Abstract
Background
In recent research, it has been shown that rs2289487 within the PLIN1 gene has different variants that have been associated with obesity, type 2 diabetes, and other diseases. However, the isochizomers such as the BsmI enzyme required for detection of this polymorphism through polymerase chain reaction‐restriction fragment length polymorphism (PCR‐RFLP) method are expensive. In this study, we aimed to explore a novel PCR‐RFLP method for identifying the single‐nucleotide polymorphism (SNP) of rs2289487 of PLIN1 gene.
Methods
A new restriction enzyme site was created through created restriction site PCR. In the forward primer, a deoxynucleotide A was substituted with C, and after PCR a new restriction enzyme site for BstUI was introduced into the PCR products. A total of 108 samples from Han Chinese were tested to evaluate this new method.
Results
Allele frequencies in the Asian population were 0.326 for allele A and 0.674 for allele G, and the genotype frequencies were 12.8% for AA, 39.5% for AG, and 47.7% for GG.
Conclusion
The PCR‐RFLP with new site for detecting the SNP of rs2289487 is an improved method with low cost and high accuracy.
Keywords: PCR‐RFLP, created restriction site, single‐nucleotide polymorphism, rs2289487, PLIN1
Introduction
In 2005, Rankinen et al. found that perilipin1 (PLIN1) gene was incorporated into the related genes of obesity, which were located on chromosome 15q26.1 1. Previous studies have suggested the area was associated with high blood pressure, high triglyceride levels, and diabetes 2, 3. Perilipin encoded by PLIN1 gene is the founding member of a family of proteins that share sequence similarity and the ability to bind lipid droplets. When first grouped as a family, these proteins were termed the “PAT family” after the first three members identified: perilipin, adipocyte differentiation‐related protein (ADRP), and tail‐interacting protein of 47 kDa (TIP47), S3‐12 and OXPAT; and perilipin was the most representative member in the PAT family 4.
Recently published work showed that the variants of rs2289487 within the PLIN1 gene are linked to obesity 5, 6, 7, 8, type 2 diabetes 9, and other diseases 10. To detect this single‐nucleotide polymorphism (SNP), several assays had been developed, for instance, fluorescently labeled probes technique 11, TaqMan assay 12, and polymerase chain reaction‐restriction fragment length polymorphism 13. Among them, PCR‐RFLP was the most popular due to high sensitivity and reliability and many restriction enzymes were cheap and commercial available. However, the current PCR‐RFLP assay was still facing practical challenges and was not suitable for high‐throughput screening. In this study, we demonstrated an improved PCR‐RFLP assay for the detection of a polymorphism of PLIN1 gene and examined its application in population.
Materials and Methods
Primer Design
The sequence of PLIN1 gene and polymorphism rs2289487 was from NCBI website (http://www.ncbi.nlm.nih.gov). The method of amplification‐created restriction site (CSR) was used to introduce a new enzyme site to screen the variant. The sequences of primer were designed with primer premier5.0 software as below:
Forward: 5′ CTGTGGGAGGGAAGGTGCG 3′
Reverse: 5′ CACTCCCCTGATACCTACTGG 3′.
The DNA sequence for primer designing is the following:
CTGTGGGAGGGAAGGTGCGcrttctaggcagtggggtcgggatacacaggcagggaggcagtaccccagaaagaccagaaaaatgtttggagaagccaccaatcaaaggtggactaaaactctagacattaacaacacttgctctctaCCAGTAGGTATCAGGGGAGTG.
The r represents polymorphism A/G. The length of target DNA sequence is 169 bp. The original “A” deoxynucleotide in the DNA sequence was substituted by a “C” in the forward primer (underlined) in order to introduce a restriction enzyme site “CGCG” for the BstUI restriction endonuclease.
Genotype of DNA
Genomic DNA was extracted from peripheral blood leukocytes according to the manufacturer's instructions. PCR was performed in a total volume of 25 μL containing 100 ng of genomic DNA, 1× PCR master Mix (Tiangen), and 5 pmol of each primer. DNA was denatured at 95℃ for 3 min, and thermal cycles were set at 95℃ for 30 s, 56℃ for 40 s, and 72℃ for 30 s for 30 cycles, followed by a final extension at 72℃ for 5 min. PCR products were electrophoresed on 3% agarose gels stained with ethidium bromide.
Enzyme digestion was conducted in a 20 μL final volume with 5U of BstUI enzyme (New England Biolabs [NEB]) and 10 μL of PCR product. The reaction was incubated at 60℃ overnight. The digested products were visualized on 3% agarose gels stained with ethidium bromide.
