Abstract
Background
Single‐nucleotide polymorphisms (SNPs) have been reported as a highly relevant point for the mechanisms of Parkinson's disease (PD). The invention of saturating dye makes it possible to identify heteroduplex DNA without redistribution during melting, which allows using high‐resolution melting (HRM) to detect SNPs. However, the HRM analysis for detection of those SNPs associated with PD was rarely applied.
Methods
Two SNPs, G2385R and R1628P, located in leucine‐rich repeat kinase 2 (LRRK2) gene were individually and multiplexedly genotyped using HRM analysis. The sequence variant observed in unexpected HRM curves was confirmed by DNA sequencing.
Results
HRM analysis identified successfully all genotypes both on R1628P and G2385R loci. The unexpected HRM curves appeared in R1628P amplicon generated from combinations of R1628P and rs11176013 loci. A multiplexed HRM assay that genotyped R1628P, rs11176013, and G2385R loci was efficiently established.
Conclusions
The present HRM assay is a reliable and rapid method for genotyping R1628P and G2385R loci in LRRK2 gene, and multiplex HRM analysis results in high throughput and has the potential to facilitate a wide range of genotyping studies on PD susceptibility genes.
Keywords: Parkinson's disease, HRM, LRRK2, SNPs, susceptibility locus, genotyping
INTRODUCTION
Leucine‐rich repeat kinase 2 (LRRK2, OMIM: 609007), encoded by LRRK2 on chromosome 12q12, contains five putative functional domains: an N‐terminal leucine‐rich repeat (LRR) domain, a Roc (Ras of complex protein) domain that shares sequence homology to the Ras‐related GTPase superfamily, a COR (C‐terminal of Roc) domain, a mitogen‐activated protein kinase kinase kinase (MAPKKK) domain, and a C‐terminal WD40 repeat domain 1. Many functional roles for LRRK2 have been suggested, including vesicular trafficking and endocytosis 2, 3, protein synthesis 4, immune response regulation 5, inflammation 6, and cytoskeleton homoeostasis 7. Also, increasing evidences are building to support a multifactorial role of LRRK2 in a number of human diseases, linking to risk of Parkinson's disease (PD), Crohn's disease, leprosy, and cancer 8, 9, 10. How the biology of LRRK2 interplays with the etiology of these disorders, and whether these links represent opposing or congruent roles for LRRK2 is, as yet, unclear 11.
Although the LRRK2 locus has been linked to some diseases in genome‐wide association studies, the strong links have been mainly observed between LRRK2 and familial and sporadic PD. Numerous recent studies have demonstrated that mutation in this gene is one of the most common causes of inherited PD. LRRK2 gene, also known as PARK8, contains 51 exons and encodes a very large protein of 2,527 amino acids. To date, more than 50 variants have been identified in LRRK2, including at least 16 pathogenic variants 12. Mutations known to cause PD are clustered within the central catalytic region including the GTPase (N1437H, R1441C, R1441G, and R1441H), COR (Y1699C), and kinase (G2019S and I2020T) domains 13, whereas the R1941H and G2385R mutations suppress autophosphorylation as well as phosphorylation of myelin basic protein and moesin 14. Similarly, G2385R mutation inhibits kinase activity 15. In addition, some association studies suggested that G2385R, R1628P, S1647T, and A419V showed evidence to contribute to disease susceptibility in Asians and G2019S as well as M1646T in White individuals 16.
Thus, not only pathogenic variants but these PD susceptibility single‐nucleotide polymorphisms (SNPs) in LRRK2 gene should be taken into account to perform a rapid and accurate genotyping. At present, as a routine examination, a few genetic factors have been analyzed by polymerase chain reaction‐restriction fragment length polymorphism (PCR‐RFLP) technique in the outpatients who are diagnosed with PD in our affiliated hospital 17. However, the genotyping method seems to be a little bit time‐consuming. High‐resolution melting (HRM) is a more sensitive technique and used fashionably in SNPs genotyping due to be a homogeneous, closed‐tube, post‐PCR technique 18. However, those pathogenic variants and PD susceptibility SNPs in LRRK2 gene were rarely detected by the HRM method.
In this study, we developed a robust HRM method to genotype the R1628P (rs33949390, 4883 G > C) and G2385R (rs34778348, 7153 G > A) loci, individually or simultaneously, which are associated with risk of PD in parts of Asian population, providing a rapid and effective diagnostic method.
