Abstract
Four species of previously known nematodes from the family Camallanidae were found from different hosts in South Africa: Batrachocamallanus xenopodis from the frog Xenopus muelleri, Paracamallanus cyathopharynx and Procamallanus pseudolaeviconchus from the catfish Clarias gariepinus and Spirocamallanus daleneae from the catfish Synodontis zambezensis. In the material collected from various marine fishes, several specimens of nematodes from the genus Camallanus clearly differed from all previously known species. Based on morphological differences these specimens are assigned to a new species, C. sodwanaensis. Molecular data of 18S and 28S rDNA and COI sequences are provided for the collected species and a phylogenetic analyses based on 28S gene fragmets are presented.
Keywords: Nematodes, Camallanidae, Fish, Amphibians, Africa, Phylogeny
Graphical abstract

Highlights
-
•
Five species of Camallanidae nematodes found from different hosts in South Africa.
-
•
Camallanus sodwanaensis n. sp. is the first species from marine fish in Southern Africa.
-
•
Molecular data of 18S and 28SrDNA and COI sequences provided for found species.
-
•
Phylogenetic analyses based on 28S gene fragmets is performed.
1. Introduction
The Camallanidae is a globally distributed group of parasitic nematodes that primarily infects the digestive tract of marine and freshwater fish and less often amphibians, turtles and snakes (Stromberg and Crites, 1974; Rigby and Rigby, 2014). These nematodes can be morphologicaly distinguished from all other groups by the presence of a well-developed buccal capsule often supported by different structures (basal ring, longitudinal or spiral ridges, tridents, etc.) (Rigby and Rigby, 2014).
Hitherto, the camallanid fauna of African vertebrates is poorly studied. Although numerous genera from the subfamilies Procamallaninae and Camallaninae were erected, most of them consist of only a few species. Of the Procamallaninae, three species of the genus Procamallanus Baylis, 1923 were described from freshwater fishes: P. laeviconchus Wedl, 1861, P. armatus Campana-Rouget et Therezien, 1965 and P. pseudolaeviconchus Moravec et Van As, 2015a. Seven species of the genus Spirocamallanus Olsen, 1952 were described from African freshwater fishes: S. daleneae Boomker, 1993, S. mazabukae Yeh, 1957, S. spiralis (Baylis, 1923), S. olseni Campana-Rouget et Razarihelissoa, 1965, S. serranochromis Moravec et Van As, 2015b, S. parachannae Moravec et Jirků, 2015 and S. pseudospiralis Moravec et Scholtz, 2017. Jackson and Tinsley (1995) established a new genus Batrachocamallanus Jackson et Tinsley, 1995 to include two species from pipid frogs, Batrachocamallanus slomei Southwell et Kirschner, 1937 (described as P. slomei) and B. xenopodis Jackson et Tinsley, 1995 (described as S. xenopodis), and also described two new species B. occidentalis Jackson et Tinsley, 1995 and B. siluranae Jackson et Tinsley, 1995. Of these, B. slomei and B. xenopodis were subsequently found from Xenopus spp. in different regions of Africa (Jackson and Tinsley, 1995; Svitin et al., 2018).
Four genera of the Camallaninae were described and subsequently found in aquatic vertebrates from Africa. Representitives of three genera, namely Paracamallanus Yorke et Maplestone, 1926, Zeylanema Yeh, 1960 and Neocamallanus Ali, 1957 were found in freshwater fishes, each represented by a single species: P. cyathopharynx (Baylis, 1923), Z. ctenopomae (Vassiliadès et Petter, 1972) (described as Camallanus ctenopomae) and N. polypteri (Kabre et Petter, 1997) (described as Camallanus polypteri). The genus Camallanus Railliet et Henry, 1915 includes two species found in freshwater fishes: C. longicaudatus Moravec (1973) and C. kirandensis Baylis (1928); four species found in frogs: C. kaapstaadi Southwell et Kirshner, 1937, C. dimitrovi Durette-Desset et Batcharov, 1974, C. xenopodis Jackson et Tinsley, 1995 and C. macrocephalus Jackson et Tinsley, 1995; and one species found in a freshwater turtle, C. chelonius Baker, 1983. It should be noted that all previously described species in South Africa were reported from freshwater hosts while nematode parasites of marine organisms are still poorly studied and no camallanin has been reported from this host group (Smit and Hadfield, 2015).
Details of the morphology of the buccal capsule were traditionally used for generic differentiation within the family. Nevertheless, reliability of some characters and, as a result, number of genera within the Camallanidae, are still debated. Moravec and co-authors (1988, 2006, 2015a, 2015b) considered five taxa of the Procamallaninae (Procamallanus; Spirocamallanus; Platicamallanus Bilqees et Akram, 1982; Punctocamallanus Moravec et Scholz, 1991 and Denticamallanus Moravec et Thatcher, 1997) as subgenera of Procamallanus. Jackson and Tinsley (1995) followed the opinion of Moravec and colleagues in considering differences of the buccal capsule structure alone as not sufficient for the generic differentiation. At the same time these authors distinguished Batrachocamallanus mostly based on the presence of the large number of mucrons (more than five) on the female tail, relatively smaller body size and specificity to the amphibian hosts. Later on Moravec et al. (2006) considered the latter proposed differences as not reliable generic characters and advocated for the reduction of Batrachocamallanus to a junior synonym of Procamallanus. Rigby and Rigby (2014) supported the synonymy of Batrachocamallanus, although recognizing the genera that Moravec et al. (2006) considered as subgenera. Within the subfamily Camallaninae, Rigby and Rigby (2014) recognized three valid genera: Camallanus with Zeylanema and Serpinema Yeh, 1960 as junior synonyms, Neocamallanus Ali, 1957 with junior synonym Neozeylanema Sinha et Sahay, 1966 and Oncophora Diesing, 1851 with Paracamallanus as a synonym. At the same time, Moravec and colleagues (2015c, 2017) considered Zeylanema as subgenus of Camallanus and Paracamallanus as a valid genus separate from Oncophora. In numerous works on camallanid nematodes different authors considered different characters (details of buccal structure, female tail morphology, male genital system, etc.) as generic, subgeneric or species differentiators. As a result, based on different opinions, the number of genera within the family Camallanidae varies from two to twelve (Moravec and Thatcher, 1997; Moravec and Sey, 1988; Moravec and Van As, 2015a; Moravec and Van As, 2015b; Moravec and Jirků, 2017; Rigby and Adamson, 1998; Anderson et al., 2009; Rigby and Rigby, 2014). Therefore, it is clear that additional and detailed studies, including molecular analyses, are necessary to revise the status of the different taxa within the Camallanidae.
Several molecular studies which included camallanid nematodes were published recently. Černotíková et al. (2011) studied the phylogenetic relationships of spirurine nematodes including members of the families Philometridae, Dracunculidae, Cysticolidae, Quimperidae, Rhabdochonidae, Cucullanidae and Camallanidae based on 18S rDNA gene data. In the tree provided by the authors the subclades within the clade represented by members of the Camallanidae received overall low support with C. carangis Olsen, 1954 appearing in the Procamallaninae subclade and P. rarus Travassos et Artigas 1928 at the basal position to the subclades consisted of Procamallanus spp. and Camallanus spp., albeit without support. Later, Sardella et al. (2017) redescribed S. macaensis Vicente et Santos, 1972 and included this species in the phylogenetic analyses of the Camallanidae based on 18S rDNA gene data. Similarly, in studies of Černotíková et al. (2011), Procamallanus, Spirocamallanus and Camallanus formed weakly supported clades. Recently, Chaudhary et al. (2017) provided a phylogenetic tree based on the 18S rDNA gene with overall weakly supported clades and the members of the Procamallaninae and Camallaninae simultaneously appeared in different clades.
Three publications dealt with genes other than 18S rRNA. Wu et al. (2008) showed the variability between two species, namely C. cotti Fujita, 1927 and C. hypophthalmichthys Dogel and Akhmerov, 1959 from fish in China using sequences of the internal transcribed spacer (ITS) regions of rDNA, ITS1 and ITS2. Kuzmin et al. (2011) showed the phylogenetic relationships of five species of Camallanus from Australian turtles based on the partial 28S rDNA alignments. Svitin et al. (2018) showed the phylogenetic relationships of two Camallanus species from African frogs with two species from Chinese fish based on the mitochondrial cytochrome c oxidase 1 (COI) gene dataset and five species from Australian turtles based on 28S rDNA dataset.
To date, phylogenetic studies based on the 18S rRNA gene contained numerous controversies and studies based on other genetic markers included very few species, therefore questions on the evolutionary relationships amongst the Camallanidae and the status of different taxa within the family are still not resolved.
During parasitological surveys in the KwaZulu-Natal Province of South Africa several species of camallanid nematodes were found: B. xenopodis in the frog Xenopus muelleri (Peters, 1844); Pa. cyathopharynx and P. pseodolaeviconchus from catfish Clarias gariepinus Burchell, 1822; S. daleneae from catfish Synodontis zambezensis (Peters, 1852); and specimens of Camallanus clearly different from previously known species from five species of marine fishes (Pempheris adusta Bleeker, 1877, Cirrhitus pinnulatus (Förster, 1801), Pomadasys furcatus (Bloch et Schneider, 1801), Terapon jarbua (Forsskal, 1775) and Trachinotus botla (Shaw, 1803)). In present study we follow Anderson et al. (2009) for generic identification of the species, with some modifications (see below). Detailed descriptions and molecular characterisation based on three genes (18S and 28S rDNA and COI) of found species followed by molecular analyses based on 28S rRNA gene are presented.
2. Materials and methods
Material was collected from different localities in KwaZulu-Natal Province in South Africa during August, October and November 2017, and August 2018. In total, three frogs, X. muelleri, 15 African sharptooth catfish, Clarias gariepinus, 25 Brown squeakers, Synodontis zambezensis, 22 Dusky sweepers, Pempheris adusta, four Stocky hawkfish, four Cirrhitus pinnulatus, four Banded grunters, Pomadasys furcatum, four Largespotted darts, Trachinotus botla and three Jarbua terapons, Terapon jarbua were examined for the presence of parasites.