Results
Through CRS‐PCR, we successfully introduced a BstUI restriction enzyme site into the PCR products. As shown in Figure 1, we found that the full length of PCR was 169 bp as we expected. After digestion with BstUI, homozygous allele A had only one 169‐bp band. The homozygous allele G showed two bands of 150 and 19 bp, respectively.
Figure 1.
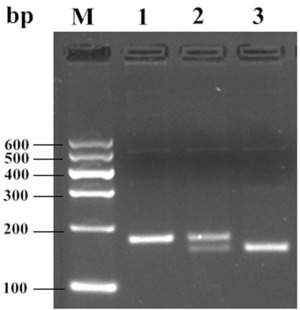
BstUI PCR‐RFLP analysis of the PLIN1 biallelic polymorphism. M, DNA ladder; Lane1, AA; Lane2, AG; Lane3, GG.
The prices of the two restriction enzymes were from NEB website. Compared to using Mn1I enzyme ($1,258/1,000 units), more than half the cost was saved by using BstUI ($619/1,000 units).
Polymorphisms of PLIN1 were detected in 108 Chinese Han samples from one enterprise in Zhengzhou University. The genotype frequencies were 14.8% for AA, 52.8% for AG, and 32.4% for GG, respectively; the allelic frequencies were 40.8% for A and 58.2% for G, respectively. The results from the new PCR‐RFLP assay were confirmed by DNA sequencing. The x 2 test showed the genotype and allele frequencies of PLIN1 do not deviate from Hardy–Weinberg equilibrium(x 2 = 0.860634, P > 0.05).
Examples of DNA sequencing of PCR product of PLIN1 gene were shown in Figure 2. The BstUI recognition site (CGCG) is indicated by a mark symbol in Figure 2A, and the vertical arrow corresponds to complementary base of the mismatched base. Three figures represent the wild type of genotypes of GG, AA, and AG, respectively, and the sequences representing the polymorphism sites are denoted in squares.
Figure 2.
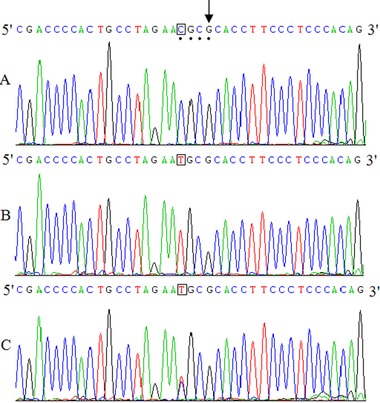
Examples of DNA sequencing of PCR product of PLIN1 gene.
Discussion
The commonly used detection methods of SNP are DNA sequencing, TaqMan assay, fluorescently labeled probes technique, molecular beacons, PCR‐RFLP, and so on. DNA sequencing is an accurate method for genotyping, but it is labor‐intensive and not suitable for high‐throughput analysis. The most prominent advantage for TaqMan probe technique is that it does not need a PCR posttreatment process such as separation or elution, so it has high detection rate. Difficulty in ensuring accurate classification is the design of accurate probe, which is often expensive and difficult to find 13. Molecular beacons are marked by fluorescent dyes of different colors to achieve the simultaneous detection of multiple SNPs. However, dyes are less for the suitable fluorescent dye‐labeled, which greatly limits the detection of flux 14. Despite the above‐mentioned methods’ disadvantages, all have their own prominent advantages, the equipments they need are often expensive, and the experiment methods are complex, so they are difficult to popularize and apply for SNP detection. The PCR‐based RFLP technique is widely used in the detection of SNP because it is easy to grasp the detection method and it also has high sensitivity and reliability.
Although many of these new techniques have been developed recently to analyze gene SNP, PCR‐RFLP method developed in 2006 15 is still a potent way to detect rs2289487 polymophism in PLIN1 gene. PCR‐RFLP was shown to be a rapid and sensitive method for the detection of gene polymorphism 16. But this method also has some limitations, such as some restriction enzymes BsmI or its isoschizomers that are very expensive and hindered the application for large‐scale screening, especially in developing countries. In order to overcome it, the PCR‐RFLP with mismatch primers and inexpensive enzyme BstUI was developed to detect PLIN1 polymorphism rs2289487 in this study.
In conclusion, we reported a simple and economical technique for analysis of PLIN1 polymorphism rs2289487. It may be used widely in the future study of the genetics of obesity, type 2 diabetes, and so on.
