Abstract
Background
Diffuse large B-cell lymphoma (DLBCL) harbors somatic hypermutation (SHM) in the immunoglobulin heavy chain and light chain variable region genes, IGHV and IGK/LV. Recent studies have revealed that IGV SHM creates neoantigens that activate T-cell responses against B-cell lymphoma.
Methods
To determine the clinical relevance of IGV SHM in DLBCL treated with standard immunochemotherapy, we performed next-generation sequencing of the immunoglobulin variable regions and complementarity determining region 3 (CDR3) for 378 patients with de novo DLBCL. The prognostic effects of IGV SHM and ongoing SHM or intra-clonal heterogeneity were analyzed in the training (192 patients), validation (186 patients), and overall DLBCL cohorts. To gain mechanistic insight, we analyzed the predicted IG-derived neoantigens’ immunogenicity potential, determined by the major histocompatibility complex-binding affinity and the frequency-of-occurrence of T cell-exposed motifs (TCEMs) in a TCEM repertoire derived from human proteome, microbiome, and pathogen databases. Furthermore, IGV SHM was correlated with molecular characteristics of DLBCL and PD-1/L1 expression in the tumor microenvironment assessed by fluorescent multiplex immunohistochemistry.
Results
SHM was commonly found in IGHV and less frequently in IGK/LV. High levels of clonal IGHV SHM (SHMhigh) were associated with prolonged overall survival in DLBCL patients, particularly those without BCL2 or MYC translocation. In contrast, long heavy chain CDR3 length, the presence of IGHV ongoing SHM in DLBCL, and high clonal IGK/LV SHM in germinal center B-cell–like (GCB)-DLBCL were associated with poor prognosis. These prognostic effects were significant in both the training and validation sets. By prediction, the SHMhigh groups harbored more potentially immune-stimulatory neoantigens with high binding affinity and rare TCEMs. PD-1/L1 expression in CD8+ T cells was significantly lower in IGHV SHMhigh than in SHMlow patients with activated B-cell–like DLBCL, whereas PD-1 expression in CD4+ T cells and PD-L1 expression in natural killer cells were higher in IGK/LV SHMhigh than in SHMlow patients with GCB-DLBCL. PD-L1/L2 (9p24.1) amplification was associated with high IGHV SHM and ongoing SHM.
Conclusions
These results show for the first time that IGV SHMhigh and ongoing SHM have prognostic effects in DLBCL and potential implications for PD-1/PD-L1 blockade and neoantigen-based immunotherapies.
Keywords: Immunoglobulin, SHM, Neoantigen, PD-1, MHC, HLA, 9p.24, BCL2, NGS, DLBCL
Background
A characteristic of mature B-cell neoplasms compared with other cancer cells is the somatic hypermutation (SHM) in genes encoding immunoglobulin (IG) heavy chain (IGH) and light chain (kappa or lambda, IGK/L) variable (V) regions. IGV SHM is acquired during antigen-based affinity maturation of activated B cells in the germinal center and mediated by activation-induced cytidine deaminase (AID) [1–4]. AID can also mediate abnormal SHM, abnormal rearrangement of D (diversity), J (joining), and V gene segments (e.g., BCL2 translocation to the IGHJ region [5, 6]), aberrant class-switch recombination (e.g., MYC translocation to the IG switch region) [5–7], and ongoing SHM in malignant B cells, implicated in the pathogenesis and evolution of B-cell neoplasms [2, 8–10].
The prognostic significance of IGV SHM has not been studied in diffuse large B-cell lymphoma (DLBCL), the most common aggressive B-cell lymphoma. In addition to the association with B-cell division and proliferation in the germinal center reaction [3] and abnormal SHM, IGV SHM may enhance the B-cell receptor (BCR) affinity and B-cell survival, suggesting unfavorable prognostic effects. Different from the tonic BCR signaling in germinal-center B-cell–like (GCB)-DLBCL [11, 12], chronic active BCR signaling [13] in activated B-cell–like (ABC)-DLBCL is driven by the self-antigen engagement of BCR and essential for B-cell survival [14]. Self-antigens can be derived from the idiotypic epitope in the BCR’s own V region and engaged with BCR [14].
On the other hand, B-cell IG-derived peptides can be processed and presented to major histocompatibility complex (MHC)-restricted CD4+/CD8+ T cells [15–18]. In mantle cell lymphoma, somatic neoantigens among all MHC-bound peptides (pMHCs) are exclusively derived from IGV and strongly biased towards MHC-II [18]. These neoantigens are mostly derived from framework region 3 (FW3) and complementarity determining region 3 (CDR3), and are created by either SHM or V-D-J recombination. In contrast, no neoantigenic pMHC were detected for somatically mutated non-IG genes, including TP53 and CCND1, despite the whole-proteomic recovery of non-neoantigenic pMHCs [18]. Similar results were found in follicular lymphoma, DLBCL, and chronic lymphoid leukemia (CLL) [19]. These results suggest that IGV SHM, but not non-IG mutations derived from aging or AID activities, has an important role in shaping the immune response against B-cell lymphomas. However, whether the positive role of IGV-derived neoantigens is significant in patients treated with immunochemotherapy and how the abundance of neoantigens affects the clinical outcome is unknown. A recent study by single-molecule imaging in live primary T cells revealed that with progressively higher pMHC densities, the set point for T-cell receptor (TCR) activation increases, and the cooperativity of pMHC:TCR binding switches from positive to negative [20]. Ii is also known that prolonged antigen exposure under suboptimal costimulatory conditions induces PD-1 expression on T cells which dampens the T-cell response [21].
Our previous in silico analysis found that IG-derived pMHCs’ T-cell exposed motifs (TCEMs), which are important determinants of the cognate interaction with the TCR, are recurrent at a wide range of frequencies in a large IGHV dataset [22]. Some TCEMs were rarely present in the TCEM repertoire built from human proteome, microbiome, and pathogenic bacteria databases [22, 23]. It is logical that T cells encountering abundant high-affinity pMHCs with germline or very common TCEMs remain in a homeostatic balance but mount an active immune response when encountering exogenous or rare TCEMs on high-affinity pMHCs.
In this study, we performed next-generation sequencing (NGS) of the IGV FW3 region and the entire CDR3 and investigated the prognostic significance of IGV SHM and ongoing SHM in 378 DLBCL patients treated with the standard immunochemotherapy regimen. In silico prediction of IG-derived pMHCs, PD-1 and PD-1-ligand 1/2 (PD-L1/2)'s cell-specific expression, BCL2/MYC/BCL6 rearrangements and mutations, and BCR signaling biomarkers were analyzed and correlated with SHM to understand the prognostic effects.
Methods
Patients
The study cohort is composed of two independent cohorts—a training set and a validation set, sequentially constructed from 21 medical centers in North America and Europe (CONSORT flow diagram in Additional file 1: Figure S1a). Included patients were diagnosed between 1999 and 2009 with de novo DLBCL according to the World Health Organization classification criteria; underwent rituximab, cyclophosphamide, doxorubicin, vincristine, and prednisone (R-CHOP) therapy; and had diagnostic biopsy specimens sufficient for NGS. Patients with transformed DLBCL, primary cutaneous DLBCL, or primary central nervous system DLBCL and HIV-positive patients were excluded. In total, 378 patients (192 training and 186 validation) were sequenced for IGH, and 269 patients also sequenced for IGK/L. The clinical features of the overall, training, and validation cohorts are in Additional file 2: Table S1. By either gene expression profiling (GEP) deposited in GSE#31312 (n = 294) or by immunohistochemistry algorithm (n = 79) [24, 25], 202 and 171 patients were classified as having GCB-DLBCL and ABC-DLBCL, respectively. Compared with GCB-DLBCL patients, ABC-DLBCL patients had significantly poorer survival (Additional file 1: Figure S1b). This study was part of the International DLBCL Rituximab-CHOP Consortium Program and conducted in accordance with the Declaration of Helsinki [24]. Material transfer agreements were established and approved by the institutional review board of each participating institution, and data collection protocols were approved as being of minimal to no risk or as exempt by the institutional review board of each participating institution.
Of the study cohort, 290 patients having a dominant clonal IG sequence identified were analyzed for prognostic impact. The median age was 63 years, the male-to-female ratio was 1.34, and the median follow-up time was 44.5 months. Molecular characteristics, including B-cell-associated gene signature [26], BCL2 and MYC translocation [27, 28], MYC and BCL6 mutation [29], and various protein expression are available for some patients, with numbers shown in Additional file 1: Figure S2.
Ultra-deep sequencing
DNA was extracted from formalin-fixed, paraffin-embedded DLBCL specimens using an Invitrogen PureLink genomic DNA kit. DNA samples that passed quantity and quality assessment were subjected to high-throughput immunosequencing of the IGH and IGK/L loci using the immunoSEQ™ platform (Adaptive Biotechnologies, Seattle, WA) [30–32]. An average of 260 ng of genomic DNA was used for each assay; the average sequencing depth of coverage was 162.08x, and the median depth of coverage was 45.57x.
For the IGH locus, a set of multiplexed forward primers matching V (CDR2/FW2) and D gene segment sequences were combined with a set of reverse primers matching J gene segment sequences to amplify both mature V-D-J and immature D-J IGH rearrangements. The reported sequence region by the immunoSEQ hsIGH assay was 130 base pairs starting from the J gene segment. The IGH CDR3 (HCDR3) sequences identified included a fraction of the V region, the complete D and J regions, and random nucleotide insertions. The average sequenced IGHV region was ~ 100 base pairs (including mostly FW3, the CDR3 V fraction, and some CDR2) covering about one-third of the IGHV gene; the median and mean HCDR3 lengths were both 48 base pairs/16 amino acids. For amplifying all possible V-D-J combinations, the assay employed a single-tube, multiplex PCR assay with 84 V and 15 D forward and 9 J reverse primers.
