA) (step C). To conjugate PEG to [P(SiDAAr)5], free acid groups of NH2−PEG−COOH were activated using EDC (coupling agent) and NHS (catalyst) in MES buffer at pH 6 for 3.5 hours at room temperature. Thereafter, 3 M activated PEG was added to the 1 M equivalent of P(SiDAAr)5 in phosphate-buffered saline (PBS) at pH 7.5 with overnight stirring (step D). The polymer was dialyzed for 2 days in 2 l of dH2O using 12K MWCO dialysis membrane to remove unreacted elements and then lyophilized.
Tetramethylrhodamine-6-isothiocyanate Conjugation to Polymer.
To determine cellular uptake of polyplex using flow cytometry, the polymer P(SiDAAr)5PEG3 was conjugated to tetramethylrhodamine-6-isothiocyanate (6-TRITC) dye (Invitrogen, Carlsbad, CA). To conjugate, 2 mg of polymer was dissolved in 1 ml of 0.1 M sodium bicarbonate buffer (pH = 9.5), and 1 ml of 6-TRITC was dissolved in 500 µl of dimethylsulfoxide (DMSO). While stirring, 100 µl of the aforementioned reactive 6-TRITC dye solution was added to the polymer solution dropwise. The reaction was continued with stirring for 1 hour at room temperature and was then stopped by adding 0.1 ml of freshly prepared 1.5 M hydroxylamine in distilled water and adjusting the pH to 8.5 with 5 M NaOH. The polymer was dialyzed overnight using the 12K MWCO dialysis membrane to remove the excess 6-TRITC dye.
Polymer Complexation and Characterization of Polyplex.
The polymer, P(SiDAAr)5PEG3 dissolved in RNase-free dH2O (Qiagen, Valencia, CA) at 1-mg/ml concentration, was filtered through a 0.22-µm sterile polyethersulfone filter (Millipore, Darmstadt, Germany). siRNA was also diluted separately to 1 mg/ml in RNase-free dH2O. The formulations of polyplex were prepared by pipetting the siRNA and polymer mixture up and down ∼30 times and incubating it for 1 hour to allow self-assembly. Three micrograms of siRNA was used per sample, and the polymer quantity was changed to form polyplex with different nucleotide/polymer (N/P) w/w ratios. Size and ζ-potential were measured in dH2O using quasi-elastic dynamic light scattering and a zeta potential analyzer (NICOMP 380 ZLS; Particle Sizing System, Santa Barbara, CA). Morphology of polyplex was characterized by transmission electron microscopy (TEM) using a Tecnai G2 TEM microscope (FEI, Hillsboro, OR). For TEM microscopy, polyplex with 1/4 N/P w/w ratio prepared as earlier in dH2O was used. Ten microliters of the polyplex dispersion was dropped on the TEM grids coated with silicon monoxide–stabilized formvar films (Electron Microscopy Sciences, Hatfield, PA). The samples were then air dried and stained with 2% uranyl acetate for 7 minutes prior to imaging.
Determination of Optimal siRNA to Polymer Complexation Ratio.
Agarose gel electrophoresis was used to determine the optimal siRNA/polymer (N/P) complexation ratio. Polyplex was prepared at N/P w/w ratios of 1/0.5, 1/1, 1/2, 1/4, and 1/6, keeping siRNA quantity constant at 1 µg while varying polymer quantity. Using naked luciferase DNA as a reference, naked siRNA and siRNA polyplex were loaded onto a 1% Gold Agarose gel (Lonza, Allendale, NJ) in Tris acetate-EDTA buffer. The samples were run at 60 V for 60 minutes on an electrophoresis system (Bio-Rad, Hercules, CA). The gel was stained using SYBR Green (Thermo Scientific, Rockford, IL) and visualized using a UV imager (FUJIFILM FLA-5100; FUJIFILM Life Science, Stamford, CT).
Protection of siRNA from RNase Degradation.
The effectiveness of the polyplex to protect siRNA from RNase degradation was determined by incubating naked siRNA and the polyplex prepared at a 1/4 N/P w/w ratio with 100 IU of RNase buffer for 30 minutes. Absorbance was measured at 260 nm using Nanodrop 1000 (Thermo Fisher Scientific, Waltham, MA).
Polyplex Stability.
Stability of the polyplex prepared at a 1/4 N/P w/w ratio was measured in the presence of heparin polyanions using a polyanion competition assay (Danielsen et al., 2005). The polyplex, consisting of 5 µg of siRNA and 20 µg of polymer in 15 µl of dH2O, was incubated for 15 minutes with various concentrations of heparin sodium solution (Sigma-Aldrich). The samples were run on a 1% Gold Agarose gel (Lonza) in Tris Acetate-EDTA buffer, and the gel was stained and visualized by the same method as described earlier.
Cell Culture.
A resistant MCF-7/Adr cell line was created through a long-term DOX treatment of sensitive MCF-7 cells. Drug resistance was maintained by incubating the cells with 100 ng/ml of DOX (Vijayaraghavalu et al., 2012). Prior to transfection, medium in the wells was changed to a drug-free medium for two passages. Both cell lines, MCF-7/Adr and MDA-MB-231-Luc-D3H2LN (Caliper Life Sciences, Hopkinton, MA), were cultured in 15% FBS-enriched DMEM with 1% penicillin-streptomycin under 5% CO2 and 95% humidity at 37°C.
Cytotoxicity of Polyplex.
