Abstract
Background
Atypical lipomatous tumor/well‐differentiated liposarcoma (ALT‐WDLPS) and dedifferentiated liposarcoma (DDLPS) are characterized cytogenetically by a 12q13–15 amplification involving the mouse double minute 2 (MDM2) oncogene. Fluorescence in situ hybridization (FISH) is used frequently to detect this amplification and aid with the diagnosis of these entities, which is difficult by morphology alone. Recently, bright‐field in situ hybridization techniques such as chromogenic in situ hybridization (CISH) have been introduced for the determination of MDM2 amplification status.
Methods
The present study compared the results of FISH and CISH for detecting MDM2 amplification in 41 cases of adipocytic tumors. Amplification was defined in both techniques as a MDM2/CEN12 ratio of 2 or greater.
Results
Eleven cases showed amplification with both FISH and CISH, and 26 cases showed no amplification with both methods. Two cases had discordant results between CISH and FISH, and two cases were not interpretable by CISH.
Conclusion
CISH is advantageous for allowing pathologists to evaluate the histologic and molecular alterations occurring simultaneously in a specimen. Moreover, CISH is found to be more cost‐ and time‐efficient when used with automation, and the signals do not quench over time. CISH technique is a reliable alternative to FISH in the evaluation of adipocytic tumors for MDM2 amplification.
Keywords: adipose tissue, human gene amplification in situ hybridization chromogenic compounds, MDM2 protein, neoplasms
INTRODUCTION
Adipocytic tumors represent a heterogeneous group of neoplasms that may be difficult to differentiate based solely on histologic examination. In addition to their subtle histologic distinctions, these tumors are each associated with a different patient prognosis and treatment regimen, making an accurate diagnosis critically important. Liposarcomas represent the single most common type of soft tissue sarcoma, and they are classified by the World Health Organization (WHO) into four main categories: atypical lipomatous tumor/well‐differentiated liposarcoma (ALT‐WDLPS), dedifferentiated liposarcoma (DDLPS), myxoid liposarcoma, and pleomorphic liposarcoma 1. A particular diagnostic challenge is distinguishing ALT‐WDLPS from benign adipocytic tumors. Similarly, DDLPS is often difficult to differentiate morphologically from other high‐grade sarcomas. In these situations, ancillary studies are often required to arrive at a correct diagnosis. Fortunately, these adipocytic tumors harbor unique cytogenetic abnormalities that can be detected with molecular techniques 2.
ALT‐WDLPS and DDLPS are characterized cytogenetically by a 12q13–15 amplification involving the mouse double minute 2 (MDM2) oncogene 3. The oncogenic properties of MDM2 are explained by its ability to inhibit the p53 tumor suppressor gene, thereby inhibiting apoptosis and enhancing cell survival 4. MDM2 is abnormally upregulated in many human cancers, with an overall frequency of MDM2 amplification estimated as 7%. The highest prevalence is documented in soft tissue tumors, followed by osteosarcoma and esophageal carcinoma 5.
Amplification of the MDM2 locus is present in the majority of ALT‐WDLPS and DDLPS, while it is not found in benign adipocytic tumors 2. This feature can thus be exploited with molecular techniques. Fluorescence in situ hybridization (FISH) has been commonly used to evaluate MDM2 status in equivocal cases of ALT‐WDLPS vs. benign lipomatous neoplasm. However, FISH requires specialized equipment to visualize fluorescence signals and needs to be interpreted with corresponding light microscopic sections. More recently, bright‐field in situ hybridization techniques such as chromogenic in situ hybridization (CISH) and silver‐enhanced in situ hybridization (SISH), which combine the general principles of immunohistochemical analysis and in situ hybridization, have been introduced for the determination of MDM2 amplification status. These new techniques use a peroxidase enzyme‐labeled probe with chromogenic detection, instead of a fluorescent‐labeled probe, allowing results to be visualized by standard bright‐field microscopy 6. They have been used successfully in the determination of amplification of other genes associated with human cancers, most notably for assessing HER2 gene status in breast carcinoma 7.
Zhang et al. describe the development and validation of an automated bright field dual‐color in situ hybridization assay to visualize both MDM2 and chromosome 12 copies within the same tumor nuclei. The assay performance was evaluated on a cohort of 100 formalin‐fixed, paraffin‐embedded soft tissue specimens, and excellent sensitivity and specificity were reported 6. Thus far, a direct comparison has not been made between FISH and CISH results in the same tumor samples. An established effectiveness of CISH for use in lieu of FISH for detecting MDM2 amplification would be beneficial for many institutions. The present study compared the results for 41 cases of adipocytic tumors that were tested for MDM2 amplification by both FISH and CISH techniques.
MATERIALS AND METHODS
A total of 41 cases of adipocytic tumors with established histopathologic diagnoses were selected to be tested for MDM2 amplification by FISH and CISH. Nineteen cases were benign, with a diagnosis of either lipoma or lipomatous neoplasm with minimal to moderate atypia. Twenty‐two cases were malignant, with diagnoses including ALT‐WDLPS, DDLPS, pleomorphic high‐grade sarcoma, myxoid liposarcoma, mixed type (myxoid/round cell) liposarcoma, and spindle cell melanoma.
FISH for assessment of MDM2 gene amplification was performed as follows. Four microns thick formalin‐fixed, paraffin‐embedded tissue slides were incubated at 56°C overnight followed by immersion in Hemo‐De solution (10 min × 3 times) at room temperature. Slides were then treated with Lugol Iodine Solution for 5 min, and rinsed in 2.5% sodium thiocyanate until clear. The slides were incubated in 100% ethanol for 2 min. Slides immersed in citrate acid were microwaved for 4 min. Slides were placed in pepsin/HCl solution (prepared by mixing 0.20 g pepsin in 50 mM HCl, pH 2.0) for 1 h at 37°C before passing them through an alcohol series (70% EtOH for 1 min, 85% EtOH for 1 min, 100% EtOH for 10 min). Hybridization was performed with MDM2/CEP12 probes (Vysis, Downers Grove, IL) overnight at 37°C in a humidified chamber. Slides were washed with saline‐sodium citrate buffer and counterstained with 4′,6‐diamidino‐2‐phenylindole (DAPI) for interpretation using a fluorescent microscope. Scoring was performed by recording the total number of green (CEP12) signals and red (MDM2) signals for each cell, with a minimum of 100 cells evaluated per sample. A MDM2/CEP12 ratio was calculated for each case. Amplification of MDM2 was defined as an MDM2/CEP12 ratio ≥2, whereas a ratio of less than 2 was defined as not amplified. In highly amplified cases, the numerous signals generate clusters which preclude quantitation of an exact numeric ratio. For these cases, the designation “amplified, cannot be quantitated (CBQ)” was used instead of an estimated ratio.
CISH for assessment of MDM2 gene amplification was performed using the BenchMark® ULTRA automated slide processing system, with all materials and probes obtained from Ventana Medical Systems, Inc., Tucson, Arizona. The protocol was adapted from that used for Ventana's INFORM HER2 Dual ISH DNA Probe Cocktail, as described in the package insert 8. The formalin‐fixed, paraffin‐embedded tissue was cut at 4 μ. The slides were incubated at 60°C for 15 min, followed by application of EZ Prep Reagent for deparaffinization. Pretreatment included incubation at 82°C and application of Cell Conditioning Solution (CC2). ISH‐Protease 3 was then applied and slides were incubated for 28 min. MDM2 DNP, and CEN12 DIG probes were applied and the slides were incubated for 8 min at 80°C. Hybridization was performed overnight at 37°C in a humidified chamber. Stringency washes (8 min × 3) were performed with Sodium Chloride Sodium Citrate buffer solution at 75°C. The metallic silver deposit for MDM2 ISH signal was developed using the ultraView SISH DNP Detection Kit, and the signal for CEN12 was visualized with a fast red and naphthol phosphate reaction using the ultraView Red ISH DIG Detection Kit, according to manufacturer's instructions. The slides were then counterstained with Hematoxylin II and Bluing Reagent for interpretation by light microscopy.
Cells were examined using a 100× objective under oil immersion. Slide adequacy was verified before enumerating each slide, using the criteria established by Ventana in the “Interpretation Guide” for the INFORM HER2 Dual ISH DNA Probe Cocktail Assay 9. The first criterion for adequacy is the presence of internal positive control staining in non‐neoplastic cells including normal adipocytes, endothelial cells, and inflammatory cells. The non‐neoplastic nuclei should contain one or two copies of both CEN12 (red signals) and MDM2 (black signals). Next, staining within the neoplastic cells must be enumerable. Due to truncation in the plane of sectioning, it is likely that not every neoplastic cell will contain signals, but the slide must contain an acceptable region of lesional tissue that is enumerable, or else the slide is considered inadequate for interpretation. Lastly, background staining must not interfere with enumeration. Once slide adequacy was confirmed, neoplastic cells were examined for the presence of red (CEN12 copies) and black (MDM2 copies) signals. For the first ten cases analyzed, two separate counts with 50 and 100 cells were performed and evaluated for equivalence. No significant difference was observed between results for counting 50 cells vs. 100 cells (see Results below), so for the remainder of cases, 50 cells were counted. Interpretation was accomplished according to Ventana's “Interpretation Guide” for the INFORM HER2 Dual ISH DNA Probe Cocktail Assay 9. The overall ratio of black to red signals was calculated for each case. An MDM2/CEN12 ratio ≥2 was defined as amplified, while a ratio of less than 2 was defined as not amplified. As with FISH, in highly amplified cases containing signal clusters, the result is reported as “amplified, cannot be quantitated (CBQ)” instead of an estimated ratio.
RESULTS
All data are reported as MDM2/CEN12 ratios. In the analysis of ten cases for MDM2 amplification by CISH, the ratios calculated from scoring 50 cells and 100 cells were not significantly different (t = −0.759, P = 0.47) (Table 1).
Table 1.
Comparison of Ratios From Scoring 50 Cells vs. 100 Cells
| Case No. | Ratio 50 Cells | Ratio 100 Cells | Difference (Ratio 100 Cells‐Ratio 50 Cells) |
|---|---|---|---|
| 1 | 1.1 | 1.21 | 0.11 |
| 2 | 1.87 | 1.84 | −0.03 |
| 3 | 1.08 | 1.12 | 0.04 |
| 4 | 0.96 | 0.90 | −0.06 |
| 5 | 0.99 | 0.97 | −0.02 |
| 6 | 1.06 | 1.01 | −0.05 |
| 7 | 1.07 | 1.03 | −0.04 |
| 8 | 1.03 | 0.98 | −0.05 |
| 9 | 1.1 | 1.05 | −0.05 |
| 10 | 1.03 | 1.05 | 0.02 |
Table 2 summarizes all of the tumors analyzed, including the patient demographics, histopathologic diagnosis, and results of MDM2 amplification by FISH and CISH. Overall, 11 cases showed amplification with both FISH and CISH, 26 cases showed no amplification with both methods, two cases showed discordant results between FISH and CISH, and two cases were noninterpretable by CISH. Table 3 displays the correlation of FISH and CISH results among the 39 interpretable cases. Results from 37 out of the 39 cases (95%) were concordant. The concordant nonamplified cases included 15 benign lipomatous neoplasms, three lipomas with borderline atypical features, five myxoid liposarcomas, two ALT‐WDLPS, and one mixed type (myxoid/round cell) liposarcoma (Fig. 1). The concordant amplified cases included seven ALT‐WDLPS, three DDLPS and one pleomorphic high‐grade sarcoma (Fig. 2). One case (undifferentiated pleomorphic high‐grade liposarcoma) showed amplification by FISH only, and one case (ALT‐WDLPS) showed amplification by CISH only. Six cases displayed weak or absent signals following application of the CISH methodology. CISH was repeated in these cases, with interpretable results in four cases while two cases remained noninterpretable.
Table 2.
Summary of Tumors Tested for MDM2 Amplification Status by FISH and CISH
| Case No. | Age | Gender | Diagnosis | Tumor Category | FISH Ratio | Status by FISH | CISH Ratio | Status by CISH |
|---|---|---|---|---|---|---|---|---|
| 1 | 26 | F | Lipoma | Benign | 1.01 | Not amplified | 0.92 | Not amplified |
| 2 | 44 | M | Lipoma | Benign | 0.98 | Not amplified | 0.98 | Not amplified |
| 3 | 38 | M | Lipoma | Benign | 0.95 | Not amplified | 0.99 | Not amplified |
| 4 | 54 | M | Lipoma | Benign | 1.03 | Not amplified | 1.04 | Not amplified |
| 5 | 71 | F | Lipoma | Benign | 0.96 | Not amplified | 1.02 | Not amplified |
| 6 | 42 | M | Lipoma | Benign | 0.97 | Not amplified | Unknown | Unknown |
| 7 | 81 | F | Benign lipomatous tumor | Benign | 0.97 | Not amplified | 0.95 | Not amplified |
| 8 | 47 | F | Benign lipomatous tissue | Benign | 0.98 | Not amplified | 1.01 | Not amplified |
| 9 | 43 | F | Hibernoma | Benign | 0.92 | Not amplified | 0.95 | Not amplified |
| 10 | 68 | F | Intramuscular lipoma | Benign | 1.02 | Not amplified | 1.05 | Not amplified |
| 11 | 60 | M | Mature fibroadipose tissue | Benign | 1.10 | Not amplified | 0.95 | Not amplified |
| 12 | 46 | F | Myolipoma | Benign | 0.94 | Not amplified | 1.03 | Not amplified |
| 13 | 25 | F | Well differentiated lipomatous tissue | Benign | 1.02 | Not amplified | 0.97 | Not amplified |
| 14 | 50 | M | Well differentiated lipomatous tumor with overlap features of spindle cell lipoma and cellular angiofibroma | Benign | 0.99 | Not amplified | 1.01 | Not amplified |
| 15 | 51 | F | Lipomatous areas with minimal atypia | Benign | 1.01 | Not amplified | 1.03 | Not amplified |
| 16 | 51 | F | Lipomatous neoplasm, minimal cytologic atypia | Benign | 1.02 | Not amplified | 1.01 | Not amplified |
| 17 | 48 | M | Atypical lipoma | Benign | 1.02 | Not amplified | 0.98 | Not amplified |
| 18 | 75 | M | Lipomatous tumor, scattered atypical cells | Benign | 1.00 | Not amplified | 1.05 | Not amplified |
| 19 | 43 | F | Lipomatous tumor, scattered moderate nuclear atypia | Benign | 1.10 | Not amplified | 1.03 | Not amplified |
| 20 | 61 | F | Myxoid LPS | Malignant | 1.01 | Not amplified | 1.21 | Not amplified |
| 21 | 40 | F | Myxoid LPS | Malignant | 0.95 | Not amplified | 1.12 | Not amplified |
| 22 | 62 | F | Myxoid LPS | Malignant | 0.98 | Not amplified | 0.90 | Not amplified |
| 23 | 65 | F | Myxoid LPS | Malignant | 1.04 | Not amplified | 0.95 | Not amplified |
| 24 | 37 | F | ALT/Well differentiated myxoid liposarcoma | Malignant | 1.00 | Not amplified | 0.99 | Not amplified |
| 25 | 75 | M | Liposarcoma, mixed type (myxoid and high grade round cell) | Malignant | 0.70 | Not amplified | 0.88 | Not amplified |
| 26 | 80 | F | ALT‐WDLPS | Malignant | 1.01 | Not amplified | 1.84 | Not amplified |
| 27 | 64 | M | ALT‐WDLPS | Malignant | 1.00 | Not amplified | 0.96 | Not amplified |
| 28 | 62 | F | ALT‐WDLPS | Malignant | 1.45 | Not amplified | CBQ | Amplified |
| 29 | 58 | F | ALT‐WDLPS | Malignant | CBQ | Amplified | CBQ | Amplified |
| 30 | 60 | M | ALT‐WDLPS | Malignant | CBQ | Amplified | CBQ | Amplified |
| 31 | 68 | F | ALT‐WDLPS | Malignant | CBQ | Amplified | CBQ | Amplified |
| 32 | 58 | M | ALT‐WDLPS | Malignant | CBQ | Amplified | CBQ | Amplified |
| 33 | 62 | M | ALT‐WDLPS | Malignant | CBQ | Amplified | CBQ | Amplified |
| 34 | 62 | F | ALT‐WDLPS | Malignant | CBQ | Amplified | CBQ | Amplified |
| 35 | 81 | M | ALT‐WDLPS | Malignant | CBQ | Amplified | CBQ | Amplified |
| 36 | 54 | M | DDLPS | Malignant | CBQ | Amplified | CBQ | Amplified |
| 37 | 53 | M | DDLPS | Malignant | CBQ | Amplified | CBQ | Amplified |
| 38 | 57 | F | Liposarcoma with low‐grade dedifferentiation | Malignant | CBQ | Amplified | CBQ | Amplified |
| 39 | 52 | M | Pleomorphic high‐grade sarcoma | Malignant | CBQ | Amplified | CBQ | Amplified |
| 40 | 55 | M | Undifferentiated pleomorphic high‐grade liposarcoma | Malignant | CBQ | Amplified | 1.05 | Not amplified |
| 41 | 50 | F | Spindle cell melanoma | Malignant | 0.9 | Not amplified | Unknown | Unknown |
CBQ, cannot be quantitated.
Table 3.
Correlation of MDM2 Amplification Status by FISH and CISH
| FISH | ||||
|---|---|---|---|---|
| Amplified | Nonamplified | Total | ||
| CISH | Amplified | 11 | 1 | 12 |
| Nonamplified | 1 | 26 | 27 | |
| Total | 12 | 27 | 39 | |
Figure 1.
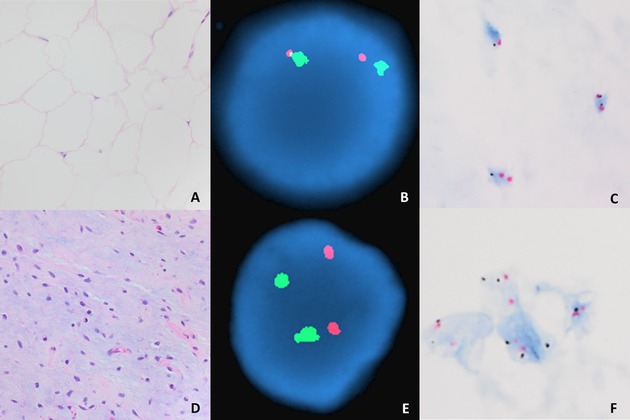
Lipoma (A) and myxoid liposarcoma (D) with negative MDM2 amplification as demonstrated by FISH (B, E) and CISH (C, F), respectively. A and D, hematoxylin, and eosin, 40×. C and F, CISH, 100×.
Figure 2.
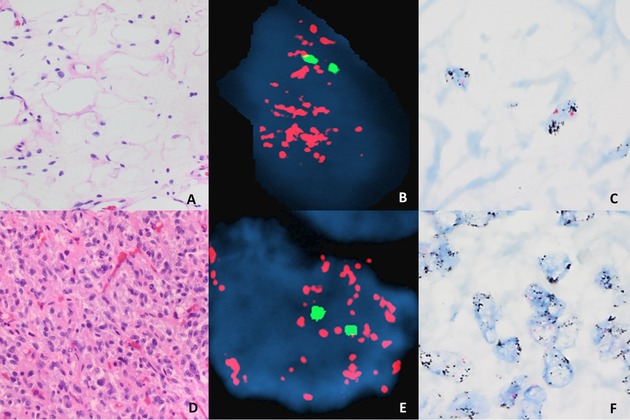
ALT‐WDLPS (A) and DDLPS (D) with positive MDM2 amplification demonstrated by FISH (B, E) and CISH (C, F), respectively. A and D, hematoxylin and eosin, 40×. C and F, CISH, 100x.
DISCUSSION
Accurate diagnosis of the various adipocytic tumors is challenging and critically important due to the impact on patient management. ALT‐WDLPS has been classically defined based on histologic aspects alone, but it is often difficult to distinguish morphologically this tumor from its benign counterparts, especially in the context of a lipomatous neoplasm in a deep seated location. The lower grade ALT‐WDLPS in particular has a very close resemblance to normal fat. Similarly, the histological complexity of DDLPS leads to a large differential diagnosis including myxofibrosarcoma, pleomorphic rhabdomyosarcoma, malignant mesenchymoma, and poorly differentiated sarcomas such as malignant fibrous histiocytoma, fibrosarcoma, and malignant hemangiopericytoma 3. Additionally, the increased use of minimally invasive biopsies for adipocytic tumors may further complicate the matter by providing limited diagnostic material. Fortunately, the presence of a unique cytogenetic abnormality in ALT‐WDLPS and DDLPS can be exploited by supplementary diagnostic tools such as immunohistochemistry, FISH, comparative genomic hybridization (CGH), and most recently CISH. The increased emphasis placed on these molecular characteristics is reflected by the current WHO classification of liposarcomas, which is based on both morphological and genetic features 1.
MDM2 is amplified in close to 100% of ALT‐WDLPS and DDLPS and is not amplified in benign lipomas 10. Myxoid liposarcomas also do not show MDM2 amplification; instead they are associated with a classical t(12;16)(q13;p11) or t(12;22)(q13;q12) translocation 11. The results of the present study are consistent with these observations. The eleven amplified cases included seven ALT‐WDLPS, three DDLPS and one pleomorphic high‐grade sarcoma. An additional ALT‐WDLPS was amplified by CISH only. The nonamplified cases included 18 lipomas, five myxoid liposarcomas, two ALT‐WDLPS and one mixed type (myxoid/round cell) liposarcoma.
The high concordance between CISH and FISH establishes the clinical utility of CISH testing for MDM2 in order to classify adipocytic tumors. CISH provides several advantages over FISH. First, CISH slides are viewed using a conventional light microscope. The crisp chromogenic signals developed in dual color CISH are supported by a hematoxylin counterstain to enhance morphological features, allowing for pathologists to evaluate the tissue architecture and molecular alterations simultaneously in a specimen. Furthermore, tumor heterogeneity is more easily appreciated by CISH 12. CISH signals are not subject to rapid fading and the slides can therefore be easily archived; in contrast, FISH slides have stricter storage requirements and are subject to quenching of the fluorescent signal. Cellular and extracellular proteins can contribute to a dull, generalized, autofluorescence that often obscures FISH signals in paraffin sections 13. Moreover, CISH is found to be more cost‐ and time‐efficient than FISH when used with automation 12.
The results from two cases were discordant between FISH and CISH. One tumor, an undifferentiated pleomorphic high‐grade liposarcoma, showed amplification by FISH but not by CISH. The CISH signals were strong and distinct, so a technical issue is not likely. Tumor heterogeneity is a possible culprit. If this tumor was genetically heterogeneous for MDM2 copy number, with a mixture of amplified and unamplified nuclei, selection of different target areas for CISH and FISH analysis would account for these discrepant results. Another case, an ALT‐WDLPS, showed amplification by CISH but not by FISH. More specifically, by FISH, 21% of nuclei examined showed MDM2 amplification, with an overall ratio of 1.45. By CISH, 70% of nuclei examined showed MDM2 amplification, and determination of an accurate ratio was not possible due to the fact that the majority of these nuclei contained clusters of black signals. Again, tumor heterogeneity could play a role in this case. CISH allows the evaluator to scan for heterogeneity at low power to ensure a localized highly amplified area is not overlooked. An additional finding by FISH in this case is that 50% of cells showed polysomy, ranging from —two to eight chromosome 12 copies. It is unclear whether or not this high polysomy caused apparent increased MDM2 expression by CISH. Certainly, this warrants further investigation and suggests the need for revised scoring guidelines to take into account the issue of polysomy.
Two cases were not interpretable, even after repeated hybridization by CISH. This compares to Zhang et al., who reported a failure of CISH in two out of 100 cases and attributed this failure to tissue inadequacy 6. In both of our cases, both black and red (control and locus‐specific) probes were weak or undetectable, in both the tumor cells and surrounding non‐neoplastic stromal cells. Furthermore, the overall staining of cells in these cases appeared uniformly pale, and this pale counterstaining was also evident on the corresponding hematoxylin and eosin stained slides. This observation is suggestive of underfixation of the tissue, but the corresponding FISH in both cases had bright interpretable signals, suggesting that a step unique to the CISH procedure was responsible for the failure with this method. According to Ventana's Interpretation Guide, there are certain steps that can be manipulated to increase signal staining intensity. The cell conditioning times and/or cycle numbers can be increased. The application time of ISH‐Protease 3 can be increased, and this extended tissue digestion time is a troubleshooting technique often used with success in FISH. However, at a certain point, extended digestion times begin to distort cell morphology, so there is a delicate balance between increasing signal intensity and sacrificing cell morphology. Lastly, signal staining intensity can be increased with extended incubation time with the Detection Kit reagents (see Materials and Methods), but this comes with a risk of causing nonspecific background staining that may obscure signal interpretation 10. Clearly, there are many variables within the CISH technique that may be adjusted when repeating failed cases, and this should be studied further to increase the overall success rate of CISH.
Determination of MDM2 gene status in liposarcomas may provide both prognostic and therapeutic significance. Increased MDM2 expression has been associated with an overall worse clinical prognosis. Not only is there an increased likelihood of distant metastases in MDM2‐amplified tumors, but there is also a decreased response to therapeutic intervention 14. Specifically, the negative regulation of p53 by MDM2 may limit the magnitude of p53 activation by DNA damaging agents, thereby limiting their therapeutic effectiveness 15. Furthermore, pharmaceuticals that block the interaction between MDM2 and p53 are currently in development 16. Therefore, testing of liposarcoma cases for MDM2 amplification status is both diagnostically helpful and clinically relevant, and CISH proves to be a viable alternative to FISH, with added advantages that make it a more attractive option.
CONFLICT OF INTEREST
Ventana Medical Systems, Inc. (Tucson, Arizona) provided our institution free‐of‐charge with the necessary probes and reagents to carry out the CISH portion of the study.
ACKNOWLEDGMENTS
This study is partially supported by a grant from Ventana Medical Systems, Inc.
Contract grant sponsor: Ventana Medical Systems, Inc.
REFERENCES
- 1. IARC . WHO Classification of Tumours of Soft Tissue and Bone. 4th ed 2013.
- 2. Nishio J. Contributions of cytogenetics and molecular cytogenetics to the diagnosis of adipocytic tumors. J Biomed Biotechnol 2011;2011:524067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Coindre JM, Pedeutour F, Aurias A. Well‐differentiated and dedifferentiated liposarcomas. Virchows Arch 2010;456:167–179. [DOI] [PubMed] [Google Scholar]
- 4. Wade M, Li YC, Wahl GM. MDM2, MDMX and p53 in oncogenesis and cancer therapy. Nat Rev Cancer 2013;13(2):83–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Momand J, Jung D, Wilczynski S, Niland J. The MDM2 gene amplification database. Nucleic Acids Res 1998;26(15):3453–3459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Zhang W, McElhinny A, Nielsen A, et al. Automated brightfield dual‐color in situ hybridization for detection of mouse double minute 2 gene amplification in sarcomas. Appl Immunohistochem Mol Morphol 2011;19(1):54–61. [DOI] [PubMed] [Google Scholar]
- 7. Nitta H, Hauss‐Wegrzyniak B, Lehrkamp M, et al. Development of automated brightfield double in situ hybridization (BDISH) application for HER2 gene and chromosome 17 centromere (CEN 17) for breast carcinomas and an assay performance comparison to manual dual color HER2 fluorescence in situ hybridization (FISH). Diagnostic Pathol 2008;3:41 Available at: http://www.diagnosticpathology.org/content/3/1/41. Accessed September 1, 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. INFORM HER2 Dual ISH DNA Probe Cocktail, package insert . Catalog/Platform Number 780–4422. Ventana Medical Systems, Inc., Tucson, Arizona, 2013. [Google Scholar]
- 9. Grogan TM, McElhinny AS, Loftin IR, et al. Interpretation Guide: Ventana INFORM HER2 Dual ISH DNA Probe Cocktail Assay. Ventana Medical Systems, Inc., Tucson, Arizona. Available at: http://www.ventana.com/product/1553?type=2013. Accessed September 1, 2013.
- 10. Conyers R, Young S, Thomas DM. Liposarcoma: Molecular genetics and therapeutics. Sarcoma 2011;2011:483154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. de Vreeze RS, de Jong D, Nederlof PM, et al. Added value of molecular biological analysis in diagnosis and clinical management of liposarcoma: A 30‐year single‐institution experience. Ann Surg Oncol 2010;17(3):686–693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Cagle PT, Allen TC. (Eds.) 2009. Basic Concepts of Molecular Pathology. New York: Springer Science and Business Media. [Google Scholar]
- 13. Wu R, Shi ZR. 2004. Comparison of chromogenic in situ hybridization, fluorescence in situ hybridization, and immunohistochemistry In Hayat MA, (ed.). Handbook of IHC and ISH of Human Carcinomas: Molecular Genetics; Lung and Breast Carcinomas. Burlington, MA: Elsevier Academic Press; p 13–21. [Google Scholar]
- 14. Rayburn E, Zhang R, He J, Wang H. MDM2 and human malignancies: expression, clinical pathology, prognostic markers, and implications for chemotherapy. Curr Cancer Drug Targets 2005;5(1):27–41. [DOI] [PubMed] [Google Scholar]
- 15. Zhang R, Wang H. MDM2 oncogene as a novel target for human cancer therapy. Curr Pharm Des 2000;6(4):393–416. [DOI] [PubMed] [Google Scholar]
- 16. Dickens MP, Fitzgerald R, Fischer PM. Small‐molecule inhibitors of MDM2 as new anticancer therapeutics. Semin Cancer Biol 2010;20(1):10–18. [DOI] [PubMed] [Google Scholar]