For the removal of potential PCR bias, every possible V-J and D-J pair was chemically synthesized as a template with specific barcodes. These templates were engineered to be recognizable as non-biologic and have universal 3′ and 5′ ends to permit amplification with universal primers and subsequent quantification by high-throughput sequencing. This synthetic immune system could then be used to calibrate the multiplex PCR assay. The multiplex pool of templates was amplified and sequenced iteratively with our IGH V/D- and J-specific primers, and the primer concentrations were adjusted to re-balance PCR amplification. Once the multiplex primer mixture amplified each V and J template nearly equivalently, residual bias was removed computationally.
A similar methodology was used for analyzing the IGK and IGL loci with the immunoSEQ hsIGKL assay, which employed 29 IGK V and 46 IGL V forward primers, plus 6 IGK J and 6 IGL J reverse primers. In addition, kappa deleting element rearrangements with the V region and the intragenic Jκ-Cκ region were also amplified. The reported sequence was ~ 130 base pairs. The median and mean lengths of light chain CDR3 were both 30 base pairs/10 amino acids.
Following high-throughput sequencing, the raw sequencing data were processed with a complexity filter and nearest neighbor algorithm to remove technical failures and correct sequencing errors. A bioinformatics pipeline clustered the sequences into distinct clonotypes based on their CDR3 sequences to determine the overall frequencies of clones. Sequences were delineated according to criteria established by the International ImMunoGeneTics (IMGT) collaboration [33] with a standard algorithm to identify V, D, and J gene segments. Sequences containing premature stop codons or out-of-frame insertions or deletions that resulted in frame shifts were classified as non-productive.
Clones that were relatively expanded with > 5% overall frequency in a sequence repertoire were identified as index trackable sequences. The dominant clones were defined as diagnostic clones representative of the malignant transformation. IGV point mutations were identified by comparing the clonal sequences with the known IMGT germline sequences and assigned as SHM events, allowing a determination of the overall SHM rate. The cutoff for SHM-positive status was > 2% deviation or < 98% identity, as used in CLL routine clinical practice and earlier studies of DLBCL [14, 34, 35].
Intra-clonal IGV variations were further analyzed in SHM-positive cases. Any sequence within the repertoire that included the same point mutations of the same germline sequence as the diagnostic sequence plus at least one additional point mutation was identified as an intra-clonal variant of the diagnostic clone. The cutoff for the presence of ongoing IGHV SHM was ≥2% accumulative frequency of intra-clonal variant sequences in the IGHV repertoire. The cutoff for high IGK/LV ongoing SHM was ≥17 intra-clonal sequence variants.
MHC-binding prediction
MHC-II binding predictions were made using neural network ensembles (NNEs) trained on MHC II binding data obtained from the IEDB repository (www.iedb.org). We used NNE methods as described previously [36] with the modification that ensembles of neural networks were used. NNE predictions of the Loge of ic50 were made for DP (13 genotypes), DQ (28 genotypes), and DR (24 genotypes). All Loge ic50 binding predictions were standardized to a common scale for all alleles using a Johnson distribution [37] to transform the raw data into zero mean, unit variance values. The threshold of high-affinity binding was set at − 1 standard deviation from the mean of the zero mean, unit variance values. This approximates the highest 16 percentiles of binding affinity. By way of reference, for the very common DRB01*0101 allele, − 1 standard deviation below the mean converts to an ic50 of approximately 50 nM.
Examining the endosomal peptidase cleavage sites indicated that a significant portion of the peptides would be expected to be excised by endosomal cathepsin B, L and S activity [22].
Frequency-of-occurrence of TCEM
MHC-II TCEMs are derived from one of two discontinuous pentamers of amino acids in the pMHC-II facing outwards and engaging the TCR [22, 38, 39]. A frequency classification (FC) metric was devised to directly index the frequency of cognate T-cell encounters of the particular TCEM, with a log base 2 transformation of the frequency-of-occurrence of 205 TCEM in approximately 50 million immunoglobulin sequences of healthy subjects [23, 40]. The scale of FC ranges from FC1 (high frequency = 1/21) to FC24 (low frequency = 1/224).
T-cell stimulation metric
For relatively rare TCEMs (FC > 16) in a high-affinity peptide, an empirical stimulation metric was computed using the principle of the additivity of variance across the entire population of allele genes [23]:
Where
and
PD-1/PD-L1/PD-L2 expression and PDL1/L2 genetic analysis
Cell type-specific expression of PD-1 and PD-L1/L2 were quantitated using the fluorescent multiplex immunohistochemistry platform MultiOmyx™; PDL1/L2 copy number alterations were evaluated by fluorescence in situ hybridization as described previously [41]. NGS RNA fusion assay was used to detect PD-L1/2 rearrangement.
Statistical analysis
Clinical and molecular features were compared using the Fisher exact test and unpaired (2-tailed) t-test. Overall survival (OS) and progression-free survival (PFS) were calculated from the date of diagnosis to the date of last follow-up or death and to the date of disease progression or death, respectively. The survival rates of two groups of patients were compared using Kaplan-Meier curves and the log-rank (Mantel-Cox) test using GraphPad Prism 7. Multivariate analyses with Cox proportional hazards regression models were performed using SPSS statistics 24. P values ≤0.05 were considered statistically significant. All comparisons were performed in the overall study cohort and the training and validation sets. The Benjamini-Hochberg procedure was performed for the multiple survival comparisons in the study cohort.
Results
High degree of clonal IGHV SHM correlates with favorable prognosis in DLBCL
IGHV index trackable sequences were identified in 224 patients, whereas no clonal sequences showed significant expansion in 65 patients, and the sequencing reads were insufficient for clonal analysis in the other 89 patients. Of the 224 patients with index trackable sequences, 145 had IMGT germline V-D-J sequences identified for diagnostic sequences (Additional file 3), whereas 79 (35%) had only reference D-J sequences resolved in IMGT (CONSORT diagram in Additional file 1: Figure S3).
The distribution of IGHD and IGHV gene usage is shown in Additional file 1: Figure S4a-b. The IGHD3 and IGHV3 families were used most frequently. Consistent with earlier studies [14, 34], IGHV4–34 was significantly overrepresented in ABC-DLBCL compared with GCB-DLBCL (Additional file 1: Figure S4c) but did not have a significant prognostic effect. The distribution of IGHV mutation degree (range, 0–20%) is shown in Additional file 1: Figure S5a; compared with ABC-DLBCL, GCB-DLBCL had a significantly higher mean mutation degree (9.6% vs 7.4%, P = 0.012). Most patients (127 of 145, 88%) were SHM-positive. The prognosis of SHM-positive and SHM-negative patients was similar.
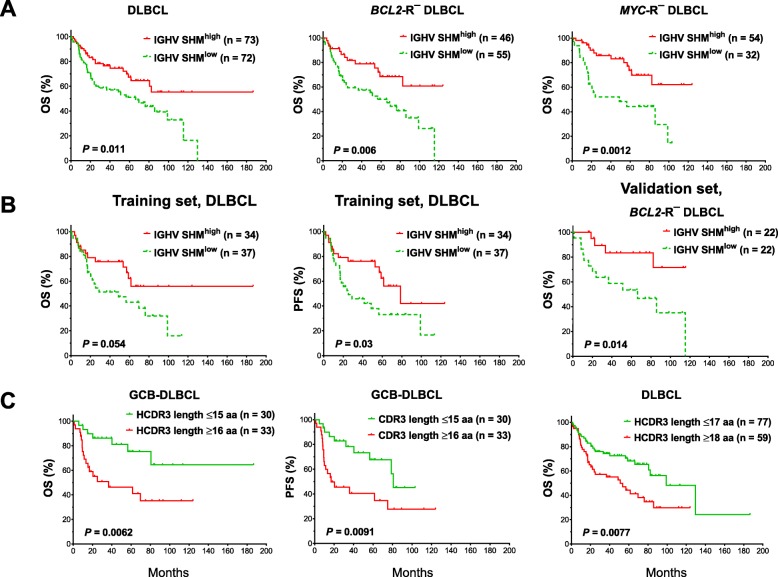
However, with the median SHM degree as the cutoff, SHMhigh was associated with significantly better OS (P = 0.011, Fig. 1a) but not PFS (P = 0.10, Additional file 1: Figure S5b). SHMhigh was associated with a significantly higher frequency of BCL2 (but not MYC) translocation (BCL2-R) in DLBCL overall (28.1%, Table 1) and in GCB-DLBCL (55%) (Additional file 2: Table S2), which may have confounded the prognostic analysis. After the exclusion of patients with BCL2-R+ DLBCL, SHMhigh was associated with significantly better OS (P = 0.006, Fig. 1a) and PFS (P = 0.012) in BCL2-R− patients. Similar favorable effects of SHMhigh were found in MYC-R− patients (for OS, P = 0.0012, Fig. 1a; for PFS, P = 0.0047). When partitioning DLBCL into GCB and ABC subtypes, the favorable prognostic effect of IGHV SHMhigh was significant in ABC-DLBCL and marginally significant in BCL2-R− and MYC-R− GCB-DLBCL (for OS, P = 0.059 and 0.066, respectively; Additional file 1: Figure S5c-d). Multivariate analysis with adjustment for clinical factors (Additional file 2: Table S2–S3) and MYC-R revealed that IGHV-SHMhigh was an independent prognostic factor for significantly longer PFS in patients with ABC-DLBCL (Additional file 2: Table S4).
Fig. 1.