MATERIALS AND METHODS
DNA Samples
The study protocol was approved by the ethical committee of China Medical University. DNA samples from 141 PD patients whose genotypes of the R1628P and G2385R loci had already been determined were used. Their genotypes were previously identified as 136 homozygous GG and 5 heterozygous GC in R1628P locus, and 121 homozygous GG, 19 heterozygous GA, and 1 homozygous AA in G2385R locus. DNA concentration was determined by NanoDrop 2000 (Thermo Fisher Scientific Inc., Waltham, MA), and then all samples were adjusted to 20∼21 ng/μl in final concentration. Informed consent was obtained from all study participants.
Primers Design and PCR/HRM Conditions
The PCR/HRM primers were designed to individually and simultaneously detect R1628P and G2385R loci. The sequence of used primers was 5′‐AAA CAC CCT AAG GGC ATT ATT TCG‐3′ (forward) and 5′‐CTA GGA GCT TAA AAT ACT GTG ACA TGT AGT TCT‐3′ (reverse) for R1628P locus amplification, and 5′‐ACA GTG GTG GTA GAC ACT GCT CTC TAT A‐3′ (forward) and 5′‐CTT AAA AAG TGC ACG CAG TCT ATT AGT C‐3′ (reverse) for G2385R locus amplification. PCR reaction and HRM assay were all performed by LightCycler® 480 Real‐Time PCR System in one same tube for each sample. Following the recommended procedure of LightCycler® 480 High‐Resolution Melting Master, we used a 20 μl standard reaction including 10 μl of LightCycler® 480 High‐Resolution Melting Master 2× containing buffer, 0.4 μl of each primer (final concentration 0.2 μm), 2.4 μl of 25 mM MgCl2 stock solution, 4.8 μl of PCR‐grade water, and 2 μl of each DNA template.
PCR program begins with a initial preincubation at 95°C for 5 min and the amplification conditions were as follows: 45 cycles of denaturation at 95°C for 10 s, annealing at 60°C for 10 s, and elongation at 72°C for 10 s (with single‐mode fluorescence acquisition). After the amplification, the melting program was followed, which included three steps: denaturation at 95°C for 1 min, renaturation at 40°C for 1 min, and a continuous reading of fluorescence from 65°C to 95°C at a rate of 25 acquisitions for each degree centigrade.
Multiplexed PCR Amplicons Melting Assay
The duplexed PCR amplicons melting assay included two fragments for genotyping R1628P and G2385R loci, and the multiplexed PCR was performed under same concentrations and volume as described above but with the adjusted ratio of concentrations of the two pairs of primers. Best image appeared in pre‐experiments when 0.4 μl of R1628P primers and 0.15 μl of G2385R primers were used. For multiplexed PCR, normalization region was adjusted to 71.77–73.21 and 81.62–82.62.
Gene Scanning by HRM Analysis
We chose one sample of each genotype for in‐run standard and set the remaining samples as unknown. HRM data were analyzed, normalized, and temperature‐shifted by the software LightCycler® 480 GeneScanning Software Version 1.5 (Roche Diagnostics, Mannheim, Germany), and then converted to a derivative plot. Normalized HRM curves were generated with the following normalization regions: R1628P, 71.77–73.21 and 78.05–79.00; G2385R, 77.11–78.12 and 81.62–82.62.
DNA Sequencing
The above PCR products from R1628P locus were purified, and then direct DNA sequencing was performed using the BigDye Terminator Cycle Sequencing Kit (Applied Biosystems, Foster City, CA).
TA‐Cloning
Because no homozygous CC in R1628P locus was previously detected, we constructed C and G alleles on the R1628P locus by TA‐clone to provide a distinguishable parameter between different genotyping methods. PCR products from genotype GC of R1628P were obtained by the same PCR assay as described above. Freshly produced amplicons were subcloned using the pGM‐T Clone® Kit (Tiangen). Plasmids of positive clones were confirmed by DNA sequencing.
RESULTS
HRM Method for Genotyping the R1628P and G2385R Loci in LRRK2 Gene
HRM analysis successfully identified all genotypes on G2385R locus (Fig. 1) and genotypes from the 141 samples matched to ones we determined in our previous study, showing a high accuracy. Normalized plots (Fig. 1A) and both normalized and temperature‐shifted difference plots (Fig. 1B) may be used to classify genotypes GG, AA, and GA on G2385R locus. When normalizing the initial and final fluorescence in all samples (Fig. 1A), it is easier to distinguish the samples between homozygotes, as well as homozygote and heterozygote. Furthermore, when shifting the temperature axis of the normalized melting curves at the point where the entire double‐stranded DNA is completely denatured, it seems to demonstrate more significant plot's differences by the different shapes of their melting curves (Fig. 1B).