Amphibian hosts were anaesthetised in 6% ethyl-3-aminobenzoate methanesulfonate (MS222) (Sigma-Aldrich Co., St. Louis, Missouri, USA) and subsequently euthanised through severing the spine and destroying the brain according to internationally accepted standard operating procedures (ethics number: NWU-00492-16-S5). Fish hosts were euthanised by cranial pithing and spinal severance (ethics number: NWU-00159-18-S5).
During the total dissection, the digestive tract was removed and placed in 9% saline. Nematodes were gently removed, washed in saline and fixed in hot 70% ethanol and subsequently stored in 70% ethanol. Prior to microscopical examination, nematodes were placed in distilled water for about 20 min and then cleared in lactophenol. Apical sections were prepared manually using a thin razor and examined en face on temporary mounts. Morphology of the nematodes was studied using Nikon E800 and Nikon ECLIPSE Ni compound microscopes equipped with DIC optics.
In total, 77 nematodes were studied of which 46 were measured. All measurements in the text are given in micrometres unless otherwise indicated. Measurements are presented as ranges followed by mean values in parentheses and measurements of type specimens are in square brackets (if applicable).
For molecular analysis, the middle fragments of the nematodes were used while anterior and posterior parts were preserved for microscopic identification. DNA was extracted using PCRBIO Rapid Extract PCR Kit following the standard protocol method recommended by the manufacturer. Polymerase chain reaction for COI was performed using the primer pair LCO1490 (5′-GGT CAA CAA ATC ATA AAG ATA TTG G-3′) and HCO2198 (5′-TAA ACT TCA GGG TGA CCA AAA AAT CA-3′). The thermocycling profile was as follows: 3 min denaturation at 94 °C, 10 cycles of 94 °C for 30 s, 45 °C for 30 s, 72 °C for 60 s and 40 cycles at 94 °C for 30 s, 51 °C for 60 s, 72 °C for 60 s for amplification, 72 °C for 10 min for extension (Folmer et al., 1994; Svitin et al., 2018). The 18S rRNA sequence fragments were amplified using the primer pair F18ScF1 (5′-ACC GCC CTA GTT CTG ACC GTA AA-3′) and F18ScR1 (5′-GGT TCA AGC CAC TGC GAT TAA AGC-3′). The thermocycling profile was as follows: 2 min denaturation at 95 °C for 30 s, 40 cycles of 95 °C for 30 s, 58 °C for 30 s and 72 °C for 90 s for amplification, 72 °C for 10 min for extinction (Lefoulon et al., 2015). The partial fragments of the 28S rRNA gene were amplified using a pair of newly designed primers: CTEf (5′-AGT GAA TGG GGA AAA GCC CA-3′) and CTEr (5′-GGA CCT CCA CCA GAG TTT CC-3′). The thermocycling profile was as follows: 3 min denaturation at 95 °C; 40 cycles of 30 s at 95 °C, 30 s at 54 °C, 2 min at 72 °C for amplification; 7 min for extension at 72 °C. Unpurified PCR products were sent to a commercial sequencing company (Inqaba Biotechnical Industries (Pty) Ltd, Pretoria, South Africa). DNA products were sequenced in both directions using the PCR primer pairs. Resulting sequences were assembled and chromatogram-based contigs were generated and trimmed using Geneious (V. 9.0) software and submitted to GenBank under the following accession numbers: COI [MN523681 – MN523683], 28S [MN525304 – MN525307], 18S [MN514768 – MN514775].
Novel partial 18S and 28S rDNA sequences and COI sequences obtained during this study were aligned with the sequences for the Camallanidae downloaded from GenBank using MUSCLE v3.7 implemented in Geneious ver. 9.1. Two alignments for the partial 28S rRNA gene were constructed. Alignment 1 was based on the longest sequences (867 nucleotide (nt)). Alignment 2 was much shorter (491 nt) in order to include the short sequences of five species of Camallanus spp. and a species of Serpinema published by Kuzmin et al. (2009) and Kuzmin et al. (2011). The final length of the alignment for 18S rDNA was 717 nt and the alignment for COI was 428 nt. The outgroup for each alignment was estimated using the basic local alignment searching tool (BLAST). The best-fitting model for each dataset was estimated prior to analyses using jModelTest (V. 2.1.2) (Guindon and Gascuel, 2003; Darriba et al., 2012). This was GTR+G for both the 28S rDNA, as well as for the 18S and COI datasets. Bayesian inference analyses were run using MrBayes (V. 3.2.2) software with the following nucleotide substitution model settings: lset nst = 6, rates = invgamma, ncat = 4, shape = estimate, inferrates = yes and basefreq = empirical. Further analyses were performed using the following parameters: mcmc ngen = 3 000 000 for 28S and COI fragments and 10 000 000 for 18S, samplefreq = 100, printfreq = 100 and diagnfreq = 1000. The maximum likelihood analyses were performed using PhyML version 3.0 (Guindon et al., 2010) run on the ATGC bioinformatics platform [http://www.atgc-montpellier.fr/ngs]. Nodal support in the maximum likelihood analyses was estimated from 100 bootstrap pseudoreplicates. Trees were visualised using the FigTree (V. 1.4.3) software (Rambaut, 2012). The p-distance and the number of difference matrix in Mega (V. 7.0) (Kumar et al., 2015) software were used for the pairwise analyses.
3. Results
In total, five species of camallanid nematodes were recovered: Batrachocamallanus xenopodis from Müller's platanna Xenopus muelleri; Paracamallanus cyathopharynx and Procamallanus pseudolaeviconchus from African sharptooth catfish Clarias gariepinus; Spirocamallanus daleneae from Brown squeaker Synodontis zambezensis; and Camallanus sodwanaensis n. sp. from five species of marine fish (Pempheris adusta, Cirrhitus pinnulatus, Pomadasys furcatus, Terapon jarbua and Trachinotus botla).
3.1. Species descriptions
Family Camallanidae Railliet et Henry, 1915.
Genus Camallanus Railliet et Henry, 1915.
Camallanus sodwanaensis n. sp.
Type host: Dusky sweeper Pempheris adusta Bleeker, 1877 (Perciformes: Pempheridae).
Other hosts: Stocky hawkfish Cirrhitus pinnulatus (Förster, 1801) (Perciformes: Cirrhitidae), Banded grunter Pomadasys furcatus (Bloch et Schneider, 1801) (Perciformes: Haemulidae), Jarbua terapon Terapon jarbua (Forsskål, 1775) (Perciformes: Terapontidae), and Largespotted dart Trachinotus botla (Shaw, 1803) (Perciformes: Carangidae).
Site of infection: Intestine.
Type locality: Sodwana Bay, KwaZulu-Natal Province, South Africa (32°40′46"E; 27°32′24"S).
Type material: Holotype (male, [NMB P509]), allotype (female, [NMB P510]), paratypes [NMB P511] deposited in the National Museum Parasite Collection (Bloemfontein, South Africa).
Intensity: Pempheris adusta: 1–15 (5.7); Cirrhitus pinnulatus: 1–5 (2.3); Terapon jarbua: 1–2 (1.5); Pomadasys furcatum: 1–1 (1); Trachinotus botla: 1; total: 1–15 (3.0).
Prevalence: Pempheris adusta – 14% (six of 22 specimens were infected); Cirrhitus pinnulatus – 75% (three of four specimens were infected); Terapon jarbua – 67%; Pomadasys furcatum – 50% (two of four specimens were infected); Trachinotus botla – 25% (one of four specimens were infected); total – 27%.
Abundance: Pempheris adusta – 0.8; Cirrhitus pinnulatus – 1.8; Terapon jarbua – 1; Pomadasys furcatum – 0.5; Trachinotus botla – 0.3; total – 0.8.
Representative DNA sequences: 28S [MN525306], 18S [MN514774].
ZooBank registration: To comply with the regulations set out in article 8.5 of the amended 2012 version of the International Code of Zoological Nomenclature (ICZN, 2012), details of the new species have been submitted to ZooBank. The Life Science Identifier (LSID) for Camallanus sodwanaensis n. sp. is urn:lsid:zoobank.org:act:BCEC589F-46B3-4645-9EC1-F6FA36B11213
Etymology: The species is named after its type locality.
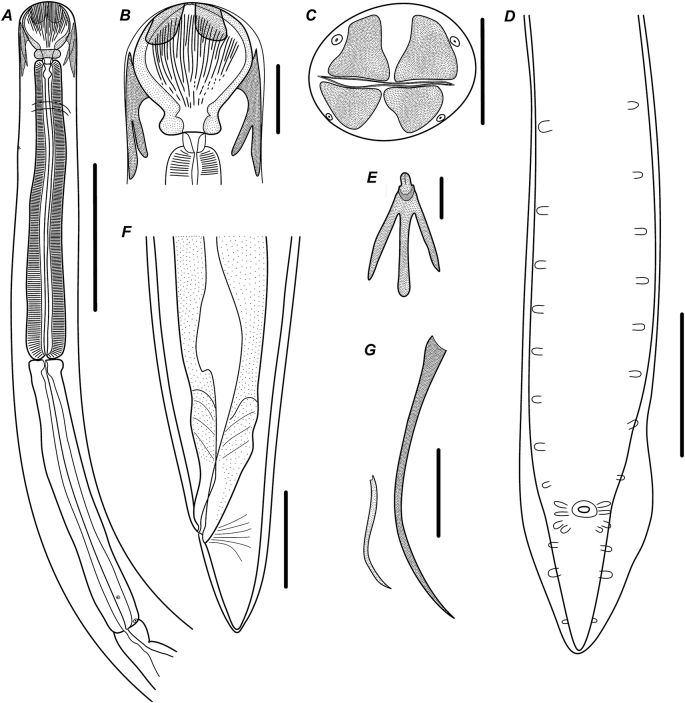
Fig. 1.
Camallanus sodwanaensis n. sp., line-drawings. A – anterior part of body, female, lateral view; B – buccal capsule, female, lateral view; C – anterior part of body, female, apical view; D – posterior part of body, male, ventral view; E – dorsal trident, male, lateral view; F – posterior part of body, female, lateral view; G – spicules, lateral view. Scale bars: A – 500; B–D, F–G – 100; E – 50.