MDA-MB-231-Luc-D3H2LN cells were seeded at 5 × 104 cells/well in a 24-well plate (BD Biosciences, San Jose, CA) in DMEM with 15% FBS. Polyplex made of 1 µg of luciferase siRNA and varying amounts of the polymer to achieve different N/P w/w ratios prepared in 15 µl of DMEM were gently added into each well and mixed with culture medium. After 16 hours of incubation, the medium in the wells was replaced with a fresh medium. The cell viability was determined after 3 days of incubation using MTT assay. For the assay, cell culture medium was removed and replaced with 150 µl of MTT reagent (0.5 mg/ml in colorless DMEM) and incubated for 3.5 hours. Thereafter, the MTT solution in culture plates was replaced with 250 µl of DMSO and incubated for another 30 minutes. One hundred and fifty microliters of the DMSO solution from the culture plates was added to a clear 96-well plate to measure absorbance at 595 nm using a plate reader (SpectraMax M2; Molecular Devices Inc., Sunnyvale, CA). Cell viability was calculated as (absorbance of the polyplex treated cells)/(absorbance of untreated cells) × 100%.
Cellular Uptake of Polyplex.
Cellular uptake of the polyplex prepared at 1/4 N/P w/w ratio was determined using TRITC-conjugated P(SiDAAr)5PEG3 in both MDA-MB-231-Luc-D3H2LN and MCF-7/Adr cell lines. Cells were seeded in six-well plates with 15% DMEM and cultured until reaching ∼80% confluency. The polyplex made of 3 µg of siRNA and 12 µg of TRITC-conjugated P(SiDAAr)5PEG3 was added to wells, gently mixed, and incubated for 4 hours. The cells were washed three times with PBS and trypsinized prior to analysis by flow cytometry using an LSR II System (BD Biosciences, Franklin Lakes, NJ) with fluorescence detector set at an excitation maximum of 530 nm and emission maximum of 569 nm.
siRNA-Mediated Gene Silencing.
For ubiquitous gene knockdown, we assayed firefly luciferase gene silencing because it has no apparent effect on cell regulatory machinery. MDA-MB-231-Luc-D3H2LN cells were seeded at a density of 2 × 105 cells/well in 24-well plates and cultured for 24 hours. The medium for all transfection studies contained DMEM with 15% FBS and 1% penicillin-streptomycin. Polyplex containing 2 µg of siRNA was added to each well and incubated for 24 hours. Thereafter, cells were washed with PBS three times and lysed with 100 µl of Reporter Lysis Buffer (Promega, Madison, WI). Twenty microliters of cell lysate and 50 µl of Luciferase Assay Reagent (Promega) were added into a Nunc F96-MicroWell White Polystyrene plate (Nunc; Thermo Scientific). Luciferase activity was measured using a Luminescence Plate Reader (Wallace 1420 VICTOR2; PerkinElmer, Waltham, MA). Cell protein levels were analyzed using BCA protein assays (Thermo Fisher Scientific). To perform the assay, 10 µl of cell lysate and 150 µl of freshly prepared BCA reagent (1 part of reagent A and 50 parts of reagent B) were added to a 96 well-plate and incubated at 37°C for 30 minutes. Absorbance was measured at a wavelength of 562 nm using a Microplate Reader (Molecular Devices). Luciferase activity was normalized to the cell protein level.
Knockdown of ABCB1 Protein via Anti-ABCB1 siRNA, Its Effect on Enhancing DOX Uptake, and Cytotoxicity in Resistant Cells.
To examine the effectiveness of the polyplex in downregulating functional siRNA against its target protein, MCF-7/Adr cells were seeded in a six-well plate and incubated with anti-ABCB1 siRNA (Dharmacon, Inc., Lafayette, CO) in 15% DMEM for 24, 48, and 72 hours. The initial cell density was adjusted to avoid cells reaching overconfluency in a long-term study. The day before transfection, the cells were seeded at 7 × 105/well for the 24-hour treatment study, 6 × 105/well for the 48-hour treatment study, and 3.6 × 105/well for the 72-hour treatment study. Polyplex with 2 µg of anti-ABCB1 siRNA was used for each condition with no change of medium during incubation time. The cells were washed, trypsinized, and incubated with 3 µl of phycoerythrin-conjugated anti-ABCB1 antibody (Thermo Fisher Scientific) at 0.1 µg/µl for 30 minutes in an ice bath. The cells were then washed twice with flow buffer (PBS with 10% FBS and 0.1% sodium azide) and kept in flow buffer on ice for flow cytometric analysis using an LSR II System (BD Biosciences).
To analyze the effect of anti-ABCB1 siRNA on cellular uptake of DOX, MCF-7/Adr cells were seeded in a six-well plate at 7 × 105 confluency. After 24 hours, the anti-ABCB1 siRNA polyplex and DOX solution at 2500 ng/ml was added to the plate and incubated for 4.5 hours. The cells were then washed, trypsinized, and collected for flow cytometry at an excitation wavelength of 470 nm and an emission wavelength of 585 nm.
To examine the effect of anti-ABCB1 siRNA on DOX cytotoxicity, MCF-7/Adr cells were seeded in a 96-well plate at 5000 cells/well. After 24 hours of cell attachment, anti-ABCB1 siRNA polyplex and DOX solution were cotreated with 0.5 µg of siRNA/well and different doses of DOX (100–10,000 ng/ml). The cells were incubated for 72 hours prior to MTT assay for cell viability.
Statistical Analysis.
Data are expressed as the mean ± S.E.M. Statistical significance was considered at P ≤ 0.05.
Results
Polymer and Polyplex Characterization.
