Abstract
We evaluated the DiversiLab (DL) system with universal primers, a semiautomated repetitive extragenic palindromic sequence‐based polymerase chain reaction (PCR) (rep‐PCR) system, for the characterization of Helicobacter pylori in Japan. All 135 isolates from Japanese patients with gastric cancer (GC, n = 55) or non‐GC (n = 80) were used and subjected to the drug susceptibility examinations (amoxicillin, AMPC; metronidazole, MNZ; and clarithromycin, CAM) by E‐test. There were 28 MNZ‐resistant (20.7%), 35 CAM‐resistant (25.9%), and 16 MNZ/CAM‐resistant (11.9%) isolates. DL rep‐PCR fingerprinting analysis at the level of 95% similarity revealed five major groups (A–E) and the other including 45 isolates. The occupation rates of GC‐derived isolates in groups B (54.2%) and E (58.8%) were higher than in the other groups: A (26.7%), C (28.6%), D (30.0%), and the other (40.0%). Relative higher occupation rates of drug resistants, such as MNZ‐, CAM‐ and double MNZ/CAM‐resistant isolates, were observed in groups B (45.8%), C (42.6%), and D (40%). Five of eight GC‐derived isolates with MNZ/CAM resistance were significantly assigned to group B (P = 0.0312, χ2‐test). These results suggest that the isolates classified in group B have a potential to contribute to the development of severe gastric disorders. The DL system, rapid and high sensitive technology, would be widely available in clinical laboratory for pathological and epidemiological analyses even in H. pylori.
Keywords: Helicobacter pylori, genotyping, rep‐PCR, DiversiLab Microbial Typing System, gastric cancer, antibiotic resistant
Helicobacter pylori colonizes the human stomach causing a variety of diseases. The isolates obtained from H. pylori associated diseases are used to establish bacterial pathogenity and/or the risk factors as bacterium–host interaction. However, these findings are insufficient to explain the development of all H. pylori associated diseases. Thus, developing a tool for interpretation of the risk factors and clinical pathology of H. pylori associated diseases is important. Repetitive sequence‐based PCR (rep‐PCR) assay determines the similarity among individual bacteria or strains at the level of genomic variation via analysis of the repetitive sequences throughout the genomes. Rep‐PCR greatly enhances a reproducibility and strain discrimination compared with other PCR‐based platforms 1, 2. Thus, rep‐PCR is recognized as an effective method 3 for epidemiological analysis and investigation of the relationship between microorganisms and their related disorders. Rep‐PCR with specific primers has grouped H. pylori isolates from duodenal ulcer or simple gastritis patients into distinct clusters 4, 5, suggesting that this method is useful to provide a strain characteristics of risk potential and/or pathogenesis of diseases. The DiversiLab (DL) system (bioMerieux, Marcy l’Etoile, France) is a rep‐PCR technique that offers semiautomated, easy‐to‐use, high‐throughput, and rapid bacterial strain typing 6, 7. However, there is no H. pylori genotyping data available using the DL system. In this study, we examined the discrimination of H. pylori isolates from gastric cancer (GC) and non‐cancer patients and evaluated whether DL system can be used to determine the potential risk factor in the clinical laboratory.
This study was retrospectively analyzed with 135 H. pylori isolates collected in different geographical locations, including 55 and 80 isolates from GC (46–86 years, average 67.7 years) and non‐GC (12–84 years, average 56.9 years), respectively. All isolates were confirmed as H. pylori by routine microbiological and genetic examinations. Helicobacter pylori was cultured on Brucella broth agar plates containing 10% horse serum and 10 μg/ml of vancomycin at 37°C under microaerobic conditions (10% CO2; 8). Genomic DNA was extracted using the UltraClean microbial DNA isolation kit (MO BIO Laboratories, Carlsbad, CA), and the regions of interest were amplified using the DL Bacterial kit with universal primers according to the manufacturer's instructions. The PCR amplicons were applied to the DL system that generated a genetic pattern using a DL Lab‐chip kit and Agilent B2100 bioanalyzer (Agilent Technologies, Germany). The genetic patterns were compared and dendrograms based on the pattern similarity were generated by the DL software (version 3.3). An interactive report consisting of the dendrogram, electropherogram, virtual gel images, scatter plots, and selectable demographic fields was automatically generated. Antibiotic susceptibility was tested by E‐test® (bioMérieux, France) for amoxicillin (AMPC), metronidazole (MNZ), and clarithromycin (CAM) via a determination of minimum inhibitory concentration (MIC). The bacterial suspension adjusted to a turbidity of 3–4 McFarland was spread on a Brucella broth agar plate containing 10% horse serum and each antibiotic stick was placed on the plates, followed by culture for 72 hr. The breakpoints used to define the resistance were as follows: AMPC > 0.5 μg/ml, MNZ > 8 μg/ml, and CAM > 1 μg/ml 9.