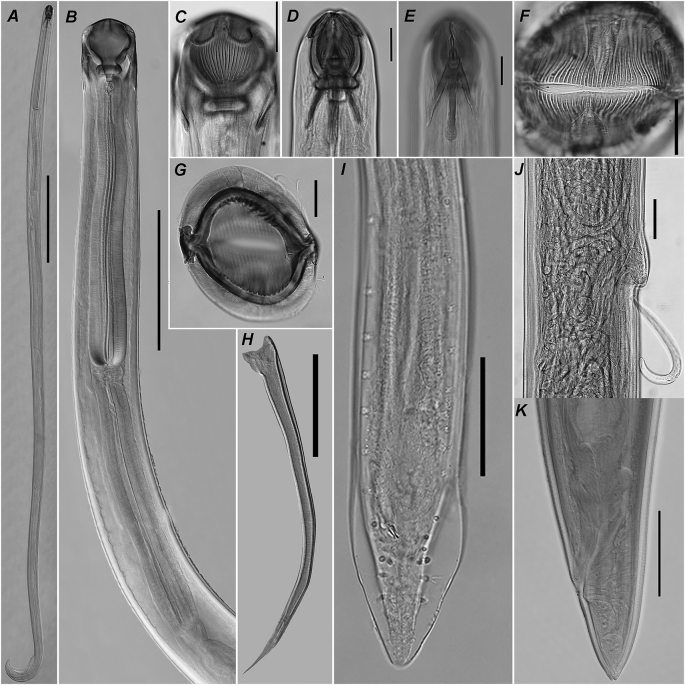
Fig. 2.
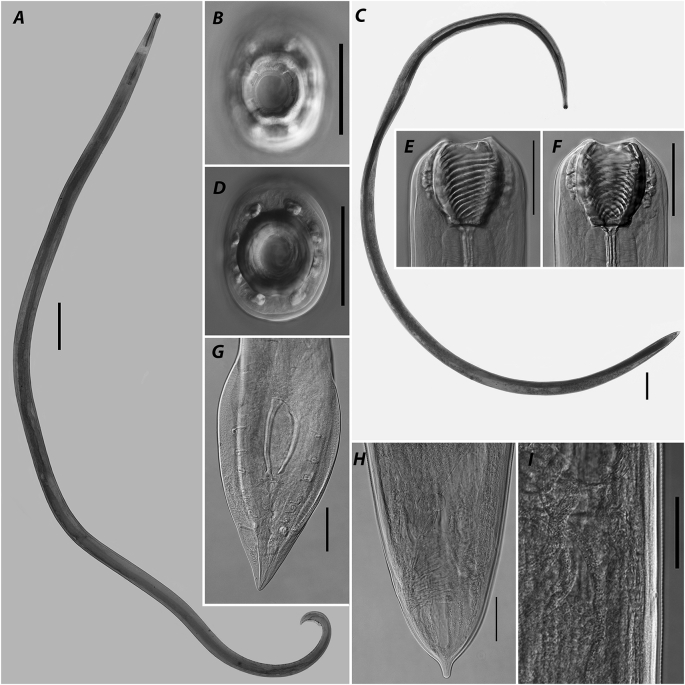
Camallanus sodwanaensis n. sp., photomicrographs. A – male, general view; B – anterior part of body, female, lateral view; C – buccal capsule, female, lateral view; D – optical section at level of buccal capsule valves mid-width, male, dorsal view; E – dorsal trident, male, dorsal view; F – anterior part of body, female, apical view; G - optical section at level of buccal capsule valves mid-length, male, apical view; H – right spicule, lateral view; I – posterior end of body, male, ventral view; J – part of body at vulva region, lateral view; K – posterior end of body, female, lateral view. Scale bars: A – 1 mm, B – 500, C–K – 100.
General. Body thin, elongated with maximum width at mid-length. Females generally larger than males. Cuticle with conspicuous transverse and fine longitudinal striations. Apical: oral opening narrow slit-like, surrounded by four conspicuous cephalic papillae (Fig. 1C; 2F). Four sclerotised plates situated on external surface of buccal capsule valves near their anterior margin. Buccal capsule with well developed valves supported by numerous ridges (Fig. 1A,B,C; 2C,D,F,G). Thick sclerotised basal ring present at base of buccal capsule. Oesophageal cup well developed. Two prominent tridents situated on ventral and dorsal sides of buccal capsule valves. Dorsal and ventral tridents equal in size and shape, each consisted of three posteriorly directed prongs. Central prong somewhat longer than sublateral ones, often reaching anterior margin of nerve ring (Fig. 1A,B,E; 2B,D,E). Muscular and glandular oesophagus almost cylindrical, slightly widening in posterior third. Nerve ring encircling oesophagus close to its anterior end. Excretory pore situated at level of nerve ring or slightly posterior to it (Fig. 1A; 2B). Deirids not observed. Intestine and rectum straight, narrow. Tail tapering with prominent phasmids situated at level of its anterior third.
Males. Measurements based on three specimens. Body 4.7–8.5 (6.6) [8.5] mm long, 112–173 (152) [173] maximum wide (Fig. 2A). Buccal capsule valves 91–101 (95) [91] long, 84–95 (92) [87] maximum wide, supported by 23–25 (25) [23] ridges, of which 9–9 (9) [9] incomplete. Basal ring 16–27 (22) [22] long, 61–69 (66) [69] wide. Oesophageal cup 13–27 (19) [16] long, 25–32 (30) [32] wide. Dorsal trident 134–160 (147) [160] long, 20–25 (23) [23] wide in lateral projection, ventral trident 131–169 (148) [160] long, 24–25 (25) [25] wide.
Muscular oesophagus 655–880 (794) [880] long, 10–14 (12) [10]% of body length; 62–79 (71) [62], 82–98 (90) [98] and 100–113 (107) [113] wide at anterior, mid-length and posterior levels, respectively. Glandular oesophagus 562–940 (767) [940] long, 11–12 (12) [12]% of body length; 67–89 (78) [89], 77–88 (83) [88] and 82–110 (96) [110] wide at anterior, mid-length and posterior levels, respectively. Nerve ring at 205–211 (207) [211], 24–31 (27) [24]% of muscular oesophagus length. Excretory pore at 210–242 (218) [242] from anterior end of body, 3–4 (4) [3]% of body length.
Caudal alae narrow, supported by papillae: eight pairs of pedunculated precloacal; two pairs of adcloacal (anterior and posterior to cloaca), five pairs of postcloacal (two pairs grouped slightly posterior to cloaca, one pair at level of tail mid-length and one close to tail end) (Fig. 1D; 2I). Spicules unequal, simple-shaped with sharpened tips (Fig. 1G; 2H). Right spicule prominent, 303–328 (313) [303] long, 4–7 (5) [4]% of body length; left one less sclerotised, poorly visible 162–205 (184) [205] long, 2–3 (3) [2]% of body length. Tail conical, tapering to rounded tip, 86–101 (94) [94] long, 1–2 (2) [2]% of body length.
Females. Measurements based on nine gravid (larvigerous) specimens. Body 6.0–11.8 (9.0) [10.9] mm long, 130–283 (222) [316] wide. Buccal capsule valves 106–173 (141) [115] long, 102–178 (141) [111] wide, supported by 19–33 (28) [28] ridges, of which 12–20 (16) [14] incomplete. Basal ring 20–30 (25) [22] long, 64–99 (83) [68] wide. Oesophageal cup 14–26 (21) [23] long, 30–40 (35) [35] wide. Dorsal trident 136–205 (171) [152] long, 20–36 (28) [20] wide in lateral projection, ventral one 134–206 (173) [151] long, 17–34 (28) [21] wide.
Muscular oesophagus 800–1173 (1053) [1087] long, 10–14 (12) [10]% of body length; 75–98 (88) [68], 89–124 (108) [112] and 104–154 (132) [113] wide at anterior, mid-length and posterior level, respectively. Glandular oesophagus 758–1152 (938) [1141] long, 9–14 (11) [10]% of body length; 86–126 (106) [63], 78–134 (111) [81] and 100–140 (123) [103] wide at anterior, mid-length and posterior level, respectively. Nerve ring at 202–287 (252) [246] from anterior end of body, 17–29 (24) [23]% of muscular oesophagus length. Excretory pore at 210–308 (263) [275], 2–4 (3) [3]% of body length. Viviparous. Vulva with distinct lips (Fig. 2J), opening posterior to small projection to body wall at 3.5–5.7 (4.8) [5.1] mm from anterior end of body, 46–58 (53) [46]% of body length. Tail 87–140 (104) [111] long, 1–2 (1) [1]% of body length. Tail tip rounded in mature females and bearing two small mucrons in immature ones (Fig. 1F; 2K).
Remarks. The species belongs to the genus Camallanus based on the presence of a well developed buccal capsule consisting of two valves, each supported by longitudinal ridges (not divided in dorsal and ventral group with a gap between), and presence of tridents on the dorsal and ventral sides of the buccal capsule valves (Anderson et al., 2009). Camallanus sodwanaensis n. sp. is the first species of the genus found in marine fish from Southern Africa. Only two species of Camallanus described from African freshwater fishes are still considered as valid and were subsequently found after the first description: C. longicaudatus and C. kirandensis. The new species can be distinguished from C. longicaudatus by the relatively smaller length of the female tail (1–2% of body length vs 12–14%) and number of postcloacal papillae in males (6 in C. longicaudatus vs 5 in C. sodwanaensis n. sp.) (Moravec, 1973). By the same characters, C. sodwanaensis n. sp. can be easily distinguised from C. kirandensis that has a comparatively long tail (868–1400 long in 8.4–20.0 mm long females, comprising approximately 7%). and only three pairs of postcloacal papillae (Baylis, 1928; Amin, 1978). Out of the species described from marine fish, C. sodwanaensis n. sp. is morphologically (size and shape of buccal capsule and tridents, morphology male spicules, general body measurements) and geographically the most closely related to C. carangis. The most reliable character to distinguish between the two species is the number of postcloacal papillae – five pairs in C. sodwanaensis n. sp. and six pairs in C. carangis (Rigby et al., 1998).
Genus Paracamallanus Yorke et Maplestone, 1926.
Paracamallanus cyathopharynx (Baylis, 1923).
Host: African sharptooth catfish Clarias gariepinus (Burchell, 1822).
Locality: Ndumo Game Reserve, KwaZulu-Natal Province, South Africa (32°30′69"E; 26°85′63"S).
Site of infection: Intestine.
