Abstract
Background
Filaggrin gene (FLG) plays an important role in skin barrier function, and loss‐of‐function mutations of FLG have been shown to be a predisposing factor for atopic dermatitis (AD). The c.3321delA mutation is the most common FLG mutation in Chinese population. We aim to develop a rapid, cost‐efficiency, and reliable closed‐tube method that has not been described for the detection of c.3321delA mutation.
Methods
Recombinant wild‐type and mutant plasmids of c.3321delA mutation were constructed, heterozygous mutant plasmids were prepared by mixing the mutant plasmids and wild‐type plasmids at 1:1 ratio. High‐resolution melting analysis (HRMA) coupled with an unlabeled DNA probe was employed to identify the shift in melting temperature of the probe–template complex, which reflects the presence of c.3321delA mutation.
Results
Unlabeled probe based HRMA was able to distinguish all three genotypes (wild‐type, heterozygote, and mutant) of c.3321delA mutation. Then, we applied this method to genotype 1,317 clinical samples. Genotyping results obtained from unlabeled probe HRMA were 100% concordant with the results from direct sequencing.
Conclusion
We developed a fast and high‐throughput method to detect the c.3321delA mutation.
Keywords: atopic dermatitis, c.3321delA mutation, filaggrin gene, skin barrier, unlabeled probe based high‐resolution melting analysis
Introduction
Filaggrin gene (FLG), one of the key genes involved in skin barrier function, is located in the epidermal differentiation complex on chromosome 1q21 1. Filaggrin encoded by FLG is an important component of the granular cell layer in the epidermis, which aggregates keratin filaments, leading to keratinocyte compaction and formation of the stratum corneum 2. Filaggrin deficiency due to loss‐of‐function mutations in FLG resulted in the loss of integrity in the stratum corneum and an associated increase in transepidermal water loss 2. This has been showed to be associated with diseases with the cardinal feature of dry skin, including atopic dermatitis (AD) 3, 4 and psoriasis 5, 6. To date, approximately 60 loss‐of‐function FLG mutations have been identified in European and Asian populations 7. FLG variant rs3126085 has been identified to correlate with an Asian‐specific FLG mutation c.3321delA, which is the most common FLG mutation in the Chinese population 7.
Several methods have been developed to genotype c.3321delA mutation, such as sequencing 4, restriction fragment length polymorphism (RFLP) 8, and real‐time PCR 9. However, a quick, cost‐effective, and reliable genotyping method has not been described for its detection. In present study, we developed an unlabeled probe high‐resolution melting analysis (HRMA) for detection of FLG mutation c.3321delA.
Materials and Methods
Patients and Sample Collection
Peripheral blood cells’ samples were obtained from 436 patients with AD, 411 patients with psoriasis, and 470 healthy donors recruited from Shenzhen Hospital, Peking University. The study was approved by the ethics committee of the Shenzhen Hospital, Peking University, and conducted according to the principles of the Declaration of Helsinki. Written informed consent was obtained from all participants.
Design of Primers and Unlabeled Probe
Primers and an unlabeled probe were designed with Primer3 software and synthesized by standard phosphoramidite chemistry (Invitrogen, Shanghai, China). Several factors were taken into account during design of the primers. Sequence variation was positioned at the center of amplicons. Primers, used in control plasmid of c.3321delA variant producing 494‐bp amplicon, was inappropriate for HRMA. Therefore, primers resulting in 112‐bp amplicon were designed for c.3321delA variant, but they failed to distinguish sample genotypes in melting analysis. Thus, we designed an unlabeled probe that was blocked on the 3’‐hydroxyl terminus with a three‐carbon (C3) alkyl group to prevent extension by Taq polymerase during PCR. Sequences of primers, probe, and amplicon lengths for c.3321delA are shown in Table 1.
Table 1.
Primers and Probe Used for PCR Amplification
| Primers/probe | Sequence 5′–3′ | Amplicon length (bp) |
|---|---|---|
| Forward primer (F1) | TGAACAGGCAAGATCAAGTC | 494 |
| Reverse primer (R1) | TATCTACCGATTGCTCATGG | |
| Middle forward primer (mF) | GTGACTGGTCGGGGGAAGGTCTGGACGTTC | |
| Middle reverse primer (mR) | CCTTCCCCCGACCAGTCACGTGCGGACTC | |
| Forward primer (F2)a | GAGTGCTCACCTGGTAGAT | 112 |
| Reverse primer (R2)a | TCAGGCCATGGACAGGA | |
| Probe | TCCAGACCTTCCCCCTGACCAGTCACGT |
Primers used only for PCR‐HRMA.
