Abstract
Background
As the second leading cause of cancer morbidity and death in women, cervical cancer remains an important public health problem worldwide. Novel biomarkers with high sensitivity and specificity for the early detection and diagnosis of cervical cancer are urgently needed. Increasing evidence shows that long noncoding RNAs (lncRNAs) are differentially expressed in cancer tissues and may serve as diagnostic markers. In multiple tumor types, exosomes harboring lncRNAs are actively released from tumor cells. In this study, we investigate the potential association of exosomal lncRNA expression with cervical cancer.
Methods
Cervicovaginal lavage specimens were collected from patients with cervical cancer and cancer‐free volunteers who are HPV‐positive or HPV‐negative. Exosomes in these specimens were isolated by ultracentrifugation and confirmed by transmission electron microscopy. The exosomal lncRNAs HOTAIR, MALAT1, and MEG3 were quantified by qRT‐PCR.
Results
Expression of HOTAIR, MALAT1 and MEG3 was predominantly observed in cervical cancer‐derived exosomes in cervicovaginal lavage samples. The expression levels of lncRNAs were significantly different in exosomes isolated from cervical cancer patients compared to normal controls.
Conclusions
Our data suggest that lncRNAs in exosomes isolated from cervicovaginal lavage are differentially expressed in cervical cancer patients and cancer‐free volunteers. Exosomal lncRNAs may have great potential to be used for the early detection and diagnosis of cervical cancer, and serve as convenient and noninvasive biomarkers.
Keywords: cervical cancer, cervicovaginal lavage, exosome, lncRNA
Introduction
Cervical cancer is the third most common cancer in women worldwide 1. It remains the second leading cause of cancer‐related death in women, despite the fact that cervical cancer incidence has greatly reduced with the application of Papanicolaou test 2. Therefore, novel biomarkers with high sensitivity and specificity for the early detection and diagnosis of cervical cancer are urgently needed to promote survival.
Advances with the development of next generation sequencing technologies revealed that the human genome is pervasively transcribed into many newly described classes of noncoding RNAs. Long noncoding RNAs (lncRNAs) are RNA transcripts greater than 200 nucleotides in length and distinct from any other known class of small and structural RNAs 3, 4. They play important regulatory roles in gene expression 3, 4. Recent data suggest that the deregulated expression of lncRNAs, as detected either in tumor tissues and/or in biological fluids, strongly correlates with the cellular functions described as the hallmark processes of cancer 5, 6.
Accumulating evidence support that lncRNAs are involved in the development and progression of cervical cancer. The expression of lncRNA MALAT1 is significantly elevated in cervical cancer tissue samples compared to adjacent normal tissues and cervical tissues from HPV‐negative healthy volunteers 7. Furthermore, MALAT1 levels correlate with tumor size, FIGO stage, invasion, and metastasis and higher MALAT1 level associates with lower overall and recurrence‐free survival 7. In vitro studies indicate that MALAT1 promotes proliferation and invasion in cervical cancer cells Hela and CaSki 7. Another study reports that MALAT1 expression in cervical epithelial tissues positively correlates with HPV infection 8. MALAT1 may modulate the epithelial‐to‐mesenchymal transition (EMT) through regulating the expression of proteins involved in EMT, including E‐cadherin, ZO‐1, β‐catenin, vimentin, and snail 8. In addition, the lncRNA Hox transcript antisense intergenic RNA (HOTAIR) has also been shown to contribute to cervical cancer progression. HOTAIR expression is increased in cervical cancer tissues than in matching nontumor tissues 9. HOTAIR expression correlates with lymph node metastasis and decreased overall survival 9. The expression of maternally expressed gene 3 (MEG3) is decreased in cervical cancer tissue samples compared to adjacent benign tissues, and is negatively correlates with FIGO stage, tumor size, HR‐HPV infection, lymph node metastasis, and miR‐21 expression 10.