Figure 1.
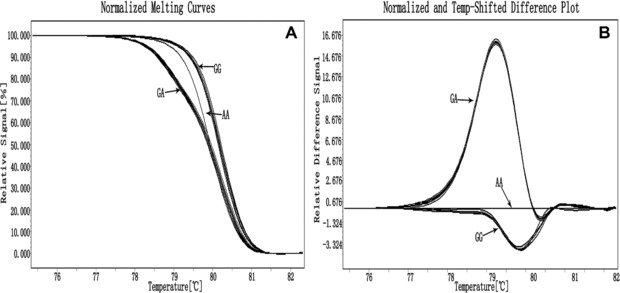
Genotyping of G2385R by LightCycler® 480. Differences between three genotypes in normalized plots (A) and normalized and temperature‐shifted plots (B).
Similarly, the R1628P locus on LRRK2 gene was also analyzed by HRM method. Two previously determined genotypes, homozygous GG and heterozygous GC, were successfully identified in all 141 samples (Fig. 2A and B). Interestingly, we observed 54 samples with GG genotype whose melting curves were obviously deviated from normal orbit. Thus, direct DNA sequencing was performed in a few samples. The results showed that the samples were not only of genotype GG of R1628P but also had another heterozygous AG mutation, which was previously designated as rs11176013 locus on SNPs online database. Normalized plots (Fig. 2A) and both normalized and temperature‐shifted difference plots (Fig. 2B) may accurately distinguish either the genotypes considered as single R1628P locus or those considered as R1628P combined with rs11176013 loci. These results suggested that even if there was another mutation in the targeted fragment, the R1628P locus could still be genotyped without mistake by HRM method.
Figure 2.
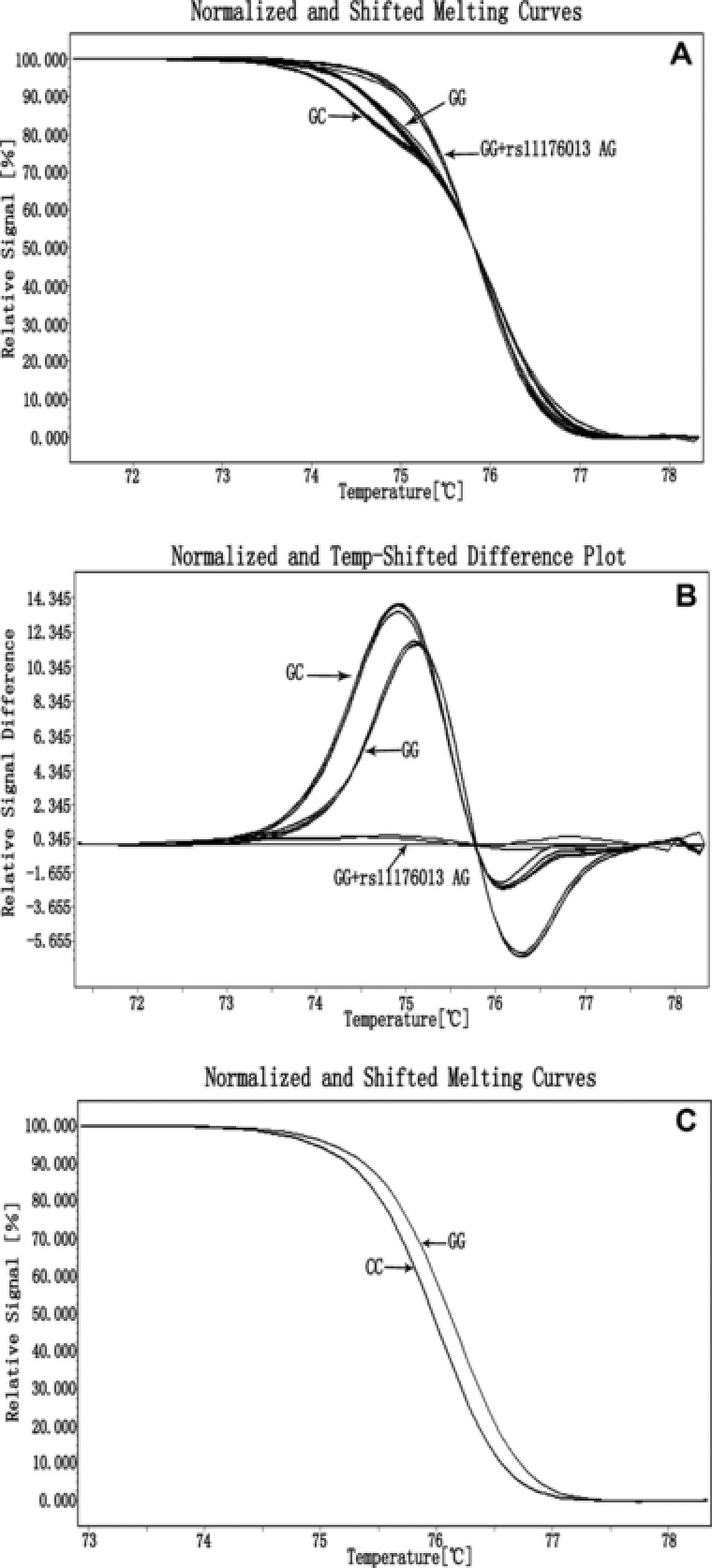
Genotyping of R1628P by LightCycler® 480. Differences between GC mutant, wild type and rs11176013 AG mutant plus R1628P wild type in normalized plots (A) and normalized and temperature‐shifted plots (B). Differences between clones in normalized plots (C).
Since we have not detected any homozygous genotype CC on R1628P locus in the previous study, to determine if the homozygous CC and GG can be distinguished by HRM method, the allelic C and G of R1628P were first prepared by TA‐cloning from PCR products of heterozygous CG sample. Subsequent HRM analysis showed that homozygous CC and GG could be genotyped by normalized and temperature‐shifted difference plots (Fig. 2C). Thus, our result provided a potential feasibility for discriminating all genotypes on the R1628P locus using HRM method.
Simultaneous Genotyping of G2385R and R1628P Loci in LRRK2 Gene