All 135 H. pylori isolates including 55 GC‐derived and 80 non‐GC‐derived isolates were clustered into five major groups (A–E) and the other including 45 individual strains by pattern typing by DL at the level of 95% similarity (Fig. 1). A total of 90 isolates belonged to the major groups consisting of 14–24 isolates in each group. We repeated rep‐PCR typing using DNAs of H. pylori ATCC 26695 and two clinical strains at least three times, and demonstrated that each similarity was ≥98.5% (data not shown). Thus, the isolates representing ≥98.5% pattern similarity could be considered as identical level in this rep‐PCR typing. We used an ATCC 26695 strain in every experiments as an internal control to monitor the reliability of the assay. The occupation rates of GC‐derived isolates in group B (54.2% [13/24]) and group E (58.8% [10/17]) were higher than those of A (26.7% [4/15]), C (28.6% [4/14]), D (30.0% [6/20]), and the other (40.0% [18/45]). The antibiotic susceptibility of all 135 isolates demonstrated 20.7% (28/135), 25.9% (35/135), and 11.9% (16/135) in MNZ, CAM, and double MNZ/CAM resistance, respectively. No AMPC‐resistant isolate was found. There was no significant relationship between drug susceptibility and rep‐PCR typing. Interestingly, as for the 16 MNZ/CAM resistance, it showed the tendency that such double drug‐resistant strains were intensively classified to groups B or D (Fig. 1). Furthermore, eight GC‐derived isolates with MNZ/CAM resistance were found, of which five isolates were assigned to group B with significance (P = 0.0312, χ2‐test).
Figure 1.
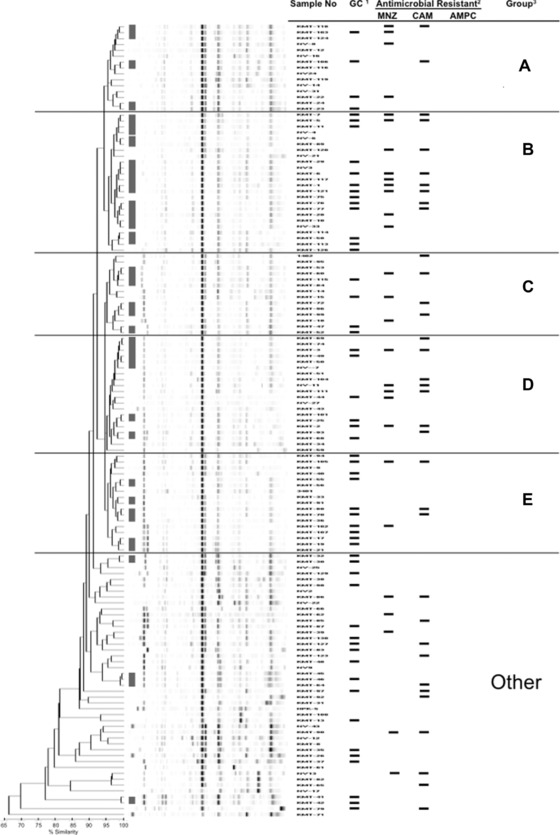
Dendrogram based on rep‐PCR pattern typing with 95% similarity level for 135 Helicobacter pylori isolates. 1Gastric cancer (GC)‐derived Helicobacter pylori isolates were shown by bar. 2Drug susceptibility examination. The isolates resist to antibiotics were shown by bar. MNZ, metronidazole; CAM, clarithromycin; AMPC, amoxicillin. 3Five major groups (A–E) and the other were provided by DL analysis. ▮: The minor groups consisted of high‐similar isolates (≥ 98.5% similarity) was depicted.