Exosomes are small membranous vesicles (30–100 nm) present in all biological fluids such as urine, blood, ascites, cervical secretions, and saliva 11, 12, 13. Exosomes carry proteins and genetic materials including DNA, mRNA, miRNA, and lncRNA, which may reveal genetic information of their parent cells 14. Moreover, it was recently reported that lncRNAs are enriched in secreted exosomes from cervical cancer HeLa cells 15, suggesting that exosomal lncRNAs can be developed into convenient and noninvasive biomarkers for the early detection, diagnosis, and prognosis of cervical cancer 16.
However, to our knowledge, the potential of using lncRNAs in exosomes in cervicovaginal lavage samples as a source for biomarkers for the early detection and diagnosis of cervical cancer has been not reported. Based on the findings from previous reports, lncRNAs MALAT1 7, HOTAIR 9, and MEG3 10 were chosen to be investigated in this study. Their expression levels in exosomes isolated from cervicovaginal lavages was examined and compared among cervical cancer patients and cancer‐free volunteers with HPV− or HPV+ status. The results of our study shed new light on the identification of new early detection and diagnostic markers for the deadly cervical cancer.
Materials and Methods
Participants
The cervicovaginal lavage samples of 30 cervical cancer patients were obtained in the Taizhou Municipal Hospital prior to definitive therapy. The normal control cervicovaginal lavage samples were collected from cancer‐free volunteers with or without HPV infection. Written consents were obtained from all subjects prior to the recruitment and the study protocol was approved by the Institutional Review Board of Hospital Ethics Committee. The clinical characteristics of the subjects are listed in Table 1.
Table 1.
Clinical characteristics of patients and volunteers
| Noncancer volunteers (HPV‐negative) | Noncancer volunteers (HPV‐positive) | Cervical cancer patients | |
|---|---|---|---|
| Case,n | 30 | 30 | 30 |
| Age (years) | 43.5 ± 5.7 | 45.0 ± 7.6 | 47.5 ± 3.1 |
| Differentiation | |||
| Well | 12 | ||
| Moderate | 8 | ||
| Poor | 10 | ||
Exosome Isolation from Cervicovaginal Lavage Samples
Exosomes from cervicovaginal lavage samples were prepared as previously described 17. Briefly, 10 ml of sterile phosphate buffered saline (PBS) was utilized for the vagina and cervix lavage. The lavage fluid converging at the posterior fornix was collected for exosome isolation by differential centrifugation. Lavage fluid was centrifuged at 3,000 × g for 10 min at 4°C to remove the cells and debris. The supernatants were transferred to microcentrifuge tubes and centrifuged at 12,000 × g for 30 min at 4°C to completely remove cellular components. Supernatants were centrifuged at 100,000 × g for 2 h at 4°C to pellet the exosomes. Exosome‐depleted supernatants were collected as well for lncRNA expression studies.
Transmission Electron Microscopy (TEM) and ELISA
The exosome pellets were resuspended in PBS and placed onto formvar carbon coated electron microscopy grids, then negatively stained with 10% uranyl acetate. The microphotographs were obtained using a JEOL 100XCII electron microscope (JEOL, Peabody, MA, USA). The presence of exosomes was further confirmed by assessing the expression of exosome marker protein CD63 using sandwich ELISA based on the manufacturer's protocol (System Biosciences, Mountain View, CA, USA).
Quantification of Exosomal lncRNAs
Total RNA was isolated from exosomes using isothiocyanate‐phenol/chloroform extraction procedures. Quantitative RT‐PCR (qRT‐PCR) was performed using SYBR® Premix DimerEraser kit (TaKaRa, Shiga, Japan) on an ABI Prism 7900HT Detection System. The primer sequences of lncRNAs were obtained from previous publications 7, 9, 18. GAPDH was used as a housekeeper gene for the qRT‐PCR reaction. Each test was done in triple replication and the 2−ΔΔCt method was used to calculate the expression of lncRNAs in all cervicovaginal Lavage samples.
Statistical Analysis
All statistical analyses were done with STATA 10.0 (StataCorp LP, College Station, TX, USA). A P‐value of <0.05 was considered significant.