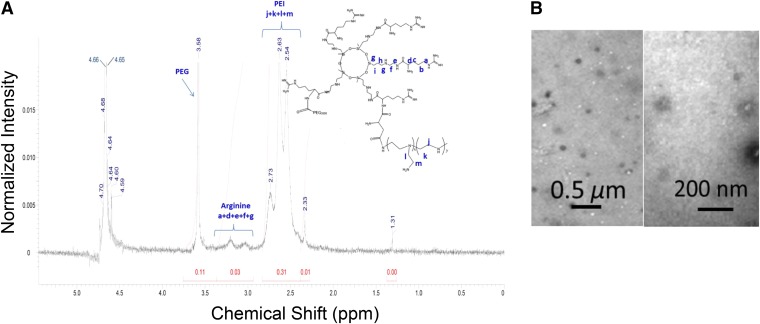
The polymer P(SiDAAr)5PEG3, prepared as per the Scheme, was characterized by 1H NMR (D2O) spectrum to confirm the presence of all components (Fig. 1A). Chemical shifts at δ = 3.4–2.9 are from SiDAAr (−HCCH2CH2CH2NH−) and at δ = 2.8–2.4 are from (−NHCH2CH2-, PEI ethylene). Here, the ethylene resonances of the SiDAAr units are clearly visible. The resonances of PEG (−OCH2CH2−) are also visualized at δ = 3.7–3.5. Branched PEI has much more abundant amino groups than the hydroxyl groups on PEG; therefore, from the moles inputted in the reaction mixture, all of the activated PEGs should react with P(SiDAAr)5. The final polymer P(SiDAAr)5PEG3 has a white sponge-like morphology as a dried polymer.
Fig. 1.
Spectral characterization of polymer and morphologic analysis of siRNA polyplex. (A) 1H NMR spectra P(SiDAAr)5PEG3 shows the polymer’s PEG (−CH2-CH2-O−) groups, as well as arginine, and PEI’s ethylene protons (−CH2−). (B) TEM images of 1/4 siRNA/P(SiDAAr)5PEG3 polyplex at low (×23,000) (left) and high (×68,000) (right) magnifications.
TEM images of the polyplex formulated at an N/P w/w ratio of 1/4 showed positively stained particulate structures with an average size of 78.3 ± 6.2 nm (n = 18) (Fig. 1B). The hydrodynamic diameter of the same formulation was 349.5 nm (polydispersity index = 0.34), and zeta potential was +30 mV. Particle size did not change significantly for the polyplex formulated with the N/P ratios of 1/2 to 1/4 w/w (327–350 nm), whereas at the N/P ratios of 1/5 and 1/6 w/w, the polyplex sizes were smaller (231–240 nm), indicating that with increasing polymer ratio, the polyplex acquired smaller size. Zeta potential for the polyplex at N/P ratio was +8.7 mV, whereas for all other polyplex combinations, it ranged from +26.2 to +42.5 mV, showing a general trend that the polyplex acquired a more cationic charge with increase in the polymeric ratio (Table 1).
TABLE 1.
Physical characteristics of polyplex formulated at different siRNA-to-polymer (N/P w/w) ratios
| siRNA/P(SiDAAr)5P3 (N/P) |
Mean Hydrodynamic Diameter |
Polydispersity Index |
Zeta Potential |
|---|---|---|---|
| nm | mV | ||
| 1/2 | 326.8 | 0.21 | +8.7 |
| 1/3 | 294.7 | 0.64 | +26.2 |
| 1/4 | 349.5 | 0.34 | +30.0 |
| 1/5 | 230.6 | 0.52 | +42.5 |
| 1/6 | 240.1 | 0.46 | +36.3 |
Complexation and siRNA Stability.
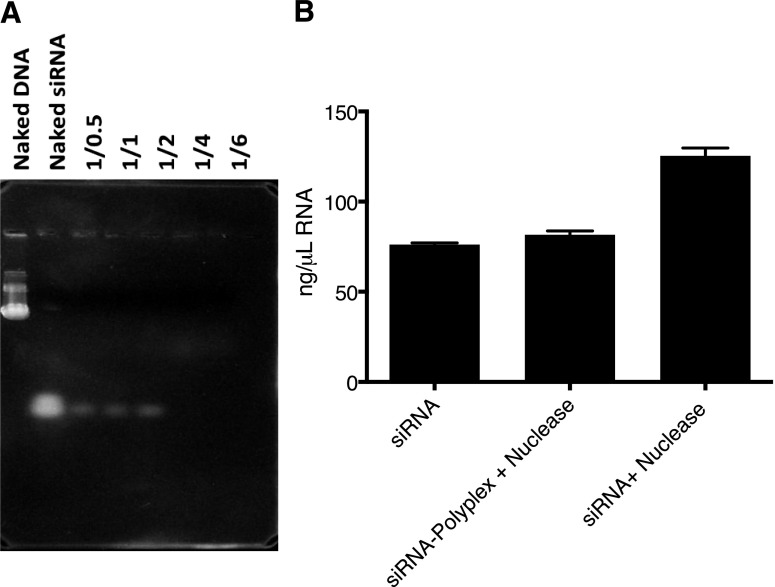
Complete siRNA complexation at 1/4 N/P w/w ratio was evident from no observable migration of free siRNA down the gel (Fig. 2A). In the presence of RNase, free siRNA fragmented, as evident from the increase in absorbance, with the degradation products contributing to this increase; however, no such increase was seen when polyplex was incubated with the nuclease (Fig. 2B).
Fig. 2.
Complexation and protection of siRNA from RNase. (A) Agarose gel electrophoresis of polyplex with different N/P w/w ratios. Uncomplexed siRNA is shown as white bands on the gel. Complete complexation took place at N/P w/w ratios of 1/4 and 1/6 w/w. (B) siRNA degradation assay showed no increase in absorbance (a measurement of free siRNA) in 1/4 P(SiDAAr)5PEG3 polyplex, indicating polymer protecting siRNA from RNase degradation. Data are the mean ± S.E.M.; n = 2 to 3. P < 0.01 between siRNA, siRNA-polyplex + nuclease vs. siRNA + nuclease.
Polyplex Stability in the Presence of Excess Polyanion.