The underlined base in the probe sequence indicates the position of the variation.
Construction of Recombinant Plasmids
Based on the sequence of FLG (GeneBank: NC_000001.11), two pairs of primers were designed for amplifying the objective fragments (Table 1). A 494‐bp region of the wild‐type genotype of FLG was amplified from human genomic DNA (gDNA) using the primers F1 and R1. Fifty microliter reaction mixtures contained 1 μl of gDNA (40–60 ng), 5 μl Pfu PCR buffer (Fermentas, 10×), 1 μl dNTPs (200 μM), 10 μl MgCl2 (2.5 mM), 1 μl primers (200 nM), 1 μl dimethylsulfoxide, and 1 μl Pfu DNA Polymerase (Fermentas, 1.25 U). DNA amplification was performed in a S1000 Thermal Cycler (Bio‐Rad) with the following program: 95˚C for 3 min, 34 cycles of 95˚C for 30 s, 55˚C for 30 s, and 72˚C for 70 s. The PCR products were cloned into the pMD‐19 T simple vector (Takara, Dalian, China). Clones were verified by sequencing. The mutant genotype of FLG c.3321delA was obtained by overlap‐PCR with the primers containing c.3321delA mutant site. DNA amplification and next cloning steps were performed as described above. Both wild‐type and mutant plasmids were quantified by spectrophotometry and diluted to 0.5 ng to generate a standard curve. By mixing the mutant plasmids and wild‐type plasmids at 1:1 ratio, we prepared heterozygous mutant plasmids.
Genotyping
gDNA was isolated from peripheral blood cells by using TIANamp genomic DNA kit (Tiangen, Beijing, China) according to manufacturer's instruction, quantified by NanoDrop 2000 spectrophotometer (ThermoScientific) and stored at –80˚C until use. Genotyping was assayed by HRMA with one pair of primers and unlabeled probe (Table 1). Briefly, asymmetric PCR reaction was performed in a volume of 20 μl containing 2 μl TrueStart HotStart Taq buffer (Fermentas, 10×), 0.4 μl dNTPs (200μM), 2 μl MgCl2 (2.5 mM), 1.6 μl forward primer F2 (80 nM), 0.8 μl (each) excess reverse primer R2 and C3‐block probe (400 nM), and 0.2 μl TrueStart HotStart Taq DNA Polymerase (Fermentas, 1.0 U). The PCR reactions were performed in a S1000 Thermal Cycler; the conditions were 95˚C for 3 min, followed by 50 cycles at 95˚C for 10 s and 68˚C for 20 s and a final extension at 72˚C for 5 min. The 10 μl of PCR products were supplied with 1.0 μl LC Green Plus (Idaho) and then subjected to HRM in LightCycler 480 SW 1.5 software (Roche) or HR‐1 high‐resolution melting instrument (Idaho). The samples were first denatured at 95˚C for 30 s and rapidly cooled to 40˚C for 30 s and then melted from 60 to 95˚C with a 0.3˚C/s ramp rate. After PCR, melting curve analysis was performed using the LightCycler 480 (Roche) or LightScanner (Idaho). For each assay, we always include positive and negative controls (homozygous wild‐type, heterozygous, and homozygous mutant).
Sequencing
To verify the outcome by the unlabeled probe HRMA, we amplified ten samples as determined by unlabeled probe HRMA using the same primer pairs of unlabeled probe HRMA for sequencing analysis in both directions (Invitrogen), in comparison with the FLG reference DNA sequence.
Results
The sensitivity and accuracy of HRMA with unlabeled probes were dramatically improved 10. In this study, we designed an unlabeled probe to complement the wild‐type genotype of FLG mutation c.3321delA. After asymmetric PCR, full‐length duplex product and excess single‐stranded product complementary to the probe were produced. As showed in Figure 1A, the entire melting profile could be divided into two regions, representing the melting of probe–product and product–product strands, respectively. With/without allele A resulted in different probe–product melting transitions based on the stability of the mismatch present. It was clear to see these transitions by plotting the negative derivative (dF/dT) of fluorescence (F) versus temperature (T, Fig. 1C).
Figure 1.

HRMA of FLG mutation c.3321delA using an unlabeled probe in control plasmids. (A) The melting peaks include both unlabeled probe and PCR products region. (B and C) The melting peaks only contain unlabeled probe region. Genotypes are indicated in the figures. △A, c.3321delA mutation.