Generally, genotyping in epidemiology and clinical pathology improves our understanding of genetic variation/evolution, leading to appropriate treatment of nosocomial infection and infectious diseases. The DL system developed, a rep‐PCR tool, is widely applied and good performance of DL system was reported for subtyping of several pathogens 10, 11. Rep‐PCR with primers specific to certain genes of H. pylori documented that there was no significant difference of genotyping of isolates from between patients with GC or duodenal ulcer 12. In contrast, the genotyping in H. pylori isolates by pulsed‐field gel electrophoresis and arbitrary primer PCR reflected the characteristics of antibiotic susceptibility 13. To the best of our knowledge, there is no assessment regard to bacterial discrimination and genotyping of H. pylori using the DL system.
In this study, all 135 H. pylori isolates were subjected to a universal kit of the DL system composed of specific fingerprint kits for several bacteria except of H. pylori. There is no universal tethering for identification of H. pylori using the universal kit of the DL system. Thus, we investigated the reliability and reproducibility of this assay using DNAs of H. pylori ATCC 26695 and two clinical isolates, which showed ≥98.5% similarity as an identical level. Rep‐PCR at the level of 95% similarity revealed that all isolates were divided into five major (A–E) and the other groups. The other group composed of 45 isolates included eight isolates with ˂80% similarity. On the other hand, high‐similar strains (≥98.5% similarity) were found in the other as well as five major groups, leading to several minor groups. These minor groups were shown in Figure 1. These high‐similar isolates were from patients with individual geography and clinical records, such as diagnosis, treatment, and hospital. Thus, it suggested that these isolates were not identical, but showed extremely high similarity in this assay. We need more investigations to clear these issues (identical or not) as well as the marginal capacity of the utility.
Go et al. reported that different pattern was obtained from duodenal ulcer isolates and gastritis isolates by rep‐PCR of H. pylori 4. The rep‐PCR fingerprints showed clustering by geographic region, but not by disease presentation 12. Interestingly, our results documented that the occupation rates of GC‐derived isolates were higher in group B (54.2%) and group E (58.8%) than other groups. CagA and VacA, major pathogens of H. pylori, are well known to relate with gastric carcinoma 14. However, both genes were conserved in all isolates by PCR (data not shown), suggesting that the peculiar situation of the rep sequences around the both genes of the isolates in two groups (B and E) may profoundly reflect the pathogenicity of CagA and VacA. In the future, we should accumulate these data and generate rep‐PCR libraries to serve as reference standards for pathogenic analysis.
Drug‐resistant isolates distributed over six groups, indicating that this genotyping could not reflect the antibiotic susceptibility (MNZ, CAM, and AMPC). However, 16 MNZ/CAM‐resistant isolates were found and these double drug‐resistant isolates seemed to be intensively divided into group B (n = 6) and group D (n = 4). Furthermore, five of eight GC‐derived isolates with double drug‐resistant were significantly assigned to group B (P = 0.0312, χ2‐test). Therefore, the isolates in group B deserved further investigation to clarify biological characteristics in term of latent pathogenesis and multidrug resistance.
Helicobacter pylori has a capacity for high genetic diversity and infection in early childhood possibly leads to persistent colonization in the stomach with adaptation to individual hosts 15, 16, 17, 18. Consequently, the H. pylori genome has been rearranged for persistent infection in the individual stomachs. Li et al. reported that the differences among the H. pylori strains exist in single gene allelic variants as well as in the conserved noncoding regions, such as rep sequences, throughout the entire bacterial genome 12. Thus, DL system may be available to detect such genetic alterations of H. pylori. DL system in laboratory will enable comprehensive genetic analysis of H. pylori and provide important information in clinical pathology.
Grant sponsor: MEXT/JSPS KAKENHI; Grant numbers: 23590677 and 24590697.