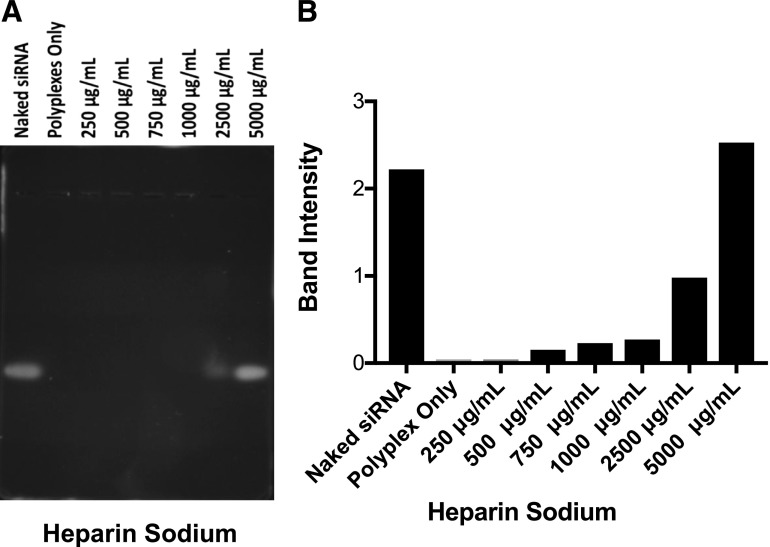
Polyanion competition assay showed that the polyplex remained intact up to a heparin concentration of 1000 µg/ml; however, at higher concentrations, siRNA was released from the polyplex due to the replacement of siRNA from the polyplex by polyanion heparin molecules. At 5000-µg/ml heparin concentration, based on band intensity, the amount of free siRNA migrating down the gel was about the same as that of free siRNA (Fig. 3).
Fig. 3.
Stability of polyplex in the presence of polyanion. Polyanion competition assay revealed siRNA polyplex stability in the presence of varying concentrations of heparin sodium. (A) Gel image. (B) Quantification of band intensity. Partial siRNA release took place at 2500 µg/ml of heparin sodium, whereas at 5000 µg/ml of heparin sodium, nearly all of the siRNA was released from the polyplex.
Cellular Uptake of siRNA Polyplex.
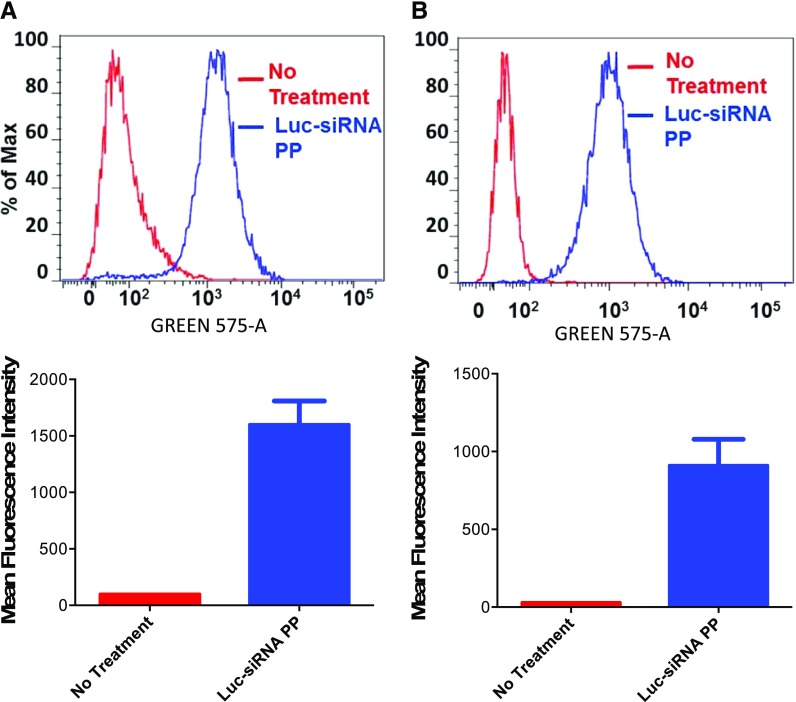
Intracellular uptake of polyplex using TRITC-conjugated P(SiDAAr)5PEG3 polymer showed uptake in both MDA-MB-231-Luc-D3H2LN and MCF-7/Adr cells. This uptake was seen at 4 hours, indicating rapid cellular uptake of the polyplex (Fig. 4).
Fig. 4.
Polyplex uptake in metastatic and resistant breast cancer cells. Uptake in MDA-MB-231-Luc-D3H2LN (A) and MCF-7/Adr cell lines (B) by flow cytometry and the respective mean fluorescence intensity following incubation of cells with polyplex for 4 hours. Luc-siRNA PP, cells that were treated with polyplex prepared with TRITC-conjugated polymer; No treatment, cells that were not treated with polyplex. y-Axis represents the data normalized to the maximum number of cells counted, whereas x-axis represents the channel used for measuring fluorescence signal. Untreated n = 1, treated n = 8 to 9 repeats of the same sample measurements.
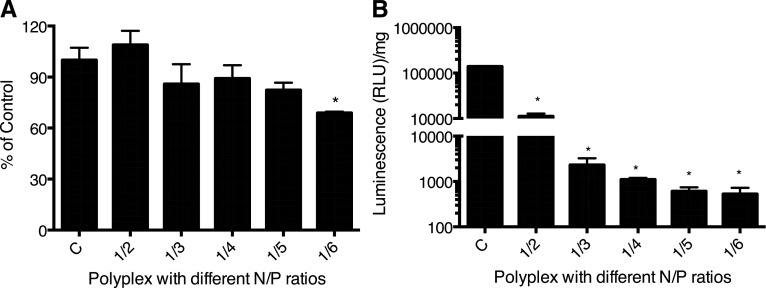
Polyplex Toxicity and Antiluciferase Transfection Efficiency.