The probe‐target melted at 73–80˚C. A perfectly matched probe‐target hybrid has a characteristic melting temperature that is higher than a mismatched hybrid. In our study, the probe was designed to complement the wild‐type c.3321A; thus, the dissociation of the probe occurs at a lower temperature when there is a mismatch between the probe and template. Analysis of this melting region clearly identified the genotype. The wild‐type plasmids (harboring the allele A) and the mutant plasmids (without the allele A) showed derivative melting peaks at 78.2˚C and 75.8˚C, respectively. The heterozygous plasmid showed two peaks, one at each temperature representing the combination of both alleles (Fig. 1C). Therefore, this unlabeled probe was able to distinguish all three genotypes (wild‐type, heterozygote, and mutant; (Fig. 1).
Then, this method was used to screen DNA from blood samples (Fig. 2). DNA samples of 436 patients with AD, 411 patients with psoriasis, and 470 healthy donors were successfully genotyped for c.3321delA mutation by unlabeled probe HRMA. As presented in Figure 2, three types of melting curves were displayed, which correlated with wild‐type (A/A), heterozygous (A/△A), and mutant homozygous (△A/△A) genotypes. Among 1,317 clinical samples, we found one homozygous mutant sample (in one AD patient), and 65 heterozygous samples (34 in AD patients, 16 in psoriasis patients, and 15 in healthy donors). To compare the concordant of unlabeled probe HRMA with previously described direct DNA sequencing method, we amplified ten samples (one homozygous mutant sample, three heterozygous samples, and six wild‐type samples) as determined by unlabeled probe HRMA for DNA sequencing analysis. As a result, genotyping results obtained with unlabeled probe HRMA were 100% concordant with that obtained from direct sequencing (Fig. 3).
Figure 2.
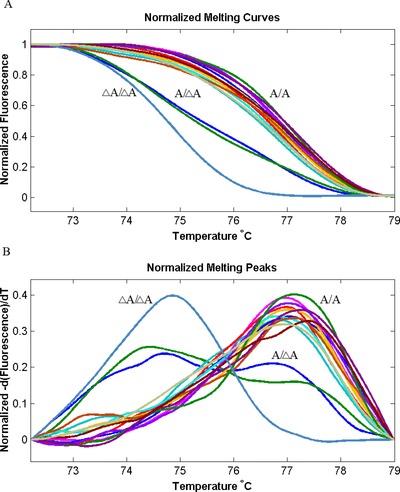
HRMA of FLG mutation c.3321delA using an unlabeled probe in samples. (A and B) The melting peaks contain unlabeled probe region. Genotypes are indicated in the figures. △A, c.3321delA mutation.
Figure 3.
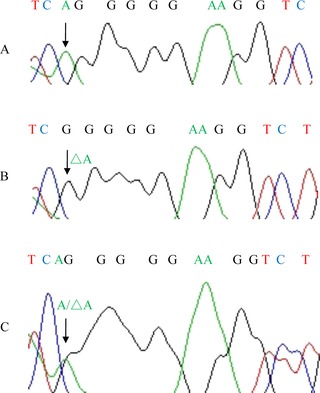
Direct sequence results of three genotypes of FLG mutation c.3321delA in samples. (A) Wild‐type; (B) mutant type; (C) heterozygote.
Discussion
HRMA is emerging as an inexpensive, accurate, and rapid method of genotyping and mutation scanning 11, 12. Unlabeled probe HRMA is a new modified HRMA 11, 12, 13. As shown in our study, unlabeled probe HRMA was proved to be a fast and cost‐effective genotyping method for FLG mutation c.3321delA. During an asymmetric PCR with both saturate dye LCGreen Plus and ∼30 bp C3‐blocked unlabeled oligonucleotide probes, the excess sense strand of the amplicon is generated, which allows the probe to anneal and form a duplex. Then LCGreen Plus binds to the duplex to generating a fluorescent signal 12. During HRMA, the probe dissociates from the amplicon, resulting in a decrease in the fluorescent signal. If the probe and the amplicon are not perfectly matched, the probe will dissociate from the DNA template at lower temperature 13. High resolution of different genotypes can be achieved with the melt‐curve profiles for the probe–amplicon complex. In this study, we developed an accurate HRMA method with unlabeled probe for detecting the c.3321delA mutation.