REFERENCES
- 1. Rademaker JL, Hoste B, Louws FJ, et al. Comparison of AFLP and rep‐PCR genomic fingerprinting with DNA‐DNA homology studies: Xanthomonas as a model system. Int J Syst Evol Microbiol 2000;50(2):665–677. [DOI] [PubMed] [Google Scholar]
- 2. Burucoa C, Lhomme V, Fauchere JL. Performance criteria of DNA fingerprinting methods for typing of Helicobacter pylori isolates: Experimental results and meta‐analysis. J Clin Microbiol 1999;37(12):4071–4080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Dombek PE, Johnson LK, Zimmerley ST, Sadowsky MJ. Use of repetitive DNA sequences and the PCR to differentiate Escherichia coli isolates from human and animal sources. Appl Environ Microbiol 2000;66(6):2572–2577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Go MF, Chan KY, Versalovic J, Koeuth T, Graham DY, Lupski JR. Cluster analysis of Helicobacter pylori genomic DNA fingerprints suggests gastroduodenal disease‐specific associations. Scand J Gastroenterol 1995;30(7):640–646. [DOI] [PubMed] [Google Scholar]
- 5. Kidd M, Atherton JC, Lastovica AJ, Louw JA. Clustering of South African Helicobacter pylori isolates from peptic ulcer disease patients is demonstrated by repetitive extragenic palindromic‐PCR fingerprinting. J Clin Microbiol 2001;39(5):1833–1839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Healy M, Huong J, Bittner T, et al. Microbial DNA typing by automated repetitive‐sequence‐based PCR. J Clin Microbiol 2005;43(1):199–207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Deplano A, Denis O, Rodriguez‐Villalobos H, et al. Controlled performance evaluation of the DiversiLab repetitive‐sequence‐based genotyping system for typing multidrug‐resistant health care‐associated bacterial pathogens. J Clin Microbiol 2011;49(10):3616–3620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Trang VT, Takeuchi H, Kudo H, et al. Antimicrobial activity of aminoreductone against Helicobacter pylori . J Agric Food Chem 2009;57(23):11343–11348. [DOI] [PubMed] [Google Scholar]
- 9. Kawai T, Kawakami K, Kataoka M, et al. Comparison of efficacies of dual therapy and triple therapy using rabeprazole in second‐line eradication of Helicobacter pylori in Japan. Aliment Pharmacol Ther 2006;24(S4):16–22. [Google Scholar]
- 10. Ben‐Darif E, De Pinna E, Threlfall EJ, Bolton FJ, Upton M, Fox AJ. Comparison of a semi‐automated rep‐PCR system and multilocus sequence typing for differentiation of Salmonella enterica isolates. J Microbiol Methods 2010;81(1):11–16. [DOI] [PubMed] [Google Scholar]
- 11. Fluit AC, Terlingen AM, Andriessen L, et al. Evaluation of the DiversiLab system for detection of hospital outbreaks of infections by different bacterial species. J Clin Microbiol 2010;48(11):3979–3989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Li L, Graham DY, Gutierrez O, et al. Genomic fingerprinting and genotyping of Helicobacter pylori strains from patients with duodenal ulcer or gastric cancer from different geographic regions. Dig Dis Sci 2002;47(11):2512–2518. [DOI] [PubMed] [Google Scholar]
- 13. Nada T, Ando T, Nobata K, et al. DNA typing for Helicobacter pylori isolates from eradication‐failed patients: Comparison of the isolates before and after therapy. Aliment Pharmacol Ther 2004;20(Suppl 1):39–47. [DOI] [PubMed] [Google Scholar]
- 14. Hatakeyama M, Higashi H. Helicobacter pylori CagA: A new paradigm for bacterial carcinogenesis. Cancer Sci 2005;96(12):835–843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Malaty HM, El‐Kasabany A, Graham DY, et al. Age at acquisition of Helicobacter pylori infection: A follow‐up study from infancy to adulthood. Lancet 2002;359(9310):931–935. [DOI] [PubMed] [Google Scholar]
- 16. Granstrom M, Tindberg Y, Blennow M. Seroepidemiology of Helicobacter pylori infection in a cohort of children monitored from 6 months to 11years of age. J Clin Microbiol 1997;35(2):468–470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Ashorn M, Maki M, Hallstrom M, et al. Helicobacter pylori infection in Finnish children and adolescents: A serologic cross‐sectional and follow‐up study. Scand J Gastroenterol 1995;30(9):876–879. [DOI] [PubMed] [Google Scholar]
- 18. Shibayama K, Takeuchi H, Wachino J, Mori S, Arakawa Y. Biochemical and pathophysiological characterization of Helicobacter pylori asparaginase. Microbiol Immunol 2011;55(6):408–417. [DOI] [PubMed] [Google Scholar]