Polyplex showed no significant cytotoxicity at all except for the polyplex prepared at an N/P w/w ratio of 1/6 (Fig. 5A). Transfection increased with increasing polymeric ratio in the polyplex as evident from increasing luciferase expression knockdown, reaching 98% at the N/P ratio of 1/4 w/w compared with control, but changed insignificantly thereafter (Fig. 5B). The polyplex with 1/4 N/P w/w ratio showed almost complete luciferase gene expression knockdown yet low toxicity; therefore, this ratio was selected for the subsequent experiment.
Fig. 5.
Cytotoxicity and transfection efficiency of siRNA/P(SiDAAr)5PEG3 polyplex in the MDA-MB-231-Luc-D3H2LN cell line. (A) Polyplex toxicity. *P < 0.05 compared with the control group, n = 6. (B) Luc-siRNA transfection with polyplex at varying N/P w/w ratios. The polymer composition of the polyplex increases and becomes highly effective at N/P ratio of 1/4 w/w, where there is a complete complexation of siRNA. *P < 0.01 compared with the control group, n = 2. RLU, relative light unit.
ABCB1 Protein Downregulation, DOX Uptake, and Its Cytotoxicity in MCF-7/Adr Cells.
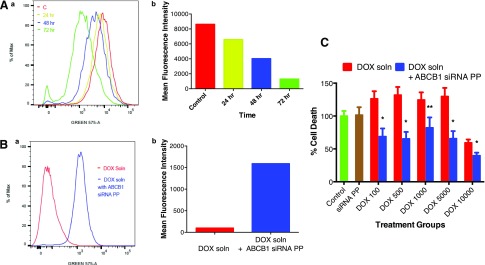
Based on the shift in the fluorescence intensity curve compared with control following 72-hour incubation, there was ∼70% downregulation of the ABCB1 protein in MCF-7/Adr cells transfected with anti-ABCB1 siRNA polyplex (Fig. 6A). MCF-7/Adr cells transfected with anti-ABCB1 siRNA polyplex also enabled greater intracellular accumulation of DOX as evident from the shift in DOX fluorescence intensity compared with the uptake in untransfected cells (Fig. 6B). Greater uptake of DOX in anti-ABCB1 siRNA polyplex–treated cells showed greater drug cytotoxicity than in untreated cells, but the effect was not DOX dose dependent (Fig. 6C).
Fig. 6.
Knockdown ABCB1 with anti-ABCB1 siRNA polyplex and its effect on DOX accumulation and cytotoxicity in MCF-7/Adr cells. (Aa) Flow cytometric analysis of suppression of ABCB1 protein expression following incubation with anti-ABCB1 siRNA polyplex. (Ab) The suppression was seen to increase with incubation time. (Ba) Cotreatment of cells with anti-ABCB1 siRNA polyplex significantly enhanced DOX uptake. (Bb) Change in mean fluorescence intensity of cells cotreated with anti-ABCB1 siRNA polyplex and DOX. Cells were treated with anti-ABCB1 siRNA polyplex and DOX solution at 2500 ng/ml for 4.5 hours prior to flow cytometric analysis of cells for DOX uptake. (C) Cell viability following treating cells with DOX solution alone or cotreatment with anti-ABCB1 siRNA polyplex at different doses DOX (numbers indicate nanograms per milliliter dose) for 72 hours prior to measuring cell viability using MTT assay. n = 6. *P < 0.05; **P = 0.053 compared with DOX alone. PP, polyplex. Flow cytometry: y-axis represents the data normalized to the maximum number of cells counted, whereas x-axis represents the channel used for measuring fluorescence signal.
Discussion
Critical issues in siRNA delivery include its effective incorporation into a delivery vector, ability to protect siRNA from nuclease degradation, and achieving effective transfection even in the presence of serum proteins. In this study, we demonstrated that the polyplex formulated using the polymer P(SiDAAr)5PEG3 meets the aforementioned criteria due to its balanced composition of PEI to condense and protect siRNA, PEG to stabilize polyplex and reduce toxicity, and arginine to facilitate cellular uptake. The spectral characterization confirms the presence of all of the aforementioned components in the polymer (Fig. 1A). The hydrodynamic diameter of the polyplex was significantly greater than the TEM diameter (Fig. 1B), but this is expected considering that the TEM diameter is in a dry state of the polyplex whereas the hydrodynamic diameter is measured in a hydrated state, and this hydration effect is expected to be greater with our polyplex because of the branched nature of the polymer with several hydrophilic hydroxy and amino groups. At an optimal 1/4 N/P w/w ratio, the polyplex demonstrated complete complexation of siRNA (Fig. 2A) and stabilized it from degradation due to nucleases (Fig. 2B). The formed polyplex showed stability in the presence of excess polyanion heparin (Fig. 3), indicating interaction of siRNA and polymer, and stability of the polyplex in the presence of anions such as serum proteins, which was also evident from the transfection data since it was carried out in culture medium containing serum. Although PEI is one of the most widely used transfection reagents, it has limited cellular uptake in the presence of serum, which translates to its limited in vivo efficacy (Wang et al., 2016). Critical for transfection is the release of RNA from the polyplex following its cellular uptake. Our data showed the uptake of the polyplex inside cells (Fig. 4), but it is also effective in transfecting cells, an indication that siRNA is released from the polyplex following its cellular uptake (Fig. 5).
Cytocompatibility of the polyplex is an equally important issue (Douglas et al., 2008). Our data showed that the polyplex at an optimized 1/4 N/P w/w ratio is quite cytocompatible, suggesting that this composition provides the right balance between PEI and PEG in masking cationic charge of PEI without influencing the polyplex cellular uptake (Fig. 4) or its transfection (Fig. 5). Previously, we have shown that PEI alone is quite toxic to cells (Lu et al., 2015).