Results
Characterization of Isolated Exosomes in Cervicovaginal Lavage Samples
To ensure the quality of exosomes isolated from cervicovaginal lavage samples, we characterized the microvesicles by electron microscopy and ELISA. Under electron microscopy, round vesicle structures with sizes varying between 30–100 nm were observed (Fig. 1), which is consistent with previously reported structural characteristics of exosomes 19. The presence of exosomes was further confirmed by the commonly used exosomal protein marker CD63 by ELISA (data not shown).
Figure 1.
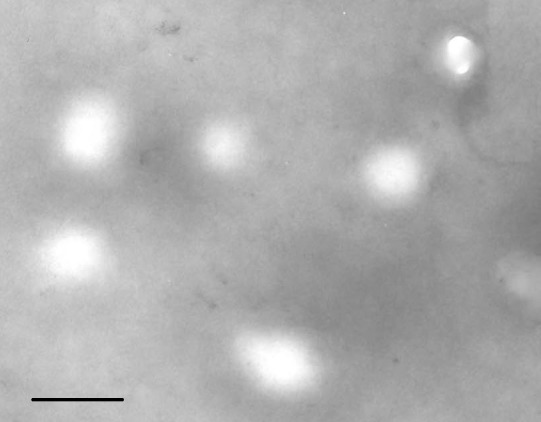
Validation of exosomes isolated from cervicovaginal lavage samples. Morphological characterization of exosomes isolated from cervicovaginal lavage samples is performed by transmission electron microscopy. Bar, 100 nm.
LncRNAs are Enriched in Exosomes Isolated from Cervicovaginal Lavage Specimens
Exosomal lncRNA expression in cervicovaginal lavage samples of cervical cancer patients is unknown. Thus, we first examined the expression of three lncRNAs with potential involvement in cervical cancer, MALAT1, HOTAIR, and MEG3, in exosomes isolated from the control cervicovaginal lavages and exosome‐depleted supernatants by qRT‐PCR. The concentrations of all three lncRNAs were significantly higher in exosomes than in exosome‐depleted supernatants (P < 0.01, Fig. 2a), indicating that these lncRNAs are enriched in exosomes.
Figure 2.
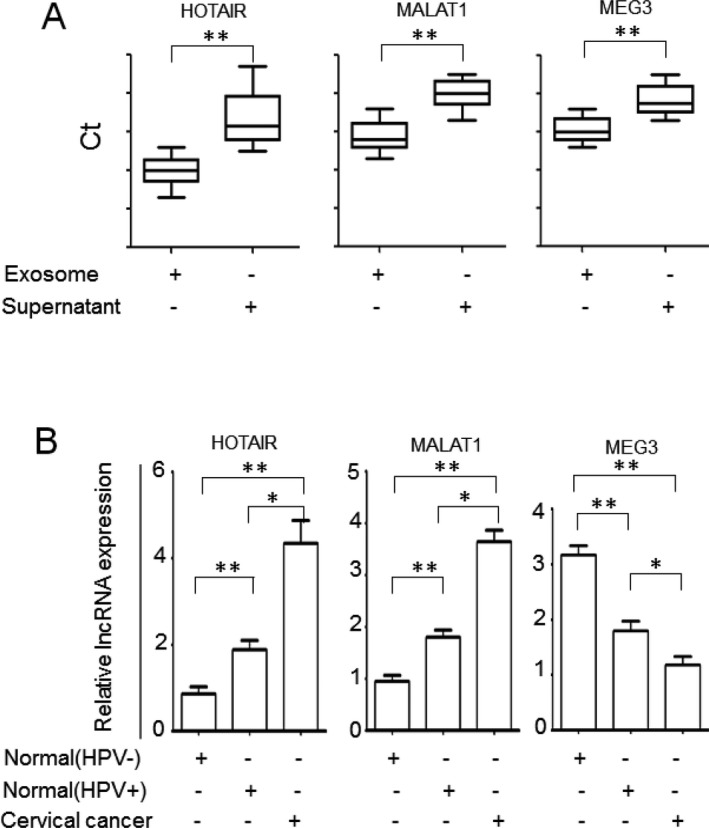
LncRNA expression in cervicovaginal lavage samples of cervical cancer. A. LncRNAs are enriched in exosomes isolated from cervicovaginal lavage samples. The levels of lncRNAs in exosome pellets and exosome‐depleted supernatants were determined by qRT‐PCR. The absolute Ct values are presented. B. The exosomal lncRNA expression in cervicovaginal lavage samples from patients with cervical cancer and cancer‐free control subjects was determined by qRT‐PCR. The samples were run in triplicate. *, P < 0.05; **, P < 0.01.