Ethical Approval
All procedures performed in studies involving human participants were in accordance with the ethical standards of the institutional and/or national research committee and with the 1964 Helsinki Declaration and its later amendments or comparable ethical standards.
Grant sponsor: National Natural Science Foundation of China; Grant number: NSFC81001239; Grant sponsor: Science and Technology Research Project of Health Department in Henan Province; Grant number: 2011020084.
References
- 1. Rankinen T, Zuberi A, Chagnon YC, et al. The human obesity gene map: The 2005 update. Obesity 2006;14(4):529–644. [DOI] [PubMed] [Google Scholar]
- 2. Mori Y, Otabe S, Dina C, et al. Genome‐wide search for type 2 diabetes in Japanese affected sib‐pails confirms susceptibility genes on 3q, 15q, and 20q and identifies two new candidate loci on 7p and 11p. Diabetes 2002;51(4):1247–1255. [DOI] [PubMed] [Google Scholar]
- 3. Duggirala R, Blangero J, Almasy L, et al. A major susceptibility locus influencing plasma triglyceride concentrations is located on chromosome 15q in Mexican Americans. Am J Hum Genet 2000;66(4):1237–1245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Bickel PE, Tansey JT, Welte MA. PAT proteins, an ancient family of lipid droplet proteins that regulate cellular lipid stores. Biochim Biophys Acta 2009;1791(6):419–440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Qi L, Shen H, Larson I, et al. Gender‐specific association of a perilipin gene haplotype with obesity risk in a white population. Obes Res 2004;12(11):1758–1765. [DOI] [PubMed] [Google Scholar]
- 6. Soenen S, Mariman ECM, Vogels N, et al. Relationship between perilipin gene polymorphisms and body weight and body composition during weight loss and weight maintenance. Physiol Behav 2009;96(4–5):723–728. [DOI] [PubMed] [Google Scholar]
- 7. Qi L, Corella D, Sorli JV, et al. Genetic variation at the perilipin (PLIN) locus is associated with obesity‐related phenotypes in White women. Clin Genet 2004;66(4):299–310. [DOI] [PubMed] [Google Scholar]
- 8. Qi L, Tai SY, Tan CE, et al. Intragenic linkage disequilibrium structure of the human perilipin gene (PLIN) and haplotype association with increased obesity risk in a multiethnic Asian population. J Mol Med 2005;83(6):448–456. [DOI] [PubMed] [Google Scholar]
- 9. Corella D, Qi L, Tai ES, et al. Perilipin gene variation determines higher susceptibility to insulin resistance in Asian women when consuming a high‐saturated fat, low‐carbohydrate diet. Diabetes Care 2006;29(6):1313–1319. [DOI] [PubMed] [Google Scholar]
- 10. Perez‐Martinez P, Yiannakouris N, Lopez‐Miranda J, et al. Postprandial triacylglycerol metabolism is modified by the presence of genetic variation at the perilipin (PLIN) locus in 2 white populations. Am J Clin Nutr 2008;87(3):744–752. [DOI] [PubMed] [Google Scholar]
- 11. Goni L, Cuervo M, Milagro FI, Martinez JA. A genetic risk tool for obesity predisposition assessment and personalized nutrition implementation based on macronutrient intake. Genes Nutr 2015;10(1):445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Qi L, Zhang CL, Greenberg A, Hu FB. Common variations in perilipin gene, central obesity, and risk of type 2 diabetes in US women. Obesity 2008;16(5):1061–1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. de Kok JB, Wiegerinck ETG, Giesendorf BAJ, Swinkels DW. Rapid genotyping of single nucleotide polymorphisms using novel minor groove binding DNA oligonucleotides (MGB probes). Hum Mutat 2002;19(5):554–559. [DOI] [PubMed] [Google Scholar]
- 14. Steemers FJ, Ferguson JA, Walt DR. Screening unlabeled DNA targets with randomly ordered fiber‐optic gene arrays. Nat Biotechnol 2000;18(1):91–94. [DOI] [PubMed] [Google Scholar]
- 15. Meirhaeghe A, Thomas S, Ancot F, et al. Study of the impact of perilipin polymorphisms in a French population. J Negat Results Biomed 2006;5:10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Fontecha GA, Garcia K, Rueda MM, Sosa‐Ochoa W, Sanchez AL, Leiva B. A PCR‐RFLP method for the simultaneous differentiation of three Entamoeba species. Exp Parasitol 2015;151:80–83. [DOI] [PubMed] [Google Scholar]