Intensity: 1–8 (3.3).
Prevalence: 46% (seven of 15 infected).
Abundance: 1.5.
Representative DNA sequences: 18S [MN514775], COI [MN523683].
Description (Fig. 3).
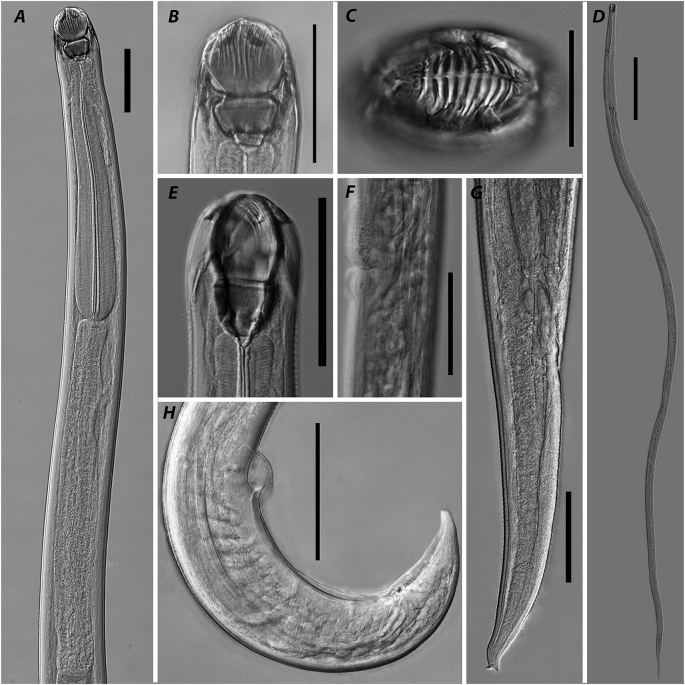
Fig. 3.
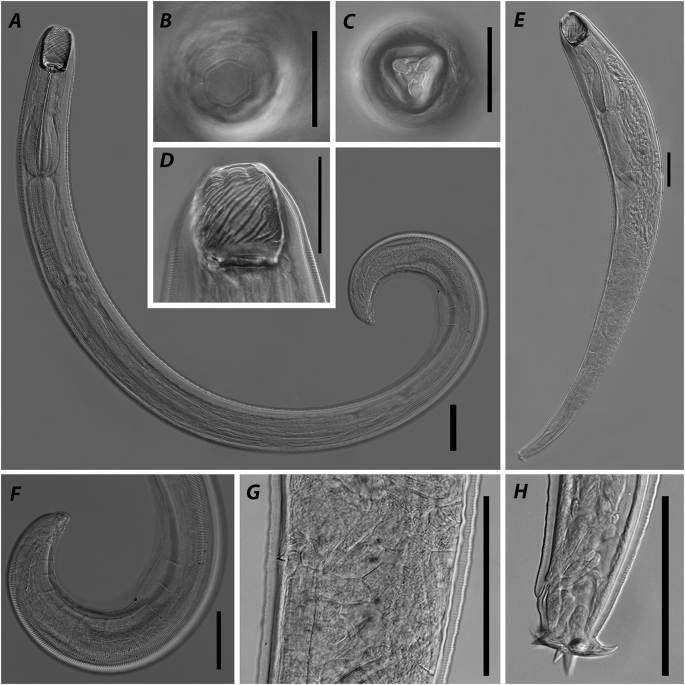
Paracamallanus cyathopgharynx, photomicrographs. A – anterior part of body, male, lateral view; B – buccal capsule, male, lateral view; C – anterior part of body, male, apical view; D – female, general view; E - optical section at level of buccal capsule valves mid-length, male, dorsal view; F - part of body at vulva region, lateral view; G – posterior end of body, female, lateral view; H – posterior end of body, male, lateral view. Scale bars: A–C, E–H – 100; D – 1 mm.
General. Medium-sized nematode, body thin with maximum width at mid-length. Cuticle with conspicuous transverse striations along entire body. Apical: oral opening slit-like, surrounded by four cephalic plates, four conspicuous outer cephalic papillae, 4 min inner cephalic papillae and two amphids (Fig. 3C). Buccal capsule well sclerotised, divided in anterior and posterior parts. Anterior part consisting of two valves, each supported by nine longitudinal ridges and two tridents on dorsal and ventral sides (Fig. 3B,E). Each trident consisted of three posteriorly directed prongs of which central one somewhat longer than sublateral. Dorsal and ventral tridents equal in size and shape, beginning at level of buccal capsule anterior quarter and ending at level of oesophageal cup. Posterior part of buccal capsule shorter and narrower than anterior one with thick well-sclerotised walls. Oesophageal cup shorter than wide, poorly sclerotised. Muscular oesophagus evenly widened from anterior to posterior part. Glandular oesophagus almost cylindrical, slightly widened in middle third. Nerve ring encircling muscular oesophagus at level of its anterior third. Excretory pore opening somewhat posterior to level of nerve ring (Fig. 3A). Intestine and rectum straight, narrow. Tail tapering.
Males. Measurements based on nine specimens. Body 1.6–6.8 (5.4) mm long, 46–124 (102) wide. Anterior part of buccal capsule 51–61 (58) long, 53–58 (55) wide. Posterior part of buccal capsule 35–41 (38) long, 38–53 (46) wide. Oesophageal cup 4–8 (6) long, 8–18 (13) wide. Dorsal trident 65–76 (71) long, 9–12 (11) wide in lateral projection, ventral one 65–77 (71) long, 9–13 (11) wide. Muscular oesophagus 204–469 (384) long, 5.8–12.8 (7.5)% of body length; 31–47 (41), 30–62 (47) and 37–73 (57) wide at anterior, mid-length and posterior level, respectively. Glandular oesophagus 210–670 (531) long, 7.8–13.2 (10.2)% of body length; 33–63 (52), 44–72 (56) and 44–73 (54) wide at anterior, mid-length and posterior level, respectively. Nerve ring at 134–165 (147) from anterior end, 32.0–65.7 (40.0)% of muscular oesophagus. Excretory pore at 149–251 (196) from anterior end, 2.6–4.9 (3.4)% of body length.
Posterior end coiled ventrally. Caudal alae narrow, supported by papillae: five pairs of precloacal pedunculated papillae, two pairs of adcloacal papillae (anterior and posterior to cloaca), six pairs of postcloacal papillae (three pairs grouped posterior to cloaca, two pairs at mid-length of tail, one pair close to tail end). Spicules unequal. Right one longer, well-sclerotised, 177–271 (223) bearing short process on its tip, 27–69 (43) long. Left spicule shorter, less sclerotised, simple-shaped with sharpened tip, 37–70 (52) long. Tail tapering with rounded tip, 59–71 (66) long (Fig. 3H).
Females. Measurements based on seven gravid specimens. Body 9.5–15.8 (11.7) mm long, 130–204 (167) wide (Fig. 3D). Anterior part of buccal capsule 71–84 (76) long, 67–84 (75) wide. Posterior part of buccal capsule 48–56 (53) long, 63–70 (66) wide. Oesophageal cup 7–10 (9) long, 12–22 (17) wide. Dorsal trident 80–112 (93) long, 11–15 (13) wide in lateral projection, ventral one 78–112 (92) long, 11–16 (14) wide. Muscular oesophagus 535–681 (581) long, 3.5–5.8 (5.1)% of body length; 48–66 (58), 54–74 (68) and 70–110 (87) wide at anterior, mid-length and posterior level, respectively. Glandular oesophagus 680–955 (793) long, 6.0–7.4 (6.8)% of body length; 61–91 (71), 70–92 (80) and 76–119 (92) wide at anterior, mid-length and posterior level, respectively. Nerve ring at 173–213 (192) from anterior end, 28.8–38.4 (33.2)% of muscular oesophagus. Excretory pore at 227–420 (275) from anterior end, 1.6–4.2 (2.4)% of body length.
Vulva with slightly elevated lips at 3.7–8.5 (6.1) mm from anterior end, 37.1–56.3 (52.1)% of body length (Fig. 3F). Tail tapering, 291–507 (362) long, bearing three small mucrons on tip (Fig. 3G).
Remarks. The species has been found in many localities throughout Africa from clariid catfishes Mwita (2011); Madanire-Moyo and Barson (2010); Ajala and Fawole (2014); Moravec and Van As (2015c); Moravec and Jirků (2017) and was reported once from Israel (Paperna, 1964). Nevertheless, the morphology of the species was illuminated only in the latest redescription provided by Moravec and Van As (2015c). In the redescription the authors described an unusual shape of the right spicule consisting of two parts: thin elongated anterior and short well sclerotised posterior that has often been confused with the gubernaculum or the left spicule. At the same time, the left spicule was described as poorly sclerotised and needle-like. In present study, we also found a clearly visible right spicule consisting of two parts and poorly sclerotised (visible only on high magnification with DIC and when dissected) left one, both with slightly wider ranges of measurement values. Also, similar to that in the latest redescription, we found eight cephalic papillae on the anterior end of nematodes. Despite that papillae of inner circle are minute and often covered with host tissue, they were clearly observed under the light microscope using high magnification and DIC.
Rigby and Rigby (2014) proposed Paracamallanus as a junior synonym of the genus Oncophora based on the similarities in their buccal capsule morphology. These authors suggested that the only difference between genera is the greater width of the female posterior to the vulva in Oncophora and assumed it as not indicative of different genera. In our opinion, significance of the characters for generic differentiation should be confirmed with sufficient molecular analyses. Therefore, in the present study we prefer to assign found species to genus Paracamallanus following Moravec and Van As (2015c) and Moravec and Scholtz (2017).
Genus Procamallanus Baylis, 1923.
Procamallanus pseudolaeviconchus Moravec et Van As, 2015.
Host: African sharptooth catfish Clarias gariepinus (Burchell, 1822).
Locality: Ndumo Game Reserve, KwaZulu-Natal Province, South Africa (32°30′69"E; 26°85′63"S).
Site of infection: Intestine.
Intensity: 1–2 (1.3).
Prevalence: 46% (seven of 15 infected).
Abundance: 0.6.
Representative DNA sequences: 18S [MN514770], 28S [MN525307], COI [MN523682].
Description (Fig. 4).