ABCB1 is a major contributor to multidrug resistance, as it actively pumps out chemotherapeutic agents, thus lowering drug concentration inside cells (Susa et al., 2010). MCF-7/Adr cells are known to overexpress the ABCB1 efflux pump (Jiang et al., 2016). It is evident from our data that MCF-7/Adr incubated with anti-ABCB1 siRNA polyplex resulted in a time-dependent increased downregulation of ABCB1 protein (Fig. 6A). This effect can be explained considering the half-life of ABCB1 protein degradation from the cell membrane ranges between 15 and 72 hours (Richert et al., 1988; Mickley et al., 1989; Chin et al., 1990; Aleman et al., 2003). Anti-ABCB1 siRNA transfection resulted in enhanced DOX cellar uptake (Fig. 6B), hence increased drug cytotoxicity (Fig. 6C), further confirming the efficacy of anti-ABCB1 siRNA polyplex. However, the cytotoxic effect of DOX was not dose dependent, suggesting that further optimization of the treatment protocol may be needed, which could involve pretreating resistant cells with anti-ABCB1 siRNA polyplex to optimize the dose and time required to maximize downregulation of the efflux protein prior to treatment with DOX. Others have also tested this approach and have shown that sustained delivery for siRNA is better for drug efficacy than its transient effect (Susa et al., 2010).
In addition to overexpression of membrane-bound drug transporter proteins, such as P-glycoprotein (ABCB1), there are other multidrug resistance–associated proteins (MRP1, ABCC1, and MRP2, ABCC2) and breast cancer resistance protein (ABCG2) (Housman et al., 2014; Issa et al., 2017) which may not have been affected by treating resistant cells with anti-ABCB1 siRNA alone. Thus, a cocktail of siRNA against all of the efflux proteins and/or other cellular targets responsible for drug resistance may be needed to optimize the treatment effect to reverse drug resistance. We have previously demonstrated that the similarly formulated polyplex is effective in delivering DNA alone or DNA and siRNA together, achieving enhanced transfections of both moieties (Morris and Sharma, 2010; Lu et al., 2015). Thus, with our polyplex, both cytoplasmic and nuclear factors (e.g., DNA damage repair mechanism, transcription factors, epigenetic modifications, etc.) responsible for drug resistance can be targeted simultaneously (Housman et al., 2014). We have also reported epigenetic modifications in MCF-7/Adr cells as a cause of drug resistance (Vijayaraghavalu and Labhasetwar, 2013); thus, a combination treatment that targets multiple mechanisms may be needed to address the complex nature of drug resistance.
Although we have not determined the effect of functional siRNA in metastatic breast cancer cells using our polyplex, there are several genes (e.g., CDK8, a cyclin-dependent kinase member of the mediator complex; Rho-associated coiled-coil-containing protein kinase, etc.) that can be targeted to inhibit cancer cell proliferation (Lee et al., 2016; Ahmadzada et al., 2018). In addition to cancer cells, our studies with similarly formulated DNA polyplex demonstrated gene transfection in neuronal cells, both in vitro and in vivo (Joshi et al., 2018), indicating broader applications of our polyplex for delivery of nucleic acids for potentially treating different disease conditions.
Conclusion
The formulated polyplex at an optimal composition is cytocompatible and effective in delivering siRNA to both resistant and metastatic breast cancer cells. With functional siRNA against anti-ABCB1, the treatment was partially effective in overcoming DOX resistance. Since polyplex showed stability in the presence of excess anion, and the transfection studies were carried out in the presence of serum, it is likely that our polyplex formulation could be effective in vivo and potentially can be explored in treating different types of cancers and other diseases.
Scheme.
for Polymer Synthesis. Steps involved in synthesis of polymer: oligo(-alkylaminosiloxanes) (A), l-arginine–modified oligo(-alkylaminosiloxanes) (SiDAAr5) (B), P(SiDAAr)5 (C), and conjugation to PEG [P(SiDAAr)5PEG3] (D).
Abbreviations
- ABCB1
multidrug resistance protein 1
- DMEM
Dulbecco’s modified Eagle’s medium
- DMSO
dimethylsulfoxide
- DOX
doxorubicin
- EDC
1-ethyl-3-(3-dimethylaminopropyl)carbodiimide hydrochloride
- FBS
fetal bovine serum
- MTT
3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide
- MWCO
molecular weight cutoff
- NHS
N-hydroxysuccinimide
- N/P ratio
nucleotide/polymer
- w/w ratio
weight/weight ratio
- PBS
phosphate-buffered saline
- PEG
polyethylene glycol
- PEI
poly(ethyleneimine)
- P(SiDAAr)5
5 l-arginine oligo(alkylaminosiloxane) grafted poly(ethyleneimine)
- P(SiDAAr)5PEG3
3 polyethylene glycol modified l-arginine oligo(alkylaminosiloxane) graft poly(ethyleneimine)
- SiDAAr
l-arginine oligo(alkylaminosiloxane)
- siRNA
small interfering RNA
- TEM
transmission electron microscopy
- 6-TRITC
tetramethylrhodamine-6-isothiocyanate
Authorship Contributions
Participated in research design: Lu, Morris, Labhasetwar.
Conducted experiments: Lu, Morris.
Performed data analysis: Lu, Morris, Labhasetwar.
Wrote or contributed to the writing of the manuscript: Lu, Morris, Labhasetwar.
Footnotes
This study was supported by the National Institutes of Health (NIH) National Cancer Institute [Grant 1R01CA14939 (to V.L.)]. The transmission electron microscope used for imaging was supported by an equipment grant from the NIH [Grant S10RR031536].