In general, the PCR‐RFLP 8 and sequencing 4, 8 are common alterations for the detection of c.3321delA mutation due to the large and repetitive structure of FLG. However, both RFLP and sequencing have limitations in sensitivity/cost and increase the risk of contamination of PCR products. Furthermore, sequencing requires special sequencing facilities and sample preparation reagents and steps. For this reason, Kono et al. established a high‐throughput FLG mutation detection system by real‐time PCR 9. However, this resulted in an increase in cost compared with any other analysis for the use of expensive reagents such as the fluorescence. In conclusion, we developed a novel unlabeled probe HRMA to detect the c.3321delA mutation with benefits including the high throughput, elimination of labor costs, and prevention of cross contamination.
Conflicts of Interest
None declared.
Grant sponsor: National Natural Scientific Foundation of China; Grant numbers: 81271755, 81371737; Grant sponsor: Guangdong Natural Scientific Foundation; Grant number: 2014A030313708; Grant sponsor: Shenzhen Research Grant; Grant numbers: CXZZ20140416144209739, JCYJ20130329110752142, KQCX20120803145850990.
Contributor Information
Wei Zhang, Email: zhangweispace@yeah.net.
Xia Dou, Email: douxia@medmail.com.cn.
Bo Yu, Email: yubomd@hotmail.com.
References
- 1. Sandilands A, Terron‐Kwiatkowski A, Hull PR, et al. Comprehensive analysis of the gene encoding filaggrin uncovers prevalent and rare mutations in ichthyosis vulgaris and atopic eczema. Nat Genet 2007;39:650–654. [DOI] [PubMed] [Google Scholar]
- 2. McGrath JA, Uitto J. The filaggrin story: Novel insights into skin‐barrier function and disease. Trends Mol Med 2008;14:20–27. [DOI] [PubMed] [Google Scholar]
- 3. Zhang H, Guo Y, Wang W, Yu X, Yao Z. Associations of FLG mutations between ichthyosis vulgaris and atopic dermatitis in Han Chinese. Allergy 2011;66:1253–1254. [DOI] [PubMed] [Google Scholar]
- 4. Zhang H, Guo Y, Wang W, Shi M, Chen X, Yao Z. Mutations in the filaggrin gene in Han Chinese patients with atopic dermatitis. Allergy 2011;66:420–427. [DOI] [PubMed] [Google Scholar]
- 5. Hu Z, Xiong Z, Xu X, Li F, Lu L, Li W, et al. Loss‐of‐function mutations in filaggrin gene associate with psoriasis vulgaris in Chinese population. Hum Genet 2012;131:1269–1274. [DOI] [PubMed] [Google Scholar]
- 6. Chang YC, Wu WM, Chen CH, Hu CF, Hsu LA. Association between P478S polymorphism of the filaggrin gene and risk of psoriasis in a Chinese population in Taiwan. Arch Dermatol Res 2008;300:133–137. [DOI] [PubMed] [Google Scholar]
- 7. Meng L, Wang L, Tang H, et al. Filaggrin gene mutation c.3321delA is associated with various clinical features of atopic dermatitis in the Chinese Han population. PloS One 2014;9:e98235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Ma L, Zhang L, Di ZH, et al. Association analysis of filaggrin gene mutations and atopic dermatitis in Northern China. Br J Dermatol 2010;162:225–227. [DOI] [PubMed] [Google Scholar]
- 9. Kono M, Nomura T, Ohguchi Y, et al. Comprehensive screening for a complete set of Japanese‐population‐specific filaggrin gene mutations. Allergy 2014;69:537–540. [DOI] [PubMed] [Google Scholar]
- 10. Cruz RE, Shokoples SE, Manage DP, Yanow SK. High‐throughput genotyping of single nucleotide polymorphisms in the Plasmodium falciparum dhfr gene by asymmetric PCR and melt‐curve analysis. J Clin Microbiol 2010;48:3081–3087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Zhou L, Wang L, Palais R, Pryor R, Wittwer CT. High‐resolution DNA melting analysis for simultaneous mutation scanning and genotyping in solution. Clin Chem 2005;51:1770–1777. [DOI] [PubMed] [Google Scholar]
- 12. Zhou L, Myers AN, Vandersteen JG, Wang L, Wittwer CT. Closed‐tube genotyping with unlabeled oligonucleotide probes and a saturating DNA dye. Clin Chem 2004;50:1328–1335. [DOI] [PubMed] [Google Scholar]
- 13. Montgomery J, Wittwer CT, Palais R, Zhou L. Simultaneous mutation scanning and genotyping by high‐resolution DNA melting analysis. Nat Protoc 2007;2:59–66. [DOI] [PubMed] [Google Scholar]