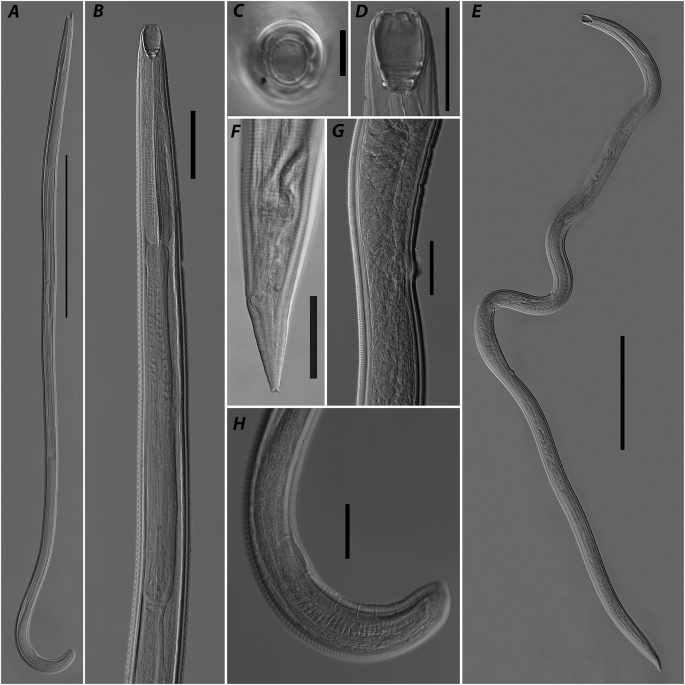
Fig. 4.
Procamallanus pseudolaeviconchus, photomicrographs. A – male, general view; B – anterior part of body, male, lateral view; C – anterior part of body, male, apical view; D – buccal capsule, female, lateral view; E – female, general view; F – posterior part of body, female, lateral view; G – part of body at vulva region, lateral view; H – posterior end of body, male, lateral view. Scale bars: A, E – 1 mm, B, D, F–H – 100; C – 25.
General. Body thin, elongated with maximum width at mid-body region. Cuticle with prominent transverse striations. Apical: oral opening rounded with unlobed peribuccal flange, surrounded by four inner submedian papillae, four outer submedian papillae and two amphids on lateral sides (Fig. 4C). Buccal capsule well sclerotised, longer than wide with two step-like folds and wide basal ring on its base (Fig. 4D). Oesophageal cup small, poorly sclerotised. Muscular oesophagus club-shaped with elongated posterior bulb. Glandular oesophagus almost two times longer than muscular one, almost cylindrical slightly widened posteriorly. Nerve ring encircling muscular oesophagus somewhat anterior to its mid-length. Excretory pore opening at level of muscular oesophagus posterior quarter (Fig. 4B). Minute deirids situated posterior to level of nerve ring. Intestine straight, narrow. Rectum straight, with thin walls. Tail tapering with rounded tip in both sexes.
Males. Measurements based on three specimens. Body 5.1–5.9 (5.5) mm long, 114–118 (116) wide (Fig. 4A). Buccal capsule 53–61 (56) long, 38–39 (39) wide. Basal ring 7–9 (8) long, 25–26 (26) wide. Oesophageal cup 7–7 (7) long, 10–12 (11) wide. Muscular oesophagus 365–388 (373) long, 6.5–7.2 (6.8)% of body length; 29–33 (31), 38–41 (40) and 48–58 (54) wide at anterior, mid-length and posterior level, respectively. Glandular oesophagus 667–824 (740) long, 13.1–13.9 (13.4)% of body length; 44–47 (45), 54–64 (58) and 51–62 (55) wide at anterior, mid-length and posterior level, respectively. Nerve ring at 173–189 (181) from anterior end of body, 47.4–49.2 (48.4)% of muscular oesophagus length. Excretory pore at 221–384 (326) from anterior end of body, 4.3–6.7 (5.8)% of body length.
Posterior end coiled ventrally with narrow caudal alae supported by papillae: nine pairs of precloacal pedunculated papillae, one pair of adcloacal papillae (anterior to cloaca) and four pairs of postcloacal papillae (Fig. 4H). Spicules unequal, simple-shaped with sharply pointed distal ends. Right spicule clearly visible, 112–126 (118) long; left one less sclerotised, 42–47 (45) long. Gubernaculum poorly sclerotised, 43 long (measured in one specimen). Tail tapering with rounded tip 46–54 (51) long.
Females. Measurements based on three gravid species. Body 4.9–9.1 (7.6) mm long, 110–186 (150) maximum width (Fig. 4E). Buccal capsule 58–67 (61) long, 54–56 (55) wide. Basal ring 9–10 (9) long, 26–35 (31) wide. Oesophageal cup 11–11 (11) long, 14–22 (18) wide. Muscular oesophagus 418–470 (448) long, 5.0–8.6 (6.3)% of body length; 27–37 (34), 32–56 (45) and 53–71 (62) wide at anterior, mid-length and posterior level, respectively. Glandular oesophagus 577–814 (711) long, 8.5–11.8 (9.8)% of body length; 37–58 (50), 53–62 (59) and 49–61 (57) wide at anterior, mid-length and posterior level, respectively. Nerve ring at 201–217 (209) from anterior end, 45.7–48.1 (46.7)% of muscular oesophagus. Excretory pore at 261–367 (315) from anterior end, 3.0–6.5 (4.5)% of body length. Vulva postequatorial, opening posterior to small projection of body wall at 3.0–6.5 (4.5) mm from anterior end, 58–63 (60)% of body length (Fig. 4G). Tail tapering with rounded tip, 89–114 (98) long, 1.0–1.8 (1.4)% of body length (Fig. 4F).
Remarks. The species was recently described by Moravec and Van As (2015a) based on material collected from the catfish Cl. gariepinus from Egypt and Botswana. The morphology and measurements of the specimens reported here from South Africa generally correspond with the original decription and the specimens represent a new geographical record.
Barson and Avenant-Oldewage (2006) reported P. leaviconchus from Cl. gariepinus in South Africa. Although, based on the provided SEM images it is clear that the peribuccal flange of the parasites is rounded, corresponding to that of P. pseudolaeviconchus (contrary to six-lobed in P. leaviconchus).
Genus Spirocamallanus Olsen, 1952.
Spirocamallanus daleneae (Boomker, 1993).
Host: Brown squeaker Synodontis zambezensis (Peters, 1852).
Locality: Ndumo Game Reserve, KwaZulu-Natal Province, South Africa (32°30′69"E; 26°85′63"S).
Site of infection: Intestine.
Intensity: 1–2 (1.6).
Prevalence: 32% (eight of 25 infected).
Abundance: 0.52.
Representative DNA sequences: 28S [MN525304], 18S [MN514771].
Description (Fig. 5).
Fig. 5.
Spirocamallanus daleneae, photomicrographs. A – male, general view; B – anterior part of body, male, apical view; C – female, genetal view; D – optical section at buccal capsule mid-length, male, apical view; E – optical sections at buccal capsule mid-width level, male, lateral view; F – optical section at buccal capsule 2/3 width level, male, lateral view; G – posterior end of body, male, ventral view; H – posterior end of body, female, lateral view; I – part of body at vulva region, lateral view. Scale bars: A, C – 1 mm; B, D–I – 100.
General. Comparatively long nematodes, body thin with maximum width at mid-length. Cuticle with conspicuous transverse striations along entire body. Apical: oral opening rounded surrounded by 6 min papillae, four inner submedian papillae, four outer submedian papillae and two amphids (Fig. 5B). Buccal capsule sclerotised, longer than wide, with 9–14 (of which anterior and posterior ones usually incomplete) spiral ridges (Fig. 5E). Basal ring short and narrow, oesophageal cup poorly developed. Buccal capsule supported by six columns each consisting of four blocks (Fig. 5D,F). Muscular oesophgus club-shaped, almost cylindrical in anterior half with elongated posterior bulb. Glandular oesophagus somewhat shorter than muscular one, almost cylindrical along whole length, slightly widening posteriorly. Nerve ring encircling muscular oesophagus at level of its mid-length. Position of excretory pore varying within level of muscular oesophagus posterior quarter. Intestine and rectum strait, narrow. Tail tapering without mucrons.
Males. Measurements based on six specimens. Body 1.6–2.0 (1.8) mm long, 246–354 (295) wide (Fig. 5A). Buccal capsule 83–107 (95) long, 77–95 (88) wide with 11–14 (13) ridges. Basal ring 9–14 (11) long, 43–55 (51) wide. Muscular oesophagus 704–737 (717) long, 3.6–4.4 (4.1)% of body length; 54–69 (64), 51–72 (62) and 84–126 (104) wide at anterior, mid-length and posterior level, respectively. Glandular oesophagus 588–690 (628) long, 3.3–3.6 (3.5)% of body length; 65–91 (82), 91–129 (107) and 77–108 (96) wide at anterior, mid-length and posterior level, respectively. Nerve ring at 348–427 (373) from anterior end, 48.6–57.9 (52.1)% of muscular oesophagus length. Excretory pore opening at 473–652 (565) from anterior end, 2.4–3.7 (3.1)% of body length. Caudal end bended ventrally with narrow caudal alae supported by papillae: three pairs pre-anal pedunculated papillae, two pairs of sessile ad-anal papillae (one anterior and one posterior to cloaca), four pairs of post-cloacal papillae (of which posterior one situated close to alae margins) (Fig. 5G). Spicules unequal, poorly sclerotised. Right spicule larger, with bifurcated tip (with one branch somewhat longer), 163–231 (205) long; left one shorter, simple-shaped with sharpened tip, 132–199 (164) long. Tail 212–271 (235), 1.0–1.3 (1.1)% of body length.
Females. Measurements based on six gravid specimens. Body 1.2–3.0 (2.1) mm long, 227–589 (383) wide (Fig. 5C). Buccal capsule 59–123 (97) long, 63–118 (93) wide, supported by 9–13 (11) ridges. Basal ring 8–14 (10) long, 40–68 (55) wide. Muscular oesophagus 605–950 (798) long, 3.2–5.4 (4.2)% of body length; 49–78 (63), 52–77 (66) and 71–142 (107) wide at anterior, mid-length and posterior level, respectively. Glandular oesophagus 427–927 (670) long, 2.8–4.0 (3.4)% of body length; 68–101 (84), 72–136 (105) and 80–113 (97) wide at anterior, mid-length and posterior level, respectively. Nerve ring at 271–479 (348) from anterior end, 37.6–50.4 (43.8)% of muscular oesophagus length. Excretory pore at 392–766 (324) from anterior end, 2.3–4.6 (3.1)% of body length. Vulva small, often poorly visible, opening around mid-body level at 5.5–17.4 (10.4) mm from anterior end, 46.9–64.3 (53.7)% of body length (Fig. 5I). Tail conical, bearing short process with rounded tip (Fig. 5H).