References
- Ahmadzada T, Reid G, McKenzie DR. (2018) Fundamentals of siRNA and miRNA therapeutics and a review of targeted nanoparticle delivery systems in breast cancer. Biophys Rev 10:69–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alameh M, Dejesus D, Jean M, Darras V, Thibault M, Lavertu M, Buschmann MD, Merzouki A. (2012) Low molecular weight chitosan nanoparticulate system at low N:P ratio for nontoxic polynucleotide delivery. Int J Nanomedicine 7:1399–1414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alemán C, Annereau JP, Liang XJ, Cardarelli CO, Taylor B, Yin JJ, Aszalos A, Gottesman MM. (2003) P-glycoprotein, expressed in multidrug resistant cells, is not responsible for alterations in membrane fluidity or membrane potential. Cancer Res 63:3084–3091. [PubMed] [Google Scholar]
- Brissault B, Kichler A, Leborgne C, Danos O, Cheradame H, Gau J, Auvray L, Guis C. (2006) Synthesis, characterization, and gene transfer application of poly(ethylene glycol-b-ethylenimine) with high molar mass polyamine block. Biomacromolecules 7:2863–2870. [DOI] [PubMed] [Google Scholar]
- Burnett JC, Rossi JJ, Tiemann K. (2011) Current progress of siRNA/shRNA therapeutics in clinical trials. Biotechnol J 6:1130–1146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calnan BJ, Tidor B, Biancalana S, Hudson D, Frankel AD. (1991) Arginine-mediated RNA recognition: the arginine fork. Science 252:1167–1171. [DOI] [PubMed] [Google Scholar]
- Caroline Diana SM, Rekha MR. (2017) Efficacy of vinyl imidazole grafted cationized pullulan and dextran as gene delivery vectors: a comparative study. Int J Biol Macromol 105:947–955. [DOI] [PubMed] [Google Scholar]
- Chin KV, Tanaka S, Darlington G, Pastan I, Gottesman MM. (1990) Heat shock and arsenite increase expression of the multidrug resistance (MDR1) gene in human renal carcinoma cells. J Biol Chem 265:221–226. [PubMed] [Google Scholar]
- Choi JS, Nam K, Park JY, Kim JB, Lee JK, Park JS. (2004) Enhanced transfection efficiency of PAMAM dendrimer by surface modification with L-arginine. J Control Release 99:445–456. [DOI] [PubMed] [Google Scholar]
- Danielsen S, Maurstad G, Stokke BT. (2005) DNA-polycation complexation and polyplex stability in the presence of competing polyanions. Biopolymers 77:86–97. [DOI] [PubMed] [Google Scholar]
- de Fougerolles A, Vornlocher HP, Maraganore J, Lieberman J. (2007) Interfering with disease: a progress report on siRNA-based therapeutics. Nat Rev Drug Discov 6:443–453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Douglas KL, Piccirillo CA, Tabrizian M. (2008) Cell line-dependent internalization pathways and intracellular trafficking determine transfection efficiency of nanoparticle vectors. Eur J Pharm Biopharm 68:676–687. [DOI] [PubMed] [Google Scholar]
- Dykxhoorn DM, Novina CD, Sharp PA. (2003) Killing the messenger: short RNAs that silence gene expression. Nat Rev Mol Cell Biol 4:457–467. [DOI] [PubMed] [Google Scholar]
- Hickerson RP, Vlassov AV, Wang Q, Leake D, Ilves H, Gonzalez-Gonzalez E, Contag CH, Johnston BH, Kaspar RL. (2008) Stability study of unmodified siRNA and relevance to clinical use. Oligonucleotides 18:345–354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Höbel S, Aigner A. (2010) Polyethylenimine (PEI)/siRNA-mediated gene knockdown in vitro and in vivo. Methods Mol Biol 623:283–297. [DOI] [PubMed] [Google Scholar]
- Housman G, Byler S, Heerboth S, Lapinska K, Longacre M, Snyder N, Sarkar S. (2014) Drug resistance in cancer: an overview. Cancers (Basel) 6:1769–1792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunter AC, Moghimi SM. (2010) Cationic carriers of genetic material and cell death: a mitochondrial tale. Biochim Biophys Acta 1797:1203–1209. [DOI] [PubMed] [Google Scholar]
- Issa ME, Takhsha FS, Chirumamilla CS, Perez-Novo C, Vanden Berghe W, Cuendet M. (2017) Epigenetic strategies to reverse drug resistance in heterogeneous multiple myeloma. Clin Epigenetics 9:17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang J, Wang X, Cheng K, Zhao W, Hua Y, Xu C, Yang Z. (2016) Psoralen reverses the P-glycoprotein-mediated multidrug resistance in human breast cancer MCF-7/ADR cells. Mol Med Rep 13:4745–4750. [DOI] [PubMed] [Google Scholar]
- Joshi CR, Raghavan V, Vijayaraghavalu S, Gao Y, Saraswathy M, Labhasetwar V, Ghorpade A. (2018) Reaching for the stars in the brain: polymer-mediated gene delivery to human astrocytes. Mol Ther Nucleic Acids 12:645–657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawakami S, Ito Y, Charoensit P, Yamashita F, Hashida M. (2006) Evaluation of proinflammatory cytokine production induced by linear and branched polyethylenimine/plasmid DNA complexes in mice. J Pharmacol Exp Ther 317:1382–1390. [DOI] [PubMed] [Google Scholar]
- Kim TI, Baek JU, Zhe Bai C, Park JS. (2007) Arginine-conjugated polypropylenimine dendrimer as a non-toxic and efficient gene delivery carrier. Biomaterials 28:2061–2067. [DOI] [PubMed] [Google Scholar]