In this study, we also developed a multiplex HRM PCR test that allows rapid and reliable SNP genotyping on multiple loci. Applying this technique, we were able to efficiently genotype R1628P, G2385R, and unexpected rs11176013 loci in all investigated samples. As shown in Figure 3, two groups of amplicon peaks from multiplex PCR products represented R1628P plus rs11176013 loci at around Tm 76°C and G2385R locus at around Tm 80°C. Thus, based on peaks’ height and Tm value, all genotypes of three loci in LRRK2 gene could be simultaneously and accurately distinguished.
Figure 3.
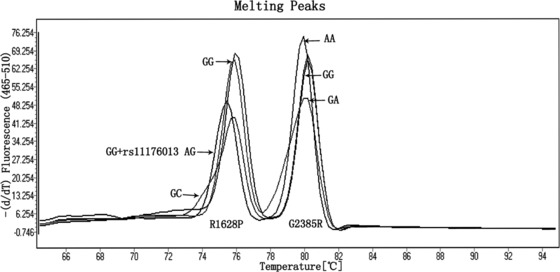
A simultaneous genotyping of R1628P and G2385R loci in LRRK2 gene. All detected genotype combinations in this study are shown.
DISCUSSION
HRM analysis of amplicons depends on DNA melting in the presence of saturating DNA‐binding dyes. As the temperature of the solution is increased, the specific sequence of the amplicon determines the melting behavior. The unique pattern of the melting curve, the derivative plot, or the difference plot may be used for amplicon analysis. Therefore, HRM analysis has been increasingly developed as a multipurpose technology in recent years. The latest developments and other applications in HRM analysis included presequence screening, SNP typing, methylation analysis, quantification (copy number variants and mosaicism), an alternative to gel‐electrophoresis, and clone characterization 19. More extensive application for HRM seemed to be mutation detection and genotyping for SNP locus. As is well known, there are many traditional scanning methods for SNP typing but almost all strategies require a separation step after PCR. HRM analysis can be performed in one closed‐tube, which does not require additional post‐PCR separation step and greatly reduces the possibility of contamination. Considering the advantages and benefits of HRM analysis, we tried to establish an HRM assay to genotype several PD susceptibility SNPs in outpatients instead of PCR‐RFLP method, which is the used strategy at present. Optimized primers design and length control of amplicons led to not only distinguish unambiguously