Remarks. The morphology of the specimens collected from the Ndumo Game Reserve corresponds to the original description of S. daleneae from the Brown squeaker Sy. zambezensis collected in South Africa's Kruger National Park (Boomker, 1993). The only difference found is that our specimens have eight columns around the buccal capsule which are not reported (probably overlooked) in the original description. Nevertheless, all other morphological (number of buccal capsule ridges, shape of tail in females, number and arrangement of papillae on male caudal region) and morphometric data, as well as host species and geographical origin, led us to assign found specimens to S. daleneae. Outside South Africa, S. daleneae has been recorded from Sy. acanthomias Boulenger, 1899 in the Central African Republic (Moravec and Jirků, 2015b) and from Sy. vanderwaali Skelton et White, 1990 in Botswana (Moravec and Van As, 2015b). The authors assigned the studied specimens to S. daleneae, but mentioned that in their material all specimens possessed a nerve ring more anterior than that in the original description (Boomker, 1993). Moreover, Moravec and Jirků (2015) described five pairs of postcloacal papillae in males contrary to the four pairs presented in the original description. These authors assumed that Boomker (1993) overlooked one pair of caudal papillae in male and the nerve ring position. Nevertheless, all specimens in our material from the type host of S. daleneae Sy. Zambezensis, from South Africa possessed a nerve encircling muscular oesophagus posterior to its mid-length and all males possessed four pairs of postcloacal papillae. In our opinion, the specimens studoed by Moravec and Van As (2015b) and Moravec and Jirků (2015) might belong to a new species while S. daleneae might be a specific parasite of Sy. zambezensis.
Moravec and Jirků (2015) and Moravec and Van As (2015b) assigned the species to the genus Procamallanus and subgenus Spirocamallanus. In the present study, we prefer to assign the species to Spirocamallanus as a separate genus due to distant phylogenetic relationships between S. daleneae and P. pseudolaeviconchus (24% (189 nt) in the 28S rDNA gene) (see Table 1).
Table 1.
Genetic divergences between different species of Camallanidae based on 28S rDNA gene alignments. Presented as percent (number of nucleotides).
| Name of species | 1 | 2 | 3 | 4 | 5 | 6 | 7 |
|---|---|---|---|---|---|---|---|
| 1. Cosmocercoides pulcher | |||||||
| 2. Spirocamallanus daleneae | 32 (253) | ||||||
| 3. Procamallanus psedolaeviconchus | 32 (254) | 24 (189) | |||||
| 4. Batrachocamallanus slomei | 32 (249) | 23 (184) | 14 (108) | ||||
| 5. Batrachocamallanus xenopodis | 32 (252) | 23 (183) | 14 (110) | 3 (23) | |||
| 6. Camallanus xenopodis | 33 (260) | 26 (201) | 20 (159) | 19 (149) | 19 (146) | ||
| 7. Camallanus kaapstaadi | 34 (263) | 25 (198) | 21 (162) | 19 (147) | 19 (146) | 4 (30) | |
| 8. Camallanus sodwanaensis n. sp. | 32 (248) | 25 (197) | 20 (157) | 20 (157) | 20 (156) | 12 (92) | 12 (96) |
Batrachocamallanus xenopodis (Baylis, 1929).
Host: Muller's platanna Xenopus muelleri (Peters, 1844).
Locality: Ndumo Game Reserve, KwaZulu-Natal Province, South Africa (32°32′34"E; 26°93′11"S).
Site of infection: Stomach.
Intensity: 1–4 (2.7).
Prevalence: 100% (three of three infected).
Abundance: 2.7.
Representative DNA sequences: 28S [MN525305], 18S [MN514768], COI [MN523681].
Description (Fig. 6).
Fig. 6.
Batrachocamallanus xenopodis, photomicrographs. A – male, general view; B – anterior part of body, male, apical view; C – optical section at base of buccal capsule level, male, apical view; D – buccal capsule, female, lateral view; E – female, general view; F – posterior part of body, male, lateral view; G – part of body at vulva region, lateral view; H – posterior part of body, female, lateral view. Scale bars: A, E, F–H – 100, B–D – 50.
General. Small nematodes, body comparatively thick with maximum width at anterior quarter. Cuticle with conspicuous transverse striations along entire body. Apical: oral opening rounded surrounded by 6 min papillae, four inner submedian papillae, four outer submedian papillae and two amphids (Fig. 6B). Buccal capsule sclerotised, longer than wide, with 12–16 (most of which incomplete) spiral ridges (Fig. 6D). Three tooth-like projections situated at base of buccal capsule (Fig. 6C). Basal ring short and narrow, oesophageal cup poorly developed. Muscular oesophgus club-shaped, almost cylindrical in anterior half with elongated posterior bulb. Glandular oesophagus as long as muscular one, almost cylindrical along whole length, slightly widening posteriorly. Nerve ring encircling muscular oesophagus somewhat anterior to its mid-length. Position of excretory pore varying within level of muscular oesophagus posterior quarter. Intestine and rectum strait, narrow. Tail tapering in males and narrowing with six mucrons in females.
Males (Fig. 6A). Posterior end coiled ventrally, caudal alae relatively long, supported by papillae: 11 precloacal pedunculated papillae, two pair of adcloacal papillae (anterior and posterior to cloaca) and four pairs of postcloacal papillae (Fig. 6F).
Females (Fig. 6E). Vulva with poorly sclerotised walls, situated at mid-body level (Fig. 6G). Tail relatively short, narrowing, bearing six mucrons (Fig. 6H).
Remarks. Due to the lack of gravid specimens in our material we could not provide measurements for this species, although all morphological characters correspond to the redescription provided by Jackson and Tinsley (1995). Moravec et al. (2006) considered the genus Batrachocamallanus as junior synonym of Procamallanus. Contrary to that opinion we prefer to assign found species to the genus Batrachocamallanus due to the distant phylogenetic relationships between B. xenopodis and P. pseudolaeviconchus (14% (110 nt) in the 28S rDNA gene) and close relationships between B. xenopodis and the type species of the genus – B. slomei (4% (30 nt) in the 28S rDNA gene) (see Table 1).
3.2. Molecular analyses
During the present study, sequences for the partial 18S and 28S rRNA genes and the mitochondrial COI gene were generated for B. xenopodis, S. daleneae and P. pseudolaeviconchus, whereas only 18S rDNA and 28S rDNA were obtained for C. sodwanaensis and only 18S rDNA and COI sequences were obtained for P. cyathopharynx. Phylogenetic analyses were performed using separate datasets according to the gene fragment amplified.
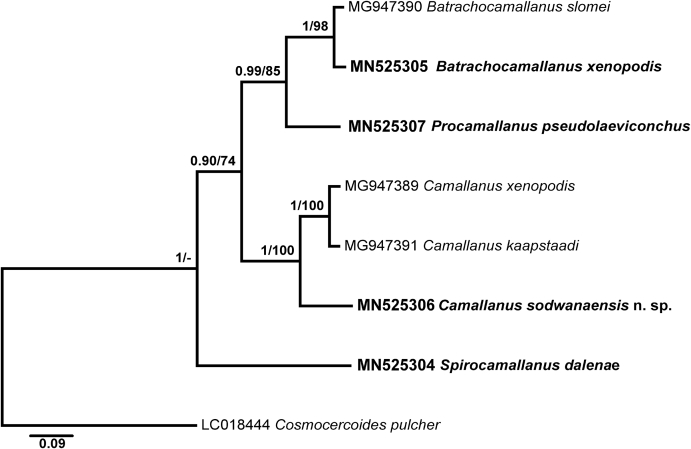
Alignment 1 of the 28S rDNA dataset comprised four newly obtained sequences, as well as two sequences for Camallanus spp. and one sequence of B. slomei retrieved from GenBank. The outgroup selected for the analyses was Cosmocercoides pulcher Wilkie, 1930 (LC018444). Bayesian inference and maximum likelihood analyses yielded similar phylogenetic topologies (Fig. 7). The novel sequence for B. xenopodis clustered with B. slomei in a highly supported clade with P. pseudolaeviconchus. The interspecific divergence between Batrachocamallanus spp. was 2.9% (23 nt) and between Batrachocamallanus spp. and P. pseudolaeviconchus it ranged from 13.8 to 14% (108–110 nt) (Table 1). The sequence of C. sodwanaensis n. sp. formed a basal branch to the clade formed by C. xenopodis and C. kaapstaadi. The genetic divergence between the new species and Camallanus spp. was 11.7–12.2% (92–96 nt) whereas differences between C. kaapstaadi and C. xenopodis was 3.8% (30 nt). The sequence for S. daleneae appeared at a basal position to the rest of the ingroup taxa.
Fig. 7.
Phylogenetic tree of Camallanidae nematodes based on 867 nucleotides long alignments of 28 rDNA gene. Nodal support presented for Bayesian Inference and Maximum Likelihood analyses (BI/ML).