- Lan H, Chen H, Liu Y, Jiang C, Mao Q, Jia D, Chen Q, Wei T. (2015) Small interfering RNA pathway modulates initial viral infection in midgut epithelium of insect after ingestion of virus. J Virol 90:917–929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee SJ, Kim MJ, Kwon IC, Roberts TM. (2016) Delivery strategies and potential targets for siRNA in major cancer types. Adv Drug Deliv Rev 104:2–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu S, Morris VB, Labhasetwar V. (2015) Codelivery of DNA and siRNA via arginine-rich PEI-based polyplexes. Mol Pharm 12:621–629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merkel OM, Beyerle A, Beckmann BM, Zheng M, Hartmann RK, Stöger T, Kissel TH. (2011) Polymer-related off-target effects in non-viral siRNA delivery. Biomaterials 32:2388–2398. [DOI] [PubMed] [Google Scholar]
- Mickley LA, Bates SE, Richert ND, Currier S, Tanaka S, Foss F, Rosen N, Fojo AT. (1989) Modulation of the expression of a multidrug resistance gene (mdr-1/P-glycoprotein) by differentiating agents. J Biol Chem 264:18031–18040. [PubMed] [Google Scholar]
- Moghimi SM, Symonds P, Murray JC, Hunter AC, Debska G, Szewczyk A. (2005) A two-stage poly(ethylenimine)-mediated cytotoxicity: implications for gene transfer/therapy. Mol Ther 11:990–995. [DOI] [PubMed] [Google Scholar]
- Morris VB, Labhasetwar V. (2015) Arginine-rich polyplexes for gene delivery to neuronal cells. Biomaterials 60:151–160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris VB, Sharma CP. (2010) Enhanced in-vitro transfection and biocompatibility of L-arginine modified oligo (-alkylaminosiloxanes)-graft-polyethylenimine. Biomaterials 31:8759–8769. [DOI] [PubMed] [Google Scholar]
- Ragelle H, Vandermeulen G, Préat V. (2013) Chitosan-based siRNA delivery systems. J Control Release 172:207–218. [DOI] [PubMed] [Google Scholar]
- Richard JP, Melikov K, Vives E, Ramos C, Verbeure B, Gait MJ, Chernomordik LV, Lebleu B. (2003) Cell-penetrating peptides. A reevaluation of the mechanism of cellular uptake. J Biol Chem 278:585–590. [DOI] [PubMed] [Google Scholar]
- Richert ND, Aldwin L, Nitecki D, Gottesman MM, Pastan I. (1988) Stability and covalent modification of P-glycoprotein in multidrug-resistant KB cells. Biochemistry 27:7607–7613. [DOI] [PubMed] [Google Scholar]
- Rietwyk S, Peer D. (2017) Next-generation lipids in RNA interference therapeutics. ACS Nano 11:7572–7586. [DOI] [PubMed] [Google Scholar]
- Scamuffa N, Siegfried G, Bontemps Y, Ma L, Basak A, Cherel G, Calvo F, Seidah NG, Khatib AM. (2008) Selective inhibition of proprotein convertases represses the metastatic potential of human colorectal tumor cells. J Clin Invest 118:352–363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shegokar R, Al Shaal L, Mishra PR. (2011) SiRNA delivery: challenges and role of carrier systems. Pharmazie 66:313–318. [PubMed] [Google Scholar]
- Susa M, Iyer AK, Ryu K, Choy E, Hornicek FJ, Mankin H, Milane L, Amiji MM, Duan Z. (2010) Inhibition of ABCB1 (MDR1) expression by an siRNA nanoparticulate delivery system to overcome drug resistance in osteosarcoma. PLoS One 5:e10764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vijayaraghavalu S, Labhasetwar V. (2013) Efficacy of decitabine-loaded nanogels in overcoming cancer drug resistance is mediated via sustained DNA methyltransferase 1 (DNMT1) depletion. Cancer Lett 331:122–129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vijayaraghavalu S, Peetla C, Lu S, Labhasetwar V. (2012) Epigenetic modulation of the biophysical properties of drug-resistant cell lipids to restore drug transport and endocytic functions. Mol Pharm 9:2730–2742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Lu Z, Wientjes MG, Au JL. (2010a) Delivery of siRNA therapeutics: barriers and carriers. AAPS J 12:492–503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S, Amornwittawat N, Juwita V, Kao Y, Duman JG, Pascal TA, Goddard WA, Wen X. (2009) Arginine, a key residue for the enhancing ability of an antifreeze protein of the beetle Dendroides canadensis. Biochemistry 48:9696–9703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Li Z, Han Y, Liang LH, Ji A. (2010b) Nanoparticle-based delivery system for application of siRNA in vivo. Curr Drug Metab 11:182–196. [DOI] [PubMed] [Google Scholar]
- Wang Z, Zou H, Wang Z, Wu J, Xia Z, Feng M. (2016) Highly stable polyglutamate derivatives/siRNA polyplex efficiently down-relegate survivin expression and augment the efficacy of cisplatin. Int J Pharm 505:24–34. [DOI] [PubMed] [Google Scholar]
- Whitehead KA, Langer R, Anderson DG. (2009) Knocking down barriers: advances in siRNA delivery. Nat Rev Drug Discov 8:129–138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu ZW, Chien CT, Liu CY, Yan JY, Lin SY. (2012) Recent progress in copolymer-mediated siRNA delivery. J Drug Target 20:551–560. [DOI] [PubMed] [Google Scholar]
- Yang X, Iyer AK, Singh A, Choy E, Hornicek FJ, Amiji MM, Duan Z. (2015) MDR1 siRNA loaded hyaluronic acid-based CD44 targeted nanoparticle systems circumvent paclitaxel resistance in ovarian cancer. Sci Rep 5:8509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan X, Naguib S, Wu Z. (2011) Recent advances of siRNA delivery by nanoparticles. Expert Opin Drug Deliv 8:521–536. [DOI] [PubMed] [Google Scholar]
- Zhu L, Mahato RI. (2010) Lipid and polymeric carrier-mediated nucleic acid delivery. Expert Opin Drug Deliv 7:1209–1226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zi Y, Zhao W, Zhou J, He H, Xie M. (2015) Silencing of TMSG1 enhances metastasis capacity by targeting V-ATPase in breast cancer. Int J Clin Exp Pathol 8:1312–1320. [PMC free article] [PubMed] [Google Scholar]