Immunoglobulin heavy chain analysis. a A high degree of IGHV SHM (SHMhigh) was associated with significantly better overall survival (OS) in DLBCL overall and in DLBCL lacking BCL2 rearrangement (BCL2-R−) or MYC rearrangement (MYC-R−). b IGHV SHMhigh was associated with significantly better OS and progression-free survival (PFS) in the training set, and significantly better OS in the BCL2-R− cases of the validation set. c Short heavy chain complementarity determining region 3 (HCDR3) length was associated with significantly better OS in the germinal center B-cell-like (GCB)-DLBCL and overall DLBCL
Table 1.
Clinicopathologic and molecular characteristics of patients with DLBCL with a low or high degree of SHM in immunoglobulin variable region genes
| IGHV SHMlow | IGHV SHMhigh | P | IGK/LV SHMlow | IGK/LV SHMhigh | P | |
|---|---|---|---|---|---|---|
| n (%) | n (%) | n (%) | n (%) | |||
| Age | ||||||
| < 60 years | 32 (44%) | 25 (36%) | .31 | 66 (40%) | 14 (37%) | .72 |
| ≥ 60 years | 40 (56%) | 45 (64%) | 97 (60%) | 24 (63%) | ||
| Sex | ||||||
| Male | 35 (49%) | 41 (59%) | .24 | 94 (58%) | 26 (68%) | .27 |
| Female | 37 (51%) | 29 (41%) | 69 (42%) | 12 (32%) | ||
| Stage | ||||||
| I - II | 29 (43%) | 34 (52%) | .39 | 72 (46%) | 14 (39%) | .58 |
| III - IV | 39 (57%) | 32 (48%) | 86 (54%) | 22 (61%) | ||
| B symptoms | ||||||
| No | 38 (56%) | 43 (63%) | .48 | 103 (66%) | 28 (78%) | .23 |
| Yes | 30 (44%) | 25 (37%) | 53 (34%) | 8 (22%) | ||
| Serum LDH level | ||||||
| Normal | 21 (33%) | 29 (45%) | .20 | 67 (46%) | 18 (55%) | .44 |
| Elevated | 43 (67%) | 35 (55%) | 78 (54%) | 15 (45%) | ||
| No. of extranodal sites | ||||||
| 0 or 1 | 51 (76%) | 53 (82%) | .53 | 126 (81%) | 26 (70%) | .18 |
| ≥ 2 | 16 (24%) | 12 (18%) | 30 (19%) | 11 (30%) | ||
| ECOG performance status | ||||||
| 0 or 1 | 47 (80%) | 44 (76%) | .66 | 113 (79%) | 26 (81%) | 1.0 |
| ≥ 2 | 12 (20%) | 14 (24%) | 30 (21%) | 6 (19%) | ||
| Largest tumor size | ||||||
| < 5 cm | 37 (66%) | 27 (53%) | .17 | 73 (57%) | 13 (43%) | .22 |
| ≥ 5 cm | 19 (34%) | 24 (47%) | 55 (43%) | 17 (57%) | ||
| IPI score | ||||||
| 0–2 | 40 (60%) | 42 (64%) | .72 | 97 (62%) | 21 (57%) | .58 |
| 3–5 | 27 (40%) | 24 (36%) | 60 (38%) | 16 (43%) | ||
| Therapy response | ||||||
| CR | 51 (71%) | 54 (77%) | .45* | 126 (77%) | 27 (73%) | .67* |
| PR | 10 | 8 | 21 | 3 | ||
| SD | 2 | 3 | 4 | 1 | ||
| PD | 9 | 5 | 12 | 6 | ||
| ABC subtype/ABC subtype | ||||||
| GCB | 27 (39%) | 39 (53%) | .094 | 78 (47%) | 21 (57%) | .36 |
| ABC | 43 (61%) | 34 (47%) | 88 (53%) | 16 (43%) | ||
| BAGS classification | ||||||
| CC/CB | 24 (52%) | 40 (75%) | .02 | 77 (64%) | 21 (81%) | .11 |
| Others | 22 (48%) | 13 (25%) | 43 (36%) | 5 (19%) | ||
| BCL2 translocation | ||||||
| No | 55 (87%) | 46 (72%) | .047 | 125 (81%) | 22 (65%) | .042 |
| Yes | 8 (13%) | 18 (28%) | 29 (19%) | 12 (35%) | ||
| MYC expression | ||||||
| < 70% | 39 (57%) | 50 (72%) | .047 | 117 (72%) | 26 (68%) | .69 |
| ≥ 70% | 29 (43%) | 19 (28%) | 46 (28%) | 12 (32%) | ||
| PI3K expression | ||||||
| < 70% | 43 (67%) | 50 (75%) | .44 | 112 (72%) | 14 (42%) | .0018 |
| ≥ 70% | 21 (33%) | 17 (25%) | 43 (28%) | 19 (58%) | ||
| p63 expression | ||||||
| < 10% | 47 (68%) | 38 (54%) | .086 | 93 (59%) | 14 (39%) | .04 |
| ≥ 10% | 22 (32%) | 33 (46%) | 65 (41%) | 22 (61%) | ||
| CD30 expression | ||||||
| < 20% | 58 (83%) | 60 (83%) | 1.0 | 135 (82%) | 37 (97%) | .012 |
| ≥ 20% | 12 (17%) | 12 (17%) | 30 (18%) | 1 (3%) | ||
| p65 expression | ||||||
| < 10% | 22 (35%) | 29 (43%) | .47 | 61 (41%) | 7 (21%) | .031 |
| ≥ 10% | 40 (65%) | 39 (57%) | 89 (59%) | 27 (79%) | ||
| CXCR4 expression | ||||||
| < 20% | 43 (73%) | 47 (70%) | .84 | 109 (72%) | 19 (53%) | .029 |
| ≥ 20% | 16 (27%) | 20 (30%) | 42 (28%) | 17 (47%) | ||
Abbreviations: IGHV Immunoglobulin heavy chain variable region gene, IGK/LV Immunoglobulin kappa or lambda light chain variable region gene, SHMlow Low degree of somatic hypermutation, SHMhigh High degree of somatic hypermutation, LDH Lactate dehydrogenase, ECOG Eastern Cooperative Oncology Group, IPI International Prognostic Index, CR Complete response, PR Partial response, SD Stable disease, PD Progressive disease, GCB germinal center B-cell–like, ABC Activated B-cell–like, BAGS B-cell–associated gene signature, CB Centroblast subtype, CC Centrocyte subtype
Note: Not all patients had data available. Significant P values (Fisher’s exact test) are in bold. *For therapy response, P values were for comparisons between CR and non-CR cases
When examining in the training and validation sets separately, in the training set, IGHV SHMhigh was associated with better OS and PFS with and without the exclusion of patients with BCL2-R+ DLBCL; in the validation set, IGHV SHMhigh was associated with significantly better OS only after the exclusion of patients with BCL2-R+ DLBCL (Fig. 1b). Together, these results confirmed the favorable effects of IGHV SHMhigh in DLBCL, although the significance may differ in DLBCL subsets.
Shorter HCDR3 length correlates with favorable prognosis in DLBCL
V-D-J resolved diagnostic sequences were rarely unproductive; only 7 patients had nonsense or out-of-frame mutations. GCB-DLBCL patients with a shorter (< median/mean) amino acid length of HCDR3 (hypervariable sequences) had significantly better OS (P = 0.0062) and PFS (P = 0.0091; Fig. 1c) despite having a significantly higher proportion of stage III/IV disease (Additional file 2: Table S5). With a cutoff of 2 amino acids higher than the median/mean, short length was associated with significantly better OS (P = 0.0077; Fig. 1c) and PFS (P = 0.002) in overall DLBCL and showed a trend towards better PFS in ABC-DLBCL (P = 0.054; Additional file 1: Figure S6a). In multivariate analysis, short HCDR3 length was a favorable prognostic factor independent of clinical parameters in only GCB-DLBCL (Additional file 2: Table S4). In line with earlier findings that CDR3 shortening is associated with SHM [42], shorter HCDR3 length was associated with higher mean IGHV SHM in GCB-DLBCL, and higher IGK/LV SHM in ABC-DLBCL (Additional file 1: Figure S6b).
In both the training and validation sets, the favorable prognostic effects of short HCDR3 length were significant. The effects in ABC- and GCB-DLBCL were significant in the training and validation set, respectively (Additional file 1: Figure S6c-d).