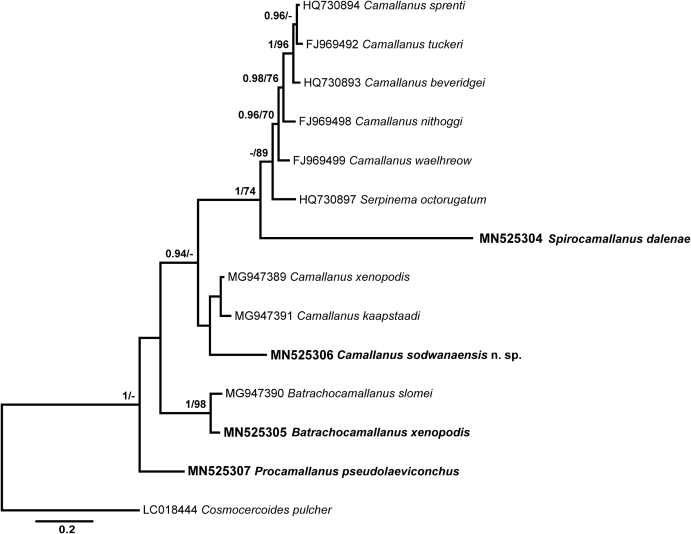
Alignment 2 of the 28S rDNA dataset comprised four newly obtained sequences and seven sequences of Camallanus spp. plus the sequences for B. slomei and Se. octorugatum retrieved from GenBank. The outgroup used in the analyses was also Co. pulcher (LC018444). Bayesian inference and maximum likelihood analyses yielded similar phylogenetic topologies (Fig. 8). The sequence for S. daleneae appeared at the basal position within the subclade consisting of Camallanus spp. and Se. octorugatum from freshwater turtles. The positions of C. sodwanaensis n. sp. and B. xenopodis were identical to the positions on the tree based on Alignment 1. The new species formed a branch close to Camallanus spp. from African frogs. The sequences for B. xenopodis and B. slomei clustered together in a strongly supported clade. Procamallanus pseudolaeviconchus, in contrast to the results obtained in the analyses based on Alignment 1, appeared at the basal position to the ingroup taxa.
Fig. 8.
Phylogenetic tree of Camallanidae nematodes based on 491 nucleotides long alignments of 28 rDNA gene. Nodal support presented for Bayesian Inference and Maximum Likelihood analyses (BI/ML).
The tree based on shorter alignments (Alignment 2) also included two well supported clades. The first clade is comprised of two larger subclades: one formed by turtle-parasitising species and another formed by C. sodwanaensis n. sp. and two species from frogs similar to the tree based on Alignment 1. The second clade includes two species of Batrachocamallanus and P. pseudolaeviconchus. The position of S. daleneae remains unresolved as it appears without nodal support separate to all species in tree based on the BI analysis and is included in the clade of turtle-parasiting species in the tree based on the ML analysis.
The phylogenetic tree based on the 18S rRNA gene obtained in the present study consists of low supported genera-level clades (Supplementary file 1). Comparative sequence analysis based on the partial 18S rRNA gene revealed the identicalness of the isolates of P. cyatopharynx collected in our study from Cl. gariepinus is South Africa and an isolate of P. cyatopharynx (DQ813445) from Werner's catfish Cl. werneri Boulenger, 1906 collected in Tanzania and confirmed the differences between P. laevionchus (JF803934) and P. pseudolaevionchus comprising 1% (7 nt), and between C. sodwanaensis n. sp. and C. carangis (DQ442664) comprising 0.5% (3 nt).
The phylogenetic tree based on the COI gene consists of low supported clades with Camallanus and Procamallanus in one subclade, thus cannot be considered as adequate for rigorous analysis (Supplementary file 2).
4. Discussion
Despite the ample morphological characters in camallanid nematodes, their application for the delineation between species and genera is still complicated. The main character used to distinguish between the genera of the Procamallaninae is the presence or absence of additional structures supporting the buccal capsule, such as spiral ridges (Spirocamallanus), small spikes (Punctocamallanus), teeth (Denticamallanus), etc. (Rigby and Rigby, 2014). However, use of these characters is complicated due to the presence of sexual dimorphism, e. g. described in P. iberingi Travassos, Artigas et Pereira, 1928, P. siluri Osmanov, 1964, P. pexatus Pinto, Fabio, Noronha et Rolas, 1976 (females with spiral ridges and males with smooth buccal capsule) and P. dentatus Moravec et Thatcher (1997) (females with spiral ridges and males with conical teeth) (see Moravec and Thatcher, 1997). Moravec and Scholz (1991), Moravec and Thatcher (1997) suggested that taxonomy based solely on the structure of the buccal capsule is more or less artificial, does not reflect phylogeny of this group, and thus needs to be revised. Therefore, these authors considered all members of the Procamallaninae as subgenera of Procamallanus “for practical reasons” (Moravec and Thatcher, 1997). At the same time, Rigby and Rigby (2014) recognized the genera that Moravec and colleagues (1991, 1997) consider to be subgenera “for the sake of tradition and simplicity”. Nevertheless, all authors agreed that only sufficient molecular studies can provide an answer to the question about the true taxonomic status and the value of the morphology of the buccal capsule for systematics.
Due to the small number of species, the present study does not support conclusions regarding the true taxonomic status of the different genera within the Camallanidae and to estimate the value of buccal capsule charachters. However, in the Procamallaninae clade, species with spiral ridges in the buccal capsule (B. xenopodis and S. daleneae) and without (B. slomei and P. pseudolaevionchus) clustered in the same clade. Absence of supported clades for Procamallanus and Spirocamallanus might be considered as evidence for the low value of buccal capsule ridges for generic differentiation. Therefore, in our opinion, division of the Procamallaninae into different genera or subgenera based on buccal capsule morphology might be equally inappropriate. However, in the present study, we prefer to follow the classification proposed by Anderson et al. (2009) recognizing most of the species in separate genera as they have been initially described. This was done due to the small number of species included in our analyses and also in order to avoid confusion in the species identification for future studies.
Other reliable characters concerned mostly the male caudal region. Several species of camallanid nematodes were described possessing only a right spicule. Although, using advanced microscopy, the inconspicuous left spicule was found in species initially described with only a right one (Moravec et al., 2006, 2016; Svitin et al., 2018). Moravec et al. (2006) also showed that the number of mucrons on the female tail can be different in larval, subgravid and gravid specimens and thus considered that this character can be used only for gravid females. In our material subgravid and even smaller larvigerous females of C. sodwanaensis n. sp. possessed two small mucrons while the largest females had rounded tail tips. Due to the high variability of some characters and inaccuracy in species descriptions, Moravec et al. (2006) stated that the most reliable character for species differentiation is the number and arrangement of caudal papillae, as none of the valid species was described with significantly varying number of caudal papillae. Despite the fact that in many descriptions phasmids were included in the number of postcloacal papillae (Moravec et al., 2016; Kuzmin et al., 2009), whereas in others the number of papillae were provided not including phasmids (Rigby et al., 1998), they can be easily found on the illustrations and text of the descriptions, thus can be compared between species. In case of C. sodwanaensis n. sp., the morphological difference between this species and C. carangis is only one pair of postcloacal papillae. Nevertheless, while most Camallanus spp. possess three anterior pairs of postcloacal papillae grouped together, C. sodwanaensis n. sp. bears only two pairs grouped, whereas the other papillae and phasmids situated similar to those of C. carangis. We agree with the opinion of Moravec et al. (2006) that the presence of one or two spicules cannot be considered as a significant character. The left spicule is often less sclerotised and poorly visible, thus might be overlooked. The left spicule of C. sodwanaensis n. sp. is almost indistinct and can be easily missed without DIC and high magnification, although it is clearly visible when dissected.
In the present study, all known species were found in the same hosts as previously reported: Pa. cyathopharynx and P. pseudolaeviconchus from Cl. gariepinus; S. daleneae from Sy. zambezensis and B. xenopodis from X. muelleri. Nonetheless, all species represent new geographical records and C. sodwanaensis n. sp. is the first Camallanus species described from marine fish in southern Africa. Unfortunately, studying the geographical distribution and host specificity of camallanid nematodes is highly complicated due to a number of species misidentifications. Therefore, in our opinion, all species records (even of well-known species) should be supported by short descriptions, illustrations and/or molecular data.
Informative phylogenetic trees were obtained only based on the partial 28S rDNA datasets. Use of partial 18S and COI sequences for analyses appeared not to be informative, probably due to the high level of conservatism of the studied fragment of 18S (630 out of 717 nt (88%) appeared to be identical for 26 species) and the variability of COI fragments (244 of 428 nt (57%) identical for nine species), respectively. In our opinion, using a combination of different nuclear (conservative) and mitochondrial (variable) genes of numerous Camallanidae species (including type species from each genus) is the only way to illuminate the real phylogenetic relationships between members of this group. Unfortunately, to date, 28S, 18S and COI sequences have been generated only for four species. Therefore, our knowledge of the phylogenetic relationships of camallanid nematodes is still at the stage of data accumulation and requires an in depth study of more species from all around the globe.
Funding
This work is based on research supported in part by the National Research Foundation (NRF) of South Africa (NRF project CPRR160429163437, grant 105979, NJ Smit, PI). This study was partially funded by the NRF-SARChI of the Department of Science and Technology (DST) (Inland Fisheries and Freshwater Ecology, Grant No. 110507). We also acknowledge use of infrastructure and equipment provided by the NRF-SAIAB Research Platforms and the funding channelled through the NRF-SAIAB Institutional Support system (M Truter). We thank Ezemvelo KZN Wildlife for research permits OP 4092/2016, OP 899/2016 and OP 1582/2018 and the Department of Agriculture, Forestry and Fisheries for permit RES2017/35. This is contribution number XXX of the North-West University (NWU) Water Research Group.
Declaration of competing interest
No conflict of interest.
Acknowledgements
The authors wish to express their sincere thanks to Dr. Ruan Gerber, Dr. Bjoern Schaeffner, Mr. Anrich Kock, Ms Coret Hoogendoorn and Ms Anneke Schoeman for their help in parasite and host species collection.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.ijppaw.2019.09.007.