LncRNAs are Differentially Expressed in Cervicovaginal Lavage Samples of Cervical Cancer
To investigate whether exosomal lncRNAs MALAT1, HOTAIR, and MEG3 are differentially expressed in cervical cancer samples, we analyzed exosomal lncRNA levels in a cohort consisting of 30 cervical cancer patients, 30 cancer‐free HPV‐positive subjects and 30 HPV‐negative normal subjects by qRT‐PCR. The results indicated that the expression of exosomal HOTAIR and MALAT1 in cervicovaginal lavage samples of the cervical cancer patients was significantly higher than that of the HPV‐positive subjects and HPV‐negative cancer‐free subjects (P < 0.01 and P < 0.05, respectively, Fig. 2b). The exosomal MEG‐3 level in cervicovaginal lavage specimens of the cervical cancer patients was significantly lower than those of the HPV‐positive subjects and HPV‐negative cancer‐free subjects (P < 0.01 and P < 0.05, respectively, Fig. 2b). Differential expression of exosomal HOTAIR, MALAT1, and MEG3 was also found between the HPV‐positive and HPV‐negative groups (P < 0.01, Fig. 2b). Higher expression levels of HOTAIR and MALAT1 and lower expression level of MEG3 were observed in HPV‐positive samples compared to HPV‐negative samples (P < 0.01, Fig. 2b). Our results indicate that lncRNAs HOTAIR, MALAT1, and MEG3 may contribute to cervical cancer development.
Discussion
Although well‐run screening programs for cervical cancer in the population at risk result in a sharp decrease in the incidence and mortality of cervical cancer, cervical cancer remains the second leading cause of cancer‐related death in women worldwide. Thus, there is an urgent need for more sensitive and accurate biomarkers for the early detection and diagnosis of cervical cancer.
Generally, the development of noncoding RNA (ncRNA) biomarkers is based on the evaluation of their expression levels in tumor tissues. However, despite reasonable correlation between ncRNA expression and patient diagnosis, this approach is limited by the required invasive procedures for tissue collection. This limitation prompted the analysis of cell‐free ncRNAs, which are present in various bodily fluids or enclosed in extracellular vesicles secreted by cells. So far, circulating ncRNAs are considered suitable biomarkers for cancer diagnosis and prognosis, because they are remarkably stable and can be detected by common techniques such as qRT‐PCR, Nevertheless, their absolute concentrations in body fluids are usually low, impacting their accurate quantification 20. Our findings that lncRNAs HOTAIR, MALAT1, and MEG3 are enriched in exosomes isolated from cervicovaginal lavage samples have opened up a new opportunity to develop extracellular ncRNA‐based biomarkers in body fluids. This approach is convenient, noninvasive, and likely to be better tolerated by patients than conventional tissue biopsies, in cervical cancer.
Exosomes are secreted by most cell types including cancer cells and are found in all body fluids 21. Recently, exosomal miRNAs have been extensively studied and well accepted as biomarkers for diagnosis and prognosis of different types of cancer. LncRNAs are present in a variety of tissues during homeostasis at much lower levels than protein‐coding genes, but their expression may be more tissue specific than protein‐coding genes 22.
Currently, plasma/serum and urine are the most commonly used body fluids for lncRNA profiling in cancer patients. However, a recent study indicated that the levels of the lncRNA AA174084 in gastric juice of gastric cancer patients were significantly higher in healthy individuals than in patients with other gastric mucosa lesions 23. In contrast, the levels of the same lncRNA in the plasma could not distinguish gastric cancer patients from healthy individuals, indicating that other body fluids may provide more suitable specimens to assess lncRNA expressions in the primary tumor tissues, particularly for epithelium‐derived cancers, such as cervical cancers.
We found that lncRNA MATAL1, HOTAIR, MEG3 are differentially expressed in exosomes isolated from cervicoviginal lavage samples compared to nontumor samples. In addition, the expression levels of these lncRNAs are different between HPV‐positive and HPV‐negative cancer‐free individuals. Our results are supported by previous findings that the HPV infection may contribute to the regulation of HOTAIR, MALAT1, and MEG3 in cervical tissues 10, 24, 25.
In summary, we observed enrichment of HOTAIR, MALAT1, and MEG3 in the exosomes of cervicovaginal lavage samples compared to exosome‐depleted supernatants. Deregulation of these lncRNAs was observed between cervical cancer and nontumor groups. The emerging use of micro‐ and nanotechnology to isolate and detect exosomes in clinical samples has promoted the research on the utility of exosomes as diagnostic and prognostic tools 26. Furthermore, screening a large cohort of lavage samples from cervical cancer patients would determine the value of exosomal lncRNAs as potential biomarkers for the early detection and diagnosis of cervical cancer.
Acknowledgments
This study was supported by the General Research Grants from Zhejiang Medicine Health Science and Technology Projects (2013KYA230).
Contributor Information
Hong Yuan, Email: yuanhonglab@163.com.
Da‐Kang Hu, Email: 89434436@qq.com.
References
- 1. International Collaboration of Epidemiological Studies of Cervical C , Appleby P, Beral V, et al. Cervical cancer and hormonal contraceptives: Collaborative reanalysis of individual data for 16,573 women with cervical cancer and 35,509 women without cervical cancer from 24 epidemiological studies. Lancet 2007;370:1609–1621. [DOI] [PubMed] [Google Scholar]
- 2. Peng L, Yuan X, Jiang B, Tang Z, Li GC. LncRNAs: Key players and novel insights into cervical cancer. Tumour Biol 2015;Dec 29. [DOI] [PubMed] [Google Scholar]
- 3. Spizzo R, Almeida MI, Colombatti A, Calin GA. Long non‐coding RNAs and cancer: A new frontier of translational research? Oncogene 2012;31:4577–4587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Mercer TR, Mattick JS. Structure and function of long noncoding RNAs in epigenetic regulation. Nat Struct Mol Biol 2013;20:300–307. [DOI] [PubMed] [Google Scholar]
- 5. Gupta RA, Shah N, Wang KC, et al. Long non‐coding RNA HOTAIR reprograms chromatin state to promote cancer metastasis. Nature 2010;464:1071–1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Hirata H, Hinoda Y, Shahryari V, et al. Long Noncoding RNA MALAT1 Promotes Aggressive Renal Cell Carcinoma through Ezh2 and Interacts with miR‐205. Cancer Res 2015;75:1322–1331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Yang L, Bai HS, Deng Y, Fan L. High MALAT1 expression predicts a poor prognosis of cervical cancer and promotes cancer cell growth and invasion. Eur Rev Med Pharmacol Sci 2015;19:3187–3193. [PubMed] [Google Scholar]
- 8. Sun R, Qin C, Jiang B, et al. Down‐regulation of MALAT1 inhibits cervical cancer cell invasion and metastasis by inhibition of epithelial‐mesenchymal transition. Mol BioSyst 2016;12:952–962. [DOI] [PubMed] [Google Scholar]
- 9. Kim HJ, Lee DW, Yim GW, et al. Long non‐coding RNA HOTAIR is associated with human cervical cancer progression. Int J Oncol 2015;46:521–530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Zhang J, Yao T, Wang Y, Yu J, Liu Y, Lin Z. Long noncoding RNA MEG3 is downregulated in cervical cancer and affects cell proliferation and apoptosis by regulating miR‐21. Cancer Biol Ther 2016;17:104–113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Skog J, Wurdinger T, van Rijn S, et al. Glioblastoma microvesicles transport RNA and proteins that promote tumour growth and provide diagnostic biomarkers. Nat Cell Biol 2008;10:1470–1476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Pisitkun T, Shen RF, Knepper MA. Identification and proteomic profiling of exosomes in human urine. Proc Natl Acad Sci U S A 2004;101:13368–13373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Lasser C, Alikhani VS, Ekstrom K, et al. Human saliva, plasma and breast milk exosomes contain RNA: Uptake by macrophages. J Transl Med 2011;9:9 Epub 2011/01/18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Xiao D, Ohlendorf J, Chen Y, et al. Identifying mRNA, microRNA and protein profiles of melanoma exosomes. PLoS ONE 2012;7:e46874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Gezer U, Ozgur E, Cetinkaya M, Isin M, Dalay N. Long non‐coding RNAs with low expression levels in cells are enriched in secreted exosomes. Cell Biol Int 2014;38:1076–1079. [DOI] [PubMed] [Google Scholar]
- 16. Nicholas J. A new diagnostic tool with the potential to predict tumor metastasis. J Natl Cancer Inst 2013;105:371–372. [DOI] [PubMed] [Google Scholar]
- 17. Coutlee F, Hankins C, Lapointe N. Comparison between vaginal tampon and cervicovaginal lavage specimen collection for detection of human papillomavirus DNA by the polymerase chain reaction. The Canadian Women's HIV Study Group. J Med Virol 1997;51:42–47. [DOI] [PubMed] [Google Scholar]
- 18. Qin R, Chen Z, Ding Y, Hao J, Hu J, Guo F. Long non‐coding RNA MEG3 inhibits the proliferation of cervical carcinoma cells through the induction of cell cycle arrest and apoptosis. Neoplasma 2013;60:486–492. [DOI] [PubMed] [Google Scholar]
- 19. Thery C, Zitvogel L, Amigorena S. Exosomes: Composition, biogenesis and function. Nat Rev Immunol 2002;2:569–579. [DOI] [PubMed] [Google Scholar]
- 20. Huang JL, Zheng L, Hu YW, Wang Q. Characteristics of long non‐coding RNA and its relation to hepatocellular carcinoma. Carcinogenesis 2014;35:507–514. [DOI] [PubMed] [Google Scholar]
- 21. Lee TH, D'Asti E, Magnus N, Al‐Nedawi K, Meehan B, Rak J. Microvesicles as mediators of intercellular communication in cancer–the emerging science of cellular ‘debris'. Semin Immunopathol 2011;33:455–467. [DOI] [PubMed] [Google Scholar]
- 22. Derrien T, Johnson R, Bussotti G, et al. The GENCODE v7 catalog of human long noncoding RNAs: Analysis of their gene structure, evolution, and expression. Genome Res 2012;22:1775–1789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Shao Y, Ye M, Jiang X, et al. Gastric juice long noncoding RNA used as a tumor marker for screening gastric cancer. Cancer 2014;120:3320–3328. [DOI] [PubMed] [Google Scholar]
- 24. Sharma S, Mandal P, Sadhukhan T, et al. Bridging Links between Long Noncoding RNA HOTAIR and HPV Oncoprotein E7 in Cervical Cancer Pathogenesis. Sci Rep 2015;5:11724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Jiang Y, Li Y, Fang S, et al. The role of MALAT1 correlates with HPV in cervical cancer. Oncol Lett 2014;7:2135–2141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Ko J, Carpenter E, Issadore D. Detection and isolation of circulating exosomes and microvesicles for cancer monitoring and diagnostics using micro‐/nano‐based devices. Analyst 2016;141:450–460. [DOI] [PMC free article] [PubMed] [Google Scholar]