IGHV SHMhigh is associated with increased predicted neoantigens with rare neoepitopes and lower PD-1 expression in CD8 T cells in ABC-DLBCL
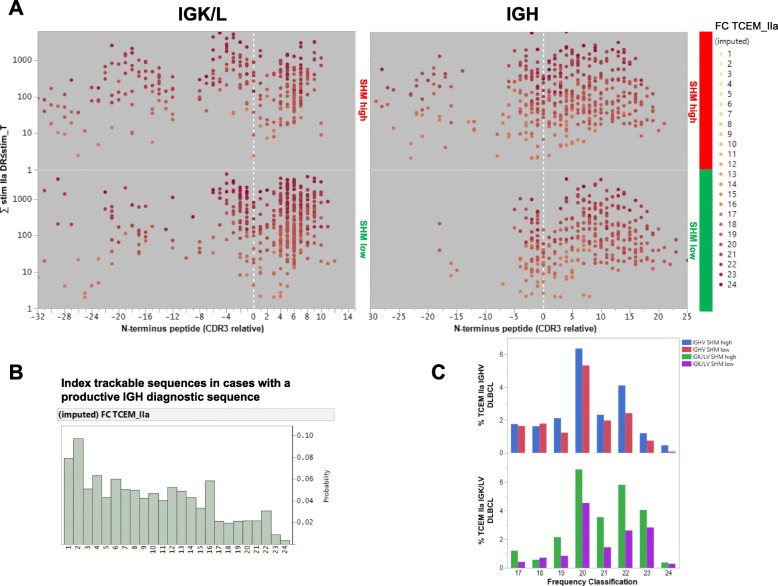
Consistent with earlier studies [18, 19], large numbers of IG-derived peptides were predicted to bind MHC-II (but not MHC-I) with high affinity in patients with a productive IGH diagnostic sequence. The IGHV-SHMhigh group Compared with the IGHV-SHMlow group had significantly more peptides with high HLA-DR-binding affinity predicted (3027 vs. 2688, ~ 16% of total peptides), with either germline (FC < 10, frequency > 1/210) or mutated TCEMs. The stimulation metric for TCEMs with an FC > 16 (relatively rare neoepitopes), which are potentially immune reactive, are plotted in Fig. 2a. These neoepitopes were a minority among patients’ TCEM repertoire identified from all index trackable sequences, as shown by the FC histogram (Fig. 2b). Compared with the IGHV-SHMlow group, the IGHV-SHMhigh group had more pMHCs with TCEM FC > 16 derived from the CDR3 (303 vs. 258) and FW3 (140 vs. 65) regions, an increased percentage of FW3 origin (4.6% vs 2.4%), and an increased percentage of rare TCEMs with an FC of 19–24 (more rare neoepitopes; Fig. 2c). A similar pattern of differences in pMHCs and neoepitopes between the SHMhigh and SHMlow groups was found in the BCL2-R−, MYC-R−, and ABC-DLBCL subcohorts as well as the training and validation sets (Additional file 1: Figure S7a-b).
Fig. 2.
Predicted MHC-binding peptides for immunoglobulin diagnostic sequences and frequency of T-cell exposed motifs (TCEMs). a Regional distribution of relatively rare neoantigens (TCEM frequency classification [FC]> > 16) derived from light chain (left) and heavy chain (right) immunoglobulin genes in DLBCL patients. Protein sequences are aligned with cysteine at the start of complementarity determining region 3 (CDR3) at the 0 of the X axis; peptides upstream of CDR3 were defined as framework region 3 (FW3). The stimulation metric was computed using the principle of the additivity of variance and is a product of the standardized MHC-II-binding affinity multiplied by the FC summed over all HLA-DR alleles. Each dot represents one peptide predicted as having high MHC-II-binding affinity (exceeding the − 1 standard deviation threshold for MHC derived from 24 HLA-DR alleles) and relatively rare TCEMs (FC > 16). The color intensities of the dots are scaled on the FC scale, which ranges from FC16 to the very rare FC24. b Histograms showing the distribution of the FC of the TCEMs in all MHC-II-binding peptides predicted for index trackable sequences. The FC scale ranges from the commonly presented FC1 to the very rare FC24. c Compared with cases without a high degree of heavy chain or light chain IGV SHM, cases with high degree of heavy chain or light chain IGV SHM had higher frequencies of relatively rare TCEMs (FC > 16)
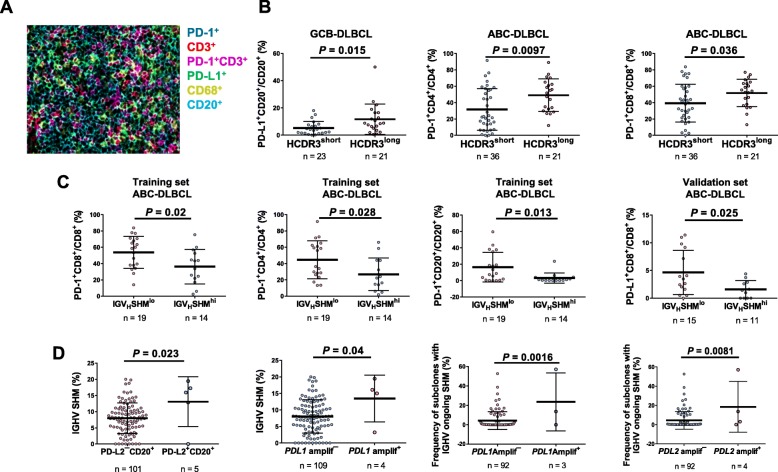
To gain insight into the immune surveillance in the tumor microenvironment, fluorescent mIHC was performed to evaluate immune cell-infiltration and cell-specific PD-1/L1/L2 expression (representative image in Fig. 3a) [41], correlating with IGHV SHM and CDR3 length. Long HCDR3 length was associated with higher PD-L1 expression in B cells in GCB-DLBCL (Fig. 3b; significant in the training set; marginally significant in the validation set) and higher PD-1 expression in CD4+/CD8+ T cells in ABC-DLBCL (Fig. 3b; significant in the validation set; strong trends in the training set). In ABC-DLBCL, IGHV-SHMhigh was associated with significantly lower PD-1 expression in T cells and B cells in the overall cohort and the training set, and significantly lower PD-L1 expression in CD8+ T cells in the overall cohort and the validation set (Fig. 3c). In the overall ABC-DLBCL cohort, IGHV SHMhigh cases compared with SHMlow cases had significantly lower mean cellularity of CD4+ T cells but similar cellularity of CD8+ T cells (Additional file 1: Figure S7c). B-cell PD-L2 expression and PD-L1/PD-L2 gene amplification (very low frequency in the study cohort, predominantly found in ABC-DLBCL) were associated with high IGHV SHM (Fig. 3d).
Fig. 3.
Comparison of PD-1 expression between groups. a A representative image of a DLBCL sample is from an ABC-DLBCL case with a low degree of IGHV SHM (2.94%) and a long (21 amino acids) heavy chain complementarity determining region 3 (HCDR3). Fluorescence multiplex immunohistochemistry detected that PD-1 was expressed in T cells and proximal to PD-L1-expressing B cells. b Long HCDR3 length was associated with high PD-L1 expression in B cells in GCB-DLBCL and high PD-1 expression in CD4+/CD8+ T cells in ABC-DLBCL. c In the training set, a high degree of IGHV SHM (SHMhi) was associated with low PD-1 expression in CD8+/CD4+ T cells and B cells in ABC-DLBCL. In the validation set, IGHV SHMhi was associated with lower PD-L1 expression in CD8+ T cells. d PD-L2 protein expression in B cells was associated with a high degree of IGHV SHM. PD-L1 gene amplification was associated with a significantly higher mean degree of SHM in the IGHV diagnostic sequence. PD-L1/L2 gene amplification was associated with a higher mean percentage of subclones with IGHV ongoing SHM in the sequence repertoire
Together, these findings suggest that the IGHV-SHMhigh group produced more T-cell stimulatory neoantigens, which may be relevant for PD-1 expression regulation and function of cognate T cells.
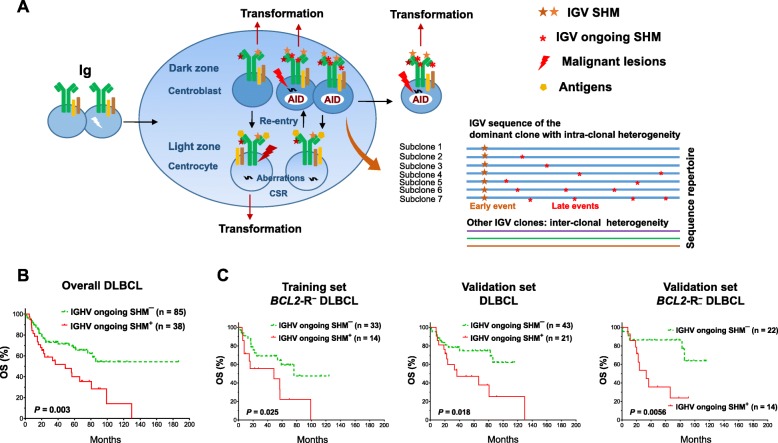
Ongoing IGHV SHM correlates with significantly poorer survival in DLBCL
Intra-clonal sequence variations (Fig. 4a) were identified in 102 (83%) of the productive IGHV SHM-positive cases (most frequently in the IGHV3 and IGHV4 families; Additional file 1: Figure S8a). With a cutoff of subclonal frequency at the 70th percentile, ongoing IGHV SHM was associated with significantly poorer OS in patients with DLBCL in the univariate analysis (P = 0.003; Fig. 4b) and poorer OS and PFS in the multivariate analysis (Additional file 2: Table S4). The adverse prognostic effect was significant regardless of GCB/ABC and MYC-R status and was significant in BCL2-R− (for OS, P = 0.007, for PFS, P = 0.01) but not BCL2-R+ patients. Similar prognostic results were found in both the training and validation cohorts (Fig. 4c).
Fig. 4.
Prognostic analysis for IGHV ongoing SHM. a Schematic illustration of the putative pathologic origins of IGV SHM and ongoing SHM in DLBCL founder clones and subclones. Transformation can occur in different stages of B-cell development. When DLBCL abnormalities are sufficient to drive lymphomagenesis, DLBCL cells exit the germinal center reaction. Predominant DLBCL clones may exhibit intra-clonal IGV variations conferred by the ongoing SHM process. b IGHV ongoing SHM was associated with significantly poorer overall survival (OS) in the overall study cohort. c IGHV ongoing SHM was associated with poorer OS in the overall validation cohort and in cases without BCL2 rearrangement (BCL2-R−) in both the training and validation sets
Ongoing IGHV SHM was associated with AICDA upregulation in overall DLBCL and the validation set. PD-L1/PD-L2 gene amplification and macrophage PD-L2 expression were associated with higher ongoing SHM (Fig. 3d, Additional file 1: Figure S8b).
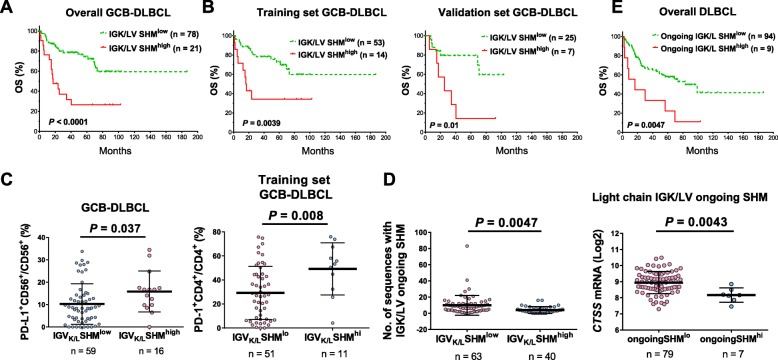
IGK/LV SHMhigh correlates with significantly poorer survival in patients with GCB-DLBCL
Light chain diagnostic sequences were identified in 205 (76%) DLBCL patients (CONSORT diagram in Additional file 1: Figure S3). Consistent with the order of rearrangement, IGL clones were seen only in patients with unproductive IGK. No prognostic difference was observed between the kappa and lambda types. Compared with IGHV, IGK/LV had significantly fewer mutations. The frequency of IGK/LV SHM-positive cases was 53.6% (105 of 205). There were many more IGK clones with no SHM than IGH or IGL clones with no SHM (Additional file 1: Figure S8c). IGLV-SHM had higher correlation with IGHV-SHM than IGKV-SHM (Additional file 1: Figure S8d).
IGK/LV SHM-positive status was not associated with prognostic effect. However, with a high cutoff close to the 80th percentile, IGK/LV SHMhigh was associated with significantly poorer OS and PFS in patients with GCB-DLBCL (P < 0.0001 for OS, Fig. 5a; P = 0.0016 for PFS); the effects were confirmed in both the training and validation cohorts (Fig. 5b, Additional file 1: Figure S9a) and by multivariate analysis (Additional file 2: Table S4). Like IGHV SHMhigh, IGK/LV SHMhigh was associated with a higher frequency of BCL2-R in DLBCL (35%, Table 1). However, the adverse prognostic effect of IGK/LV SHMhigh was independent of BCL2-R and MYC-R status and was strongest in BCL2-R+ GCB-DLBCL (Additional file 1: Figure S9b-c).
Fig. 5.
Prognostic and correlative analyses for light chain IGK/LV SHM. a A high degree of IGK/LV SHM (SHMhigh) was associated with significantly worse overall survival (OS) in GCB-DLBCL. b The adverse prognostic effect of IGK/LV SHMhigh in GCB-DLBCL was significant in both the training and validation sets. c IGK/LV SHMhigh was associated with higher PD-L1 expression in CD56+ natural killer cells in overall GCB-DLBCL cases and with high PD-1 expression in CD4+ T cells in the training set. d There was a negative correlation between light chain IGK/LV ongoing SHM and IGK/LV SHM. High IGK/LV ongoing SHM was associated with low CTSS mRNA expression. e High numbers (≥17) of subclones with IGK/LV ongoing SHM were associated with significantly poorer OS in DLBCL
A short K/LCDR3 length (≤12 aa) was associated with significantly better OS in DLBCL overall and in ABC-DLBCL (P = 0.026 and 0.012, respectively; Additional file 1: Figure S9d). However, the prognostic effect was only significant in the validation set (P = 0.015; it showed a nonsignificant trend in the training set of ABC-DLBCL, P = 0.15), and the number of cases with long K/LCDR3 length was small (4 and 3 in the training and validation sets, respectively).
IGK/LV SHMhigh is associated with increased rare neoepitopes and PD-1 expression on CD4+ T cells in GCB-DLBCL
The T-cell stimulation metric for predicted MHC-II neoantigens derived from productive IGK/L diagnostic sequences is shown in Fig. 2a. Because the IGK/L SHMhigh and SHMlow groups had unbalanced numbers of patients, the groups’ mean numbers of predicted pMHC-II were compared. IGK/LV SHMhigh patients had a larger mean number (8.4 vs 4.5 per patient) and percentage (FW3-origin, 10% vs 2.7%; CDR3-origin, 9.1% vs 7.2%) of predicted pMHC-II with FC > 16 TCEMs, but not total predicted pMHC-II (44 vs 46 per patient). The association of IGK/L SHMhigh with more pMHC-II with FC > 16 TCEMs per patient was observed in both the training and validation sets.
Compared with IGK/LV SHMlow patients, IGK/LV SHMhigh patients had significantly higher PD-L1 expression in natural killer cells (P = 0.037; Fig. 5c) and higher CTSL1 (lysosomal protease genes cathepsin L [43]) mRNA expression in GCB-LDBCL (P = 0.038; Additional file 1: Figure S9e), but significantly lower B-cell PD-1 expression (P = 0.03) in ABC-DLBCL (Additional file 1: Figure S9f). In contrast, IGHV SHMhigh was associated with lower CTSF expression in GCB-DLBCL (P = 0.048; Additional file 1: Figure S9e). In the training but not the validation set, IGK/LV SHMhigh patients had higher PD-1 expression in CD4+ T cells in GCB-DLBCL (P = 0.008, Fig. 5c) and higher AICDA mRNA in ABC-DLBCL (P = 0.047).
Because the correlation findings were differential in the training/validation sets and in the GCB/ABC subtypes, these subsets/subtypes were compared. Compared with the validation set, the training set had significantly higher mean mRNA levels of several MHC-II genes (HLA-DPA1, HLA-DPB1, HLA-DRA, HLA-DRB1/4) and lysosomal protease genes (CTSH, ASNS, and GILT) (expression data were extracted from the GEP #31312 deposit; Additional file 1: Figure S10a). These differences were largely attributable to the validation set’s MYC-R+ cases (Additional file 1: Figure S10b), and there were no significant expression differences (except for CTSH) between validation set’s MYC-R− cases and the training set. In both the training and validation sets, MYC-R was associated with downregulation of HLA-F, CTSH, and CTSK in DLBCL and GCB-DLBCL.
In both the training and validation sets, ABC-DLBCL compared with GCB-DLBCL had higher macrophage and CD8+ T-cell infiltration, higher PD-L1+ expression in B cells (Additional file 1: Figure S10c for the overall cohort), higher HLA-C/E, CTSZ, and CTSC mRNA, and lower HLA-DQB2, HLA-DRB4, and CTSK mRNA expression. In the training set only, ABC compared with GCB subtype had significantly higher CTSB, CTSL1, and CTSS expression, and in the validation set only, significantly higher CTSL3 expression and lower CTSF Expression.
High intra-clonal IGK/LV diversity is associated with unfavorable prognosis
Of the 103 productive IGK/LV SHM-positive cases, 91 (88%) had intra-clonal IGK/L variants (ongoing SHM). The numbers of sequences with ongoing IGK/LV SHM showed negative association with IGV SHM (Fig. 5d, Additional file 1: Figure S11a) and CTSS (a cathepsin with an essential role in proteolytic processing of MHC class II-associated invariant-chain peoptide fragments [43]) mRNA levels (Fig. 5d). PD-L1 polyploidy, exclusively found in GCB-DLBCL, was associated with ongoing IGK/LV SHM (Additional file 2: Table S6).
High intra-clonal IGK/L diversity (≥17 subclones), present in only 9 patients (8 were GCB-DLBCL), was associated with unfavorable clinical parameters, significantly poorer OS/PFS, and distinct gene signatures in DLBCL and GCB-DLBCL (Fig. 5e, Additional file 1: Figure S11b-c, Additional file 2: Table S6–S7). However, the prognostic effects were significant only in the training set (Additional file 1: Figure S11d) and not significant in the multivariate analysis.
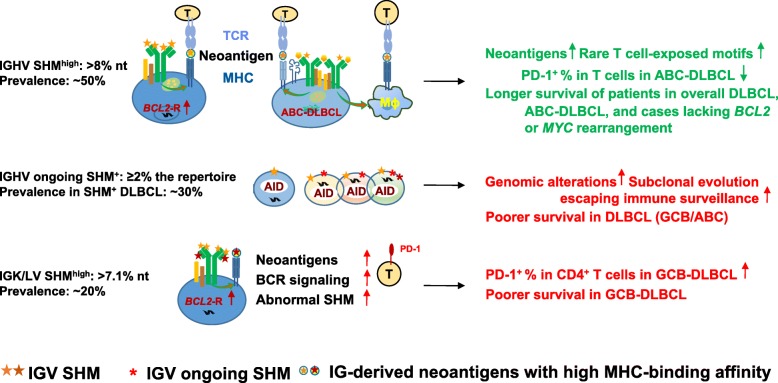
Multiple comparison correction was performed (Additional file 2: Table S8) and the validated prognostic effects with potential underlying mechanisms are illustrated in Fig. 6.
Fig. 6.
Schematic summary of the prognostic effects of IGV clonal SHM and ongoing SHM in DLBCL and putative underlying mechanisms suggested by in silico analysis and fluorescent multiplex immunohistochemistry and conventional chromogenic immunohistochemistry experiments. Abbreviations: Ig, immunoglobulin protein; AID, activation-induced cytidine deaminase; CSR, class-switch recombination; TCR, T-cell receptor; MHC, major histocompatibility complex; BCR, B-cell receptor; Mɸ, macrophage
Discussion
IGV SHM, which is distinguished from scattered genome-wide aging-associated non-IG somatic mutations by high mutation density and protein expression [44], has an essential role in neoantigen presentation [18, 44]. However, the clinical relevance of IGV SHM is less studied than that of non-IG mutations, likely owing to technical and interpretive difficulties. In this study, IGV SHMhigh and ongoing SHM identified through NGS showed prognostic significance in a large cohort of patients with de novo DLBCL treated with R-CHOP, which was validated in the training and validation sets.
First, IGHV SHMhigh was associated with significantly longer OS in DLBCL patients and longer OS and PFS in DLBCL patients without MYC/BCL2 translocations, which is reminiscent of the favorable PFS and OS incrementally associated with IGHV% deviation in CLL patients [45]. Consistent with the favorable prognostic effect, IGHV-SHMhigh patients had more enriched MHC-II neoantigens with rare neoepitopes by in silico prediction [22] but lower T-cell PD-1 expression in ABC-DLBCL. The implications of IGHV SHM for T-cell response activation and regulation warrants future study for functional validation and therapeutic exploration. A study showed that treatment with CpG, a TLR9 agonist, promoted MHC-II presentation of IG-derived neoantigens of mantle cell lymphoma cells [19].
Second, compared with IGHV, IGK/LV had less SHM, but IGK/LV SHMhigh was associated with significantly poorer OS and PFS and high PD-1 expression in CD4+ T cells and PD-L1 in natural killer cells in GCB-DLBCL, even though FW3-derived MHC-II neoantigens with rare neoepitopes were significantly higher in IGK/LV SHMhigh DLBCL compared with IGHV SHMhigh DLBCL (4.4 vs 2 per patient) and IGH/K/LV SHMlow DLBCL (1 per patient). These results appeared to suggest that the excessive neoantigens in IGK/LV SHMhigh patients with GCB-DLBCL had a negative role in T-cell response by inducing PD-1. In addition, IGK/LV SHMhigh in GCB-DLBCL could be a biomarker for stronger BCR affinity and higher B-cell proliferation propensity [3, 14], therefore synergizing with unfavorable BCL2-R which enhanced cell survival. This is supported by the mutually exclusive pattern of IGK/LV SHMhigh and IGK/LV ongoing SHMhigh, suggesting a survival advantage of the expanded IGK/LV-SHMhigh clone, leading to intra-clonal homogeneity.
Third, the presence of IGHV ongoing SHM or intra-clonal heterogeneity had an adverse prognostic effect in SHM-positive patients. Whether the adverse prognosis resulted from subclonal evolution, such as the selection of clones with less immunogenicity [46], loss of MHC expression, or enhanced cell survival, could be revealed by collecting serial tumor biopsy specimens during and after therapy in future prospective studies and subjecting them to longitudinal NGS and flow cytometry experiments to monitor the clonal evolution. The higher ongoing SHM in DLBCL patients than in CLL patients and its adverse prognostic effect in IGHV SHM-positive case, may explain why SHM-positivity status lacks a favorable prognostic effect in DLBCL but not CLL [45, 47].
In addition, as chromosome 9p24.1 amplification has been correlated with the efficacy of PD-1 blockade in Hodgkin lymphoma [48], it would be interesting to investigate the biomarker value of IGHV SHMhigh and IGV ongoing SHM for clinical response to PD-1 blockade immunotherapy in DLBCL, which showed associations with 9p24.1 amplification and PD-1 expression in the current study. In melanoma patients treated with anti-PD-1 immunotherapy, high tumor clonal mutation load was associated with improved overall survival and higher TCR-clonality (less diverse repertoire) predicted response to anti-PD-1 immunotherapy [49, 50].
Conclusions
In summary, clonal IGHV SHMhigh had favorable prognostic effect in patients with DLBCL without BCL2/MYC translocation, whereas IGHV ongoing SHM and clonal IGK/LV SHMhigh had adverse prognostic effects in DLBCL and GCB-DLBCL patients, respectively. Neoantigen loads, PD-1/PD-L1 immune checkpoint, and BCR affinity and signaling may contribute to these prognostic effects. IGV SHM evaluation has implications for the selection of PD-1/PD-L1 inhibitors, BCR-targeted agents, and effective vaccines in DLBCL patients. Because NGS is available in clinical practice, the application of IG NGS with immunoSEQ is feasible and can improve risk stratification at diagnosis and identification of dominant tumor clones in lymphoma. Future studies are warranted to determine the value of IG NGS in tracking resistant clones expanded at relapse and in indicating response to immunotherapy and to investigate the therapeutic potential of IG-based vaccines and how IG-derived neoantigens shape the immune response.
Supplementary information
Additional file 1: Fig S1.. Construction and clinical outcome of the diffuse large B-cell lymphoma (DLBCL) cohort. Fig. S2. Diagram showing numbers of cases in this mutation study that have been characterized by various biomarker studies, and survival rates of patients whose sequencing results were correlated with prognosis. Fig. S3. CONSORT flow diagram illustrating the number of cases performed for high-throughput IG sequencing and clonal sequence analysis. Fig. S4. Molecular characterization for immunoglobulin heavy chain (IGH) gene usage in the study cohort. Fig. S5. Immunoglobulin heavy chain V gene (IGHV) somatic hypermutation (SHM) analysis. Fig. S6. Analysis for length of heavy chain CDR3. Fig. S7. Prediction of MHC-binding peptides and frequency of T-cell exposed motifs (TCEM) for immunoglobulin diagnostic sequences in the training set and validation set. (a) Regional distribution of relatively rare neoantigens derived from heavy chain and light chain immunoglobulin genes in DLBCL patients in the training set (top) and validation set (bottom). (b) Cases with high degree of heavy chain or light chain IGV SHM compared with cases without had higher frequency of relatively rare TCEM in the training (left) and validation sets (right). (c) In ABC-DLBCL, high IGV SHM was associated with lower tissue cellularity of CD4+ T cells. Fig. S8. Moleclar analysis for immunoglobulin heavy chain ongoing SHM and light chain SHM. Fig. S9. Immunoglobulin light chain SHM and CDR3 analysis. Fig S10. Comparison between different subsets of DLBCL. Fig S11. Light chain IGK/LV ongoing SHM analysis.
Additional file 2: Table S1. Clinical features of 378 patients in the training and validation cohort whose DLBCL biopsies were sequenced and 290 patients whose sequencing results showed sufficient sequence reads. Table S2. Comparisons of clinicopathologic and molecular characteristics between patients with germinal-center B-cell–like (GCB) DLBCL with a low or high degree of somatic hypermutation (SHM) in immunoglobulin variable region genes. Table S3. Comparisons of clinicopathologic and molecular characteristics between patients with activated B-cell-like (ABC) subtype of DLBCL with a low or high degree of SHM in immunoglobulin variable region genes. Table S4. Significant prognostic effects of immunoglobulin molecular characteristics in DLBCL patients treated with R-CHOP by multivariate survival analysis. Table S5. Clinicopathologic and molecular characteristics of patients with DLBCL with a short or long immunoglobulin heavy/light chain CDR3 length. Table S6. Clinicopathologic and molecular characteristics of patients with DLBCL with ongoing SHM in immunoglobulin variable region genes. Table S7. Gene signatures associated with SHM in immunoglobulin sequences of DLBCL samples. Table S8. Multiple testing corrections for prognostic effects found in the overall cohort of DLBCL treated with R-CHOP by the Benjamini-Hochberg method with a false discovery rate of 0.10
Additional file 3. Diagnostic immunoglobulin heavy chain gene sequences
Acknowledgments
We thank Joseph A. Munch from MD Anderson’s Department of Scientific Publications for providing editorial assistance during the preparation of this manuscript.
Abbreviations
- ABC
Activated B-cell–like
- AID
Activation-induced cytidine deaminase
- BCR
B-cell receptor
- CDR
Complementarity determining region
- CLL
Chronic lymphoid leukemia
- CSR
Class-switch recombination
- D
Diversity
- DLBCL
Diffuse large B-cell lymphoma
- FC
Frequency classification
- FW3
Framework region 3
- GCB
Germinal-center B-cell–like
- GEP
Gene expression profiling
- HCDR3
Heavy chain CDR3
- HLA
Histocompatibility antigen
- IG
Immunoglobulin
- IGH
Immunoglobulin heavy chain
- IGK/L
Immunoglobulin kappa or lambda light chain
- IGV
Immunoglobulin variable region gene
- IMGT
International ImMunoGeneTics Information System
- J
Joining
- MHC
Major histocompatibility complex
- mIHC
Multiplex immunohistochemistry
- MYC/BCL2-R
MYC/BCL2 translocation
- NGS
Next-generation sequencing
- NNE
Network ensembles
- OS
Overall survival
- PD-1
Programmed cell death protein 1
- PD-L1
PD-1-ligand 1
- PFS
Progression-free survival
- pMHC
MHC-bound peptide
- SHM
Somatic hypermutation
- TCEM
T-cell exposed motif
- TCR
T-cell receptor
- Th2
Type 2 helper T cells
- TLR9
Toll-like receptor 9
Authors’ contributions
ZYX-M, BC, RDB, TS, IK, and KHY designed the study, conducted the research, and performed the analysis. ZYX-M, JL, YX, BC, RDB, YM, MX, TS, GCM, XT, HZ, CV, AT, KD, GB, WT, HY, EDH, JHvK, JH, MP, AJMF, MBM, MAP, JNW, BX, YL, IK and KHY collected clinical and follow-up data with the approval of the institutional review boards and the material transfer agreement or contributed vital new reagents, resources, technology, and/or analytical tools. ZYX-M, JL, YX, BC, RDB, TS, AT, JHvK, BX, IK, and KHY wrote or edited the manuscript. All authors contributed vital strategies, participated in discussions, provided scientific input, and proved the manuscript. All authors read and approved the final manuscript.
Funding
The study is supported by NIH/National Cancer Institute (grants R01CA233490 [to KHY], R01CA138688, R01CA187415 and 1RC1CA146299 [to KHY and YL]), The University of Texas MD Anderson Cancer Center Institutional Research and Development Fund, the Gundersen Lutheran Medical Foundation, the Hagemeister Lymphoma Foundation, and the University Cancer Foundation via the Sister Institution Network Fund at The University of Texas MD Anderson Cancer Center. The work of the authors is also partially supported by NIH/National Cancer Institute grants P50CA136411 and P50CA142509, and by the MD Anderson Cancer Center Support Grant CA016672.
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request based on the condition that IRB and MTA could be approved from the institutions.
Ethics approval and consent to participate
The study was conducted in accordance with the Declaration of Helsinki as part of the International DLBCL Rituximab-CHOP Consortium Program. Data collection protocols were approved as being of minimal to no risk or as exempt by the institutional review board of each participating institution.
Consent for publication
Not applicable.
Competing interests
B.C. and I.K. are employees of Adaptive Biotechnologies. R.D.B. is a co-founder of ioGenetics LLC. T.S. is a former employee of Adaptive Biotechnologies. K.H.Y. receives research support from Adaptive Biotechnologies, Roche Molecular Diagnostics, Gilead Sciences, Seattle Genetics, Daiichi Sankyo, Incyte Corporation, and HTG Molecular Diagnostics. Other authors declare no conflicts of interest.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary information accompanies this paper at 10.1186/s40425-019-0730-x.
References
- 1.Kuppers R, Rajewsky K, Hansmann ML. Diffuse large cell lymphomas are derived from mature B cells carrying V region genes with a high load of somatic mutation and evidence of selection for antibody expression. Eur J Immunol. 1997;27(6):1398–1405. doi: 10.1002/eji.1830270616. [DOI] [PubMed] [Google Scholar]
- 2.Pasqualucci L, Neumeister P, Goossens T, Nanjangud G, Chaganti RS, Kuppers R, et al. Hypermutation of multiple proto-oncogenes in B-cell diffuse large-cell lymphomas. Nature. 2001;412(6844):341–346. doi: 10.1038/35085588. [DOI] [PubMed] [Google Scholar]
- 3.De Silva NS, Klein U. Dynamics of B cells in germinal centres. Nat Rev Immunol. 2015;15(3):137–148. doi: 10.1038/nri3804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rebhandl S, Huemer M, Greil R, Geisberger R. AID/APOBEC deaminases and cancer. Oncoscience. 2015;2(4):320–333. doi: 10.18632/oncoscience.155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lieber MR. Mechanisms of human lymphoid chromosomal translocations. Nat Rev Cancer. 2016;16(6):387–398. doi: 10.1038/nrc.2016.40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Willis TG, Dyer MJ. The role of immunoglobulin translocations in the pathogenesis of B-cell malignancies. Blood. 2000;96(3):808–822. doi: 10.1182/blood.V96.3.808. [DOI] [PubMed] [Google Scholar]
- 7.Lenz G, Nagel I, Siebert R, Roschke AV, Sanger W, Wright GW, et al. Aberrant immunoglobulin class switch recombination and switch translocations in activated B cell-like diffuse large B cell lymphoma. J Exp Med. 2007;204(3):633–643. doi: 10.1084/jem.20062041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cowan G, Weston-Bell NJ, Bryant D, Seckinger A, Hose D, Zojer N, et al. Massive parallel IGHV gene sequencing reveals a germinal center pathway in origins of human multiple myeloma. Oncotarget. 2015;6(15):13229–13240. doi: 10.18632/oncotarget.3644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Huemer M, Rebhandl S, Zaborsky N, Gassner FJ, Hainzl S, Weiss L, et al. AID induces intraclonal diversity and genomic damage in CD86(+) chronic lymphocytic leukemia cells. Eur J Immunol. 2014;44(12):3747–3757. doi: 10.1002/eji.201344421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lossos IS, Alizadeh AA, Eisen MB, Chan WC, Brown PO, Botstein D, et al. Ongoing immunoglobulin somatic mutation in germinal center B cell-like but not in activated B cell-like diffuse large cell lymphomas. Proc Natl Acad Sci U S A. 2000;97(18):10209–10213. doi: 10.1073/pnas.180316097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Srinivasan L, Sasaki Y, Calado DP, Zhang B, Paik JH, DePinho RA, et al. PI3 kinase signals BCR-dependent mature B cell survival. Cell. 2009;139(3):573–586. doi: 10.1016/j.cell.2009.08.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Havranek O, Xu J, Kohrer S, Wang Z, Becker L, Comer JM, et al. Tonic B-cell receptor signaling in diffuse large B-cell lymphoma. Blood. 2017;130(8):995–1006. doi: 10.1182/blood-2016-10-747303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Erdmann T, Klener P, Lynch JT, Grau M, Vockova P, Molinsky J, et al. Sensitivity to PI3K and AKT inhibitors is mediated by divergent molecular mechanisms in subtypes of DLBCL. Blood. 2017;130(3):310–322. doi: 10.1182/blood-2016-12-758599. [DOI] [PubMed] [Google Scholar]
- 14.Young RM, Wu T, Schmitz R, Dawood M, Xiao W, Phelan JD, et al. Survival of human lymphoma cells requires B-cell receptor engagement by self-antigens. Proc Natl Acad Sci U S A. 2015;112(44):13447–13454. doi: 10.1073/pnas.1514944112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Weiss S, Bogen B. MHC class II-restricted presentation of intracellular antigen. Cell. 1991;64(4):767–776. doi: 10.1016/0092-8674(91)90506-T. [DOI] [PubMed] [Google Scholar]
- 16.Macmillan H, Strohman MJ, Ayyangar S, Jiang W, Rajasekaran N, Spura A, et al. The MHC class II cofactor HLA-DM interacts with Ig in B cells. J Immunol. 2014;193(6):2641–2650. doi: 10.4049/jimmunol.1400075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chakrabarti D, Hosh SK. Induction of syngeneic cytotoxic T lymphocytes against a B cell tumor. III. MHC class I-restricted CTL recognizes the processed form(s) of idiotype. Cell Immunol. 1992;69(5):455–464. doi: 10.1016/0008-8749(92)90259-R. [DOI] [PubMed] [Google Scholar]
- 18.Khodadoust MS, Olsson N, Wagar LE, Haabeth OA, Chen B, Swaminathan K, et al. Antigen presentation profiling reveals recognition of lymphoma immunoglobulin neoantigens. Nature. 2017;543(7647):723–727. doi: 10.1038/nature21433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Khodadoust MS, Olsson N, Chen B, Sworder B, Shree T, Liu CL, et al. B-cell lymphomas present immunoglobulin neoantigens. Blood. 2019;133(8):878–881. doi: 10.1182/blood-2018-06-845156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pielak RM, O'Donoghue GP, Lin JJ, Alfieri KN, Fay NC, Low-Nam ST, et al. Early T cell receptor signals globally modulate ligand:receptor affinities during antigen discrimination. Proc Natl Acad Sci U S A. 2017;114(46):12190–12195. doi: 10.1073/pnas.1613140114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Xu-Monette ZY, Zhang M, Li J, Young KH. PD-1/PD-L1 blockade: have we found the key to unleash the antitumor immune response? Front Immunol. 2017;8:1597. doi: 10.3389/fimmu.2017.01597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bremel RD, Homan EJ. Frequency patterns of T-cell exposed amino acid motifs in immunoglobulin heavy chain peptides presented by MHCs. Front Immunol. 2014;5:541. doi: 10.3389/fimmu.2014.00541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bremel RD, Homan EJ. Extensive T-cell epitope repertoire sharing among human proteome, gastrointestinal microbiome, and pathogenic bacteria: implications for the definition of self. Front Immunol. 2015;6:538. doi: 10.3389/fimmu.2015.00538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Visco C, Li Y, Xu-Monette ZY, Miranda RN, Green TM, Li Y, et al. Comprehensive gene expression profiling and immunohistochemical studies support application of immunophenotypic algorithm for molecular subtype classification in diffuse large B-cell lymphoma: a report from the international DLBCL rituximab-CHOP consortium program study. Leukemia. 2012;26(9):2103–2113. doi: 10.1038/leu.2012.83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Xu-Monette ZY, Wu L, Visco C, Tai YC, Tzankov A, Liu WM, et al. Mutational profile and prognostic significance of TP53 in diffuse large B-cell lymphoma patients treated with R-CHOP: report from an international DLBCL rituximab-CHOP consortium program study. Blood. 2012;120(19):3986–3996. doi: 10.1182/blood-2012-05-433334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dybkaer K, Bogsted M, Falgreen S, Bodker JS, Kjeldsen MK, Schmitz A, et al. Diffuse large B-cell lymphoma classification system that associates normal B-cell subset phenotypes with prognosis. J Clin Oncol. 2015;33(12):1379–1388. doi: 10.1200/JCO.2014.57.7080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tzankov A, Xu-Monette ZY, Gerhard M, Visco C, Dirnhofer S, Gisin N, et al. Rearrangements of MYC gene facilitate risk stratification in diffuse large B-cell lymphoma patients treated with rituximab-CHOP. Mod Pathol. 2014;27(7):958–971. doi: 10.1038/modpathol.2013.214. [DOI] [PubMed] [Google Scholar]
- 28.Visco C, Tzankov A, Xu-Monette ZY, Miranda RN, Tai YC, Li Y, et al. Patients with diffuse large B-cell lymphoma of germinal center origin with BCL2 translocations have poor outcome, irrespective of MYC status: a report from an international DLBCL rituximab-CHOP consortium program study. Haematologica. 2013;98(2):255–263. doi: 10.3324/haematol.2012.066209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Xu-Monette ZY, Deng Q, Manyam GC, Tzankov A, Li L, Xia Y, et al. Clinical and biologic significance of MYC genetic mutations in De novo diffuse large B-cell lymphoma. Clin Cancer Res. 2016;22(14):3593–3605. doi: 10.1158/1078-0432.CCR-15-2296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Larimore K, McCormick MW, Robins HS, Greenberg PD. Shaping of human germline IgH repertoires revealed by deep sequencing. J Immunol. 2012;189(6):3221–3230. doi: 10.4049/jimmunol.1201303. [DOI] [PubMed] [Google Scholar]
- 31.Wu D, Emerson RO, Sherwood A, Loh ML, Angiolillo A, Howie B, et al. Detection of minimal residual disease in B lymphoblastic leukemia by high-throughput sequencing of IGH. Clin Cancer Res. 2014;20(17):4540–4548. doi: 10.1158/1078-0432.CCR-13-3231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wood B, Wu D, Crossley B, Dai Y, Williamson D, Gawad C, et al. Measurable residual disease detection by high-throughput sequencing improves risk stratification for pediatric B-ALL. Blood. 2018;131(12):1350–1359. doi: 10.1182/blood-2017-09-806521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lefranc MP. IMGT, the international ImMunoGeneTics information system. Cold Spring Harb Protoc. 2011;2011(6):595–603. doi: 10.1101/pdb.top115. [DOI] [PubMed] [Google Scholar]
- 34.Sebastian E, Alcoceba M, Balanzategui A, Marin L, Montes-Moreno S, Flores T, et al. Molecular characterization of immunoglobulin gene rearrangements in diffuse large B-cell lymphoma: antigen-driven origin and IGHV4-34 as a particular subgroup of the non-GCB subtype. Am J Pathol. 2012;181(5):1879–1888. doi: 10.1016/j.ajpath.2012.07.028. [DOI] [PubMed] [Google Scholar]
- 35.Duke VM, Gandini D, Sherrington PD, Lin K, Heelan B, Amlot P, et al. V(H) gene usage differs in germline and mutated B-cell chronic lymphocytic leukemia. Haematologica. 2003;88(11):1259–1271. [PubMed] [Google Scholar]
- 36.Bremel RD, Homan EJ. An integrated approach to epitope analysis II: a system for proteomic-scale prediction of immunological characteristics. ImmunomeRes. 2010;6(1):8. doi: 10.1186/1745-7580-6-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Johnson NL. Systems of frequency curves generated by methods of translation. Biometrika. 1949;36(Pt. 1–2):149–176. doi: 10.1093/biomet/36.1-2.149. [DOI] [PubMed] [Google Scholar]
- 38.Rudolph MG, Stanfield RL, Wilson IA. How TCRs bind MHCs, peptides, and coreceptors. Annu Rev Immunol. 2006;24:419–466. doi: 10.1146/annurev.immunol.23.021704.115658. [DOI] [PubMed] [Google Scholar]
- 39.Weiss S, Bogen B. B-lymphoma cells process and present their endogenous immunoglobulin to major histocompatibility complex-restricted T cells. Proc Natl Acad Sci U S A. 1989;86(1):282–286. doi: 10.1073/pnas.86.1.282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.DeWitt WS, Lindau P, Snyder TM, Sherwood AM, Vignali M, Carlson CS, et al. A public database of memory and naive B-cell receptor sequences. PLoS One. 2016;11(8):e0160853. doi: 10.1371/journal.pone.0160853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Xu-Monette ZY, Xiao M, Au Q, Padmanabhan R, Xu B, Hoe N, et al. Immune profiling and quantitative analysis decipher the clinical role of immune-checkpoint expression in the tumor immune microenvironment of DLBCL. Cancer Immunol Res. 2019;7(4):644–657. doi: 10.1158/2326-6066.CIR-18-0439. [DOI] [PubMed] [Google Scholar]
- 42.Rosner K, Winter DB, Tarone RE, Skovgaard GL, Bohr VA, Gearhart PJ. Third complementarity-determining region of mutated VH immunoglobulin genes contains shorter V, D, J, P, and N components than non-mutated genes. Immunology. 2001;103(2):179–187. doi: 10.1046/j.1365-2567.2001.01220.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Adler LN, Jiang W, Bhamidipati K, Millican M, Macaubas C, Hung SC, et al. The other function: class II-restricted antigen presentation by B cells. Front Immunol. 2017;8:319. doi: 10.3389/fimmu.2017.00319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kasar S, Kim J, Improgo R, Tiao G, Polak P, Haradhvala N, et al. Whole-genome sequencing reveals activation-induced cytidine deaminase signatures during indolent chronic lymphocytic leukaemia evolution. Nat Commun. 2015;6:8866. doi: 10.1038/ncomms9866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Jain P, Nogueras Gonzalez GM, Kanagal-Shamanna R, Rozovski U, Sarwari N, Tam C, et al. The absolute percent deviation of IGHV mutation rather than a 98% cut-off predicts survival of chronic lymphocytic leukaemia patients treated with fludarabine, cyclophosphamide and rituximab. Br J Haematol. 2018;180(1):33–40. doi: 10.1111/bjh.15018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Riaz N, Havel JJ, Makarov V, Desrichard A, Urba WJ, Sims JS, et al. Tumor and microenvironment evolution during immunotherapy with Nivolumab. Cell. 2017;171(4):934–949. doi: 10.1016/j.cell.2017.09.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Dyer MJ, Oscier DG. The configuration of the immunoglobulin genes in B cell chronic lymphocytic leukemia. Leukemia. 2002;16(6):973–984. doi: 10.1038/sj.leu.2402528. [DOI] [PubMed] [Google Scholar]
- 48.Xu-Monette ZY, Zhou J, Young KH. PD-1 expression and clinical PD-1 blockade in B-cell lymphomas. Blood. 2018;131(1):68–83. doi: 10.1182/blood-2017-07-740993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tumeh PC, Harview CL, Yearley JH, Shintaku IP, Taylor EJ, Robert L, et al. PD-1 blockade induces responses by inhibiting adaptive immune resistance. Nature. 2014;515(7528):568–571. doi: 10.1038/nature13954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Roh W, Chen PL, Reuben A, Spencer CN, Prieto PA, Miller JP, et al. Integrated molecular analysis of tumor biopsies on sequential CTLA-4 and PD-1 blockade reveals markers of response and resistance. Sci Transl Med. 2017;9(379). [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1: Fig S1.. Construction and clinical outcome of the diffuse large B-cell lymphoma (DLBCL) cohort. Fig. S2. Diagram showing numbers of cases in this mutation study that have been characterized by various biomarker studies, and survival rates of patients whose sequencing results were correlated with prognosis. Fig. S3. CONSORT flow diagram illustrating the number of cases performed for high-throughput IG sequencing and clonal sequence analysis. Fig. S4. Molecular characterization for immunoglobulin heavy chain (IGH) gene usage in the study cohort. Fig. S5. Immunoglobulin heavy chain V gene (IGHV) somatic hypermutation (SHM) analysis. Fig. S6. Analysis for length of heavy chain CDR3. Fig. S7. Prediction of MHC-binding peptides and frequency of T-cell exposed motifs (TCEM) for immunoglobulin diagnostic sequences in the training set and validation set. (a) Regional distribution of relatively rare neoantigens derived from heavy chain and light chain immunoglobulin genes in DLBCL patients in the training set (top) and validation set (bottom). (b) Cases with high degree of heavy chain or light chain IGV SHM compared with cases without had higher frequency of relatively rare TCEM in the training (left) and validation sets (right). (c) In ABC-DLBCL, high IGV SHM was associated with lower tissue cellularity of CD4+ T cells. Fig. S8. Moleclar analysis for immunoglobulin heavy chain ongoing SHM and light chain SHM. Fig. S9. Immunoglobulin light chain SHM and CDR3 analysis. Fig S10. Comparison between different subsets of DLBCL. Fig S11. Light chain IGK/LV ongoing SHM analysis.
Additional file 2: Table S1. Clinical features of 378 patients in the training and validation cohort whose DLBCL biopsies were sequenced and 290 patients whose sequencing results showed sufficient sequence reads. Table S2. Comparisons of clinicopathologic and molecular characteristics between patients with germinal-center B-cell–like (GCB) DLBCL with a low or high degree of somatic hypermutation (SHM) in immunoglobulin variable region genes. Table S3. Comparisons of clinicopathologic and molecular characteristics between patients with activated B-cell-like (ABC) subtype of DLBCL with a low or high degree of SHM in immunoglobulin variable region genes. Table S4. Significant prognostic effects of immunoglobulin molecular characteristics in DLBCL patients treated with R-CHOP by multivariate survival analysis. Table S5. Clinicopathologic and molecular characteristics of patients with DLBCL with a short or long immunoglobulin heavy/light chain CDR3 length. Table S6. Clinicopathologic and molecular characteristics of patients with DLBCL with ongoing SHM in immunoglobulin variable region genes. Table S7. Gene signatures associated with SHM in immunoglobulin sequences of DLBCL samples. Table S8. Multiple testing corrections for prognostic effects found in the overall cohort of DLBCL treated with R-CHOP by the Benjamini-Hochberg method with a false discovery rate of 0.10
Additional file 3. Diagnostic immunoglobulin heavy chain gene sequences
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request based on the condition that IRB and MTA could be approved from the institutions.