Appendix A. Supplementary data
The following is the supplementary data to this article:
References
- Ajala O., Fawole O. Multiple infections of helminths in the alimentary system of Clarias gariepinus (Burchell, 1822) in a tropical reservoir. Int. J. Fish. Aquac. 2014;6:62–70. [Google Scholar]
- Anderson R.C., Chabaud A.G., Willmott S. CABI; Wallingford: 2009. Keys to the Nematode Parasites of Vertebrates: Archival Volume; p. 463. [Google Scholar]
- Amin O.M. Intestinal helminths of some nile fishes near Cairo, Egypt with redescriptions of Camallanus kirandensis Baylis, 1928 (Nematoda) and botrichocephalus aegyptiacus Rysavy and Moravec, 1975 (Cestoda) J. Parasitol. 1978;64(1):93–101. [PubMed] [Google Scholar]
- Barson M., Avenant-Oldewage A. Nematode parasites of Clarias gariepinus (Burchell, 1822) from the Rietvlei dam, South Africa. Onderstepoort J. Vet. Res. 2006;73:87–94. doi: 10.4102/ojvr.v73i2.152. [DOI] [PubMed] [Google Scholar]
- Baylis H.A. Some parasitic worms, mainly from fishes, from Lake Tanganyika. Ann. Mag. Nat. Hist. 1928;12:233–236. [Google Scholar]
- Boomker J. Parasites of South African freshwater fish. IV. Spirocamallanus daleneae n. sp. (Nematoda: Camallanidae) from Synodontis zambezensis Peters, 1852 (Mochokidae) with comments on Spirocamallanus spiralis (Baylis, 1923) Onderstepoort J. Vet. Res. 1993;60:131–137. [PubMed] [Google Scholar]
- Chaudhary A., Verma C., Tomar V., Singh H. Procamallanus spiculogubernaculus Agarwal, 1958 (Nematoda: Camallanidae) from Stinging catfish, Heteropneustes fossilis in India: morphological characterization and molecular data. Helminthologia. 2017;54:68–76. [Google Scholar]
- Černotíková E., Hor ák A., Moravec F. Phylogenetic relationships of some spirurine nematodes (Nematoda: Chromadorea: Rhabditida: Spirurina) parasitic in fishes inferred from SSU rRNA gene sequences. Folia Parasitol. 2011;58:135–148. [PubMed] [Google Scholar]
- Darriba D., Taboada G., Doallo R., posada D. jModelTest 2: more models, new heuristics and high-performance computing. Nat. Methods. 2012;9:772. doi: 10.1038/nmeth.2109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Folmer O., Black M., Hoeh W., Lutz R., Vrijenhoek R. DNA primers for amplification of mitochondrial cytochrome c oxidase subunit I from diverse metazoan invertebrates. Mol. Mar. Biol. Biotechnol. 1994;3:294–299. [PubMed] [Google Scholar]
- Guindon S., Gascuel O. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst. Biol. 2003;52:696–704. doi: 10.1080/10635150390235520. [DOI] [PubMed] [Google Scholar]
- Guindon S., Dufayard J.F., Lefort V., Anisimova M., Hordijk W., Gascuel O. New algorithms and methods to estimate Maximum-Likelihood phylogenies: assessing the performance of PhyML 3.0. Syst. Biol. 2010;59:307–321. doi: 10.1093/sysbio/syq010. [DOI] [PubMed] [Google Scholar]
- Jackson J.A., Tinsley R.C. Representatives of Batrachocamallanus n. g. (Nematoda: Procamallaninae) from Xenopus spp. (Anura: Pipidae): geographical distribution, host range and evolutionary relationships. Syst. Parasitol. 1995;31:159‒188. [Google Scholar]
- Kumar S., Stecher G., Tamura K. MEGA7: molecular evolutionary genetics analysis version 7.0. Mol. Biol. Evol. 2015 doi: 10.1093/molbev/msw054. https://www.megasoftware.net/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuzmin Y., Tkach V.V., Snyder S.D., Maier M.D. Camallanus tuckeri n. sp. (Nematoda, Camallanidae) from freshwater turtles (Pleurodira: Chelidae), in Kimberley, Western Australia. Comp. Parasitol. 2009;76:133–140. [Google Scholar]
- Kuzmin Y., Tkach V.V., Snyder S.D., Bell J.A. Camallanus Railliet et Henry, 1915 (Nematoda, Camallanidae) from Australian freshwater turtles with descriptions of two new species and molecular differentiation of known taxa. Acta Parasitol. 2011;56:213‒226. [Google Scholar]
- Lefoulon E., Bain O., Bourret J., Junker K., Guerrero R., Cañizales I., Kuzmin Y., Satoto T., Cardenas-Callirgos J., Lima S., Raccurt C., Mutafchiev Y., Gavotte L., Martin C. Shaking the tree: multi-locus sequence typing usurps current onchocercid (filarial nematode) phylogeny. PLoS Neglected Trop. Dis. 2015;9(11) doi: 10.1371/journal.pntd.0004233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madanire-Moyo G., Barson M. Diversity of metazoan parasites of the African catfish Clarias gariepinus (Burchell, 1822) as indicators of pollution in a subtropical African river system. J. Helminthol. 2010;84:216–227. doi: 10.1017/S0022149X09990563. [DOI] [PubMed] [Google Scholar]
- Moravec F. On the nematode Camallanus longicaudatus sp. n. from the Nile fish, Labeo horie Heck. Rev. Zool. Bot. Afr. 1973;87:165–173. [Google Scholar]
- Moravec F., Scholz T. Observations on some nematodes parasitic in freshwater fishes in Laos. Folia Parasitol. 1991;38:163–178. [PubMed] [Google Scholar]
- Moravec F., Sey O. Nematodes of freshwater fishes from North Vietnam. Part 1. Camallanoidea and habronematoidea. Acta Soc. Zool. Bohemoslov. 1988;52:128–148. [Google Scholar]
- Moravec F., Thatcher V. Procamallanus (Denticamallanus subgen. n.) dentatus n. sp. (Nematoda: Camallanidae) from the characid fish, Bryconops alburnoides, in the Brazilian Amazon. Parasite. 1997;4:239–243. [Google Scholar]
- Moravec F., Justine J.-L., Rigby M. Some camallanid nematodes from marine perciform fishes off New Caledonia. Folia Parasitol. 2006;53:223–239. doi: 10.14411/fp.2006.029. [DOI] [PubMed] [Google Scholar]
- Moravec F., Van As L. Procamallanus (Procamallanus) spp. (Nematoda: Camallanidae) in fishes of the Okavango river, Botswana, including the description of P. (P.) pseudolaeviconchus n. sp. parasitic in Clarias spp. (Clariidae) from Botswana and Egypt. Syst. Parasitol. 2015;90:137–149. doi: 10.1007/s11230-014-9541-0. [DOI] [PubMed] [Google Scholar]
- Moravec F., Van As L. Procamallanus (Spirocamallanus) spp. (Nematoda: Camallanidae) from fishes of the Okavango river, Botswana, including P. (S.) serranochromis n. sp. parasitic in serranochromis spp. (Cichlidae) Syst. Parasitol. 2015;90:151–164. doi: 10.1007/s11230-014-9542-z. [DOI] [PubMed] [Google Scholar]
- Moravec F., Van As L. Studies on some spirurids (Nematoda: Spiruridae) from fishes of the Okavango river, Botswana. Syst. Parasitol. 2015;91:119–138. doi: 10.1007/s11230-015-9565-0. [DOI] [PubMed] [Google Scholar]
- Moravec F., Jirků M. Two Procamallanus (Spirocamallanus) species (Nematoda: Camallanidae) from freshwater fishes in the lower Congo river. Acta Parasitol. 2015;60:226–233. doi: 10.1515/ap-2015-0032. [DOI] [PubMed] [Google Scholar]
- Moravec F., Gey D., Justine J.-L. Nematode parasites of four species of Carangoides (Osteichthyes: Carangidae) in new Caledonian waters, with a description of Philometra dispar n. sp. (Philometridae) Moravec & Thatcher 1997. Parasite. 2016;23:40. doi: 10.1051/parasite/2016049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moravec F., Jirků M. Some nematodes from freshwater fishes in central Africa. Folia Parasitol. 2017;64:033. doi: 10.14411/fp.2017.033. [DOI] [PubMed] [Google Scholar]
- Moravec F., Scholtz T. Some nematodes, including two new species, from freshwater fishes in the Sudan and Ethiopia. Folia Parasitol. 2017;64 doi: 10.14411/fp.2017.010. [DOI] [PubMed] [Google Scholar]
- Mwita C. Host-parasites relationships for Lake Victoria clariid fishes and their parasite fauna. Tanzan. J. Sci. 2011;37:179–185. [Google Scholar]
- Paperna I. The metazoan parasite fauna of israel inland water fishes. Bamidgeh. 1964;16:13–20. [Google Scholar]
- Rambaut A. University of Edinburgh, Institute of Evolutionary Biology; Edinburgh, UK: 2012. FigTree V. 1.4. Molecular Evolution, Phylogenetics and Epidemiology.http://tree.bio.ed.ac.uk/software/figtree/ [Google Scholar]
- Rigby M.C., Adamson M.L., Deardorff T.L. Camallanus carangis olsen, 1954 (Nematoda: Camallanidae) reported from French polynesia and Hawai'i with a redescription of the species. J. Parasitol. 1998;84:158–162. [PubMed] [Google Scholar]
- Rigby M., Rigby E. Order Camallanida: Superfamilies Anguillicoloidea and Camallanoidea. In: Schmidt-Rhaesa A., editor. vol. 2. Nematoda. Walter De Gruyter GmbH; Berlin/Boston: 2014. pp. 637–659. (Handbook of Zoology – Gastrotricha, Cycloneuralia and Gnathifera). [Google Scholar]
- Sardella C.J., Pereira F.B., Luque J.L. Redescription and first genetic characterisation of Procamallanus (Spirocamallanus) macaensis Vicente & Santos, 1972 (Nematoda: Camallanidae), including reevaluation of the species of Procamallanus (Spirocamallanus) from marine fishes off Brazil. Syst. Parasitol. 2017;94:657–668. doi: 10.1007/s11230-017-9728-2. [DOI] [PubMed] [Google Scholar]
- Smit N., Hadfield K. Marine fish parasitology in South Africa: history of discovery and future direction. Afr. Zool. 2015;50:79–92. [Google Scholar]
- Stromberg P., Crites J. Specialization, body volume, and geographical distribution of Camallanidae (Nematoda) Syst. Zool. 1974;23:189–201. [Google Scholar]
- Svitin R., Schoeman A., Du Preez L. New information on morphology and molecular data of camallanid nematodes parasitising Xenopus laevis (Anura: Pipidae) in South Africa. Folia Parasitol. 2018;65 doi: 10.14411/fp.2018.003. [DOI] [PubMed] [Google Scholar]
- Wu S., Wang G., Xi B., Gao D., Nie P. Molecular charachteristics of Camallanus spp. (Spirurida: Camallanidae) in fishes from China based on ITS rDNA sequences. J. Parasitol. 2008;94:731–736. doi: 10.1645/GE-1219.1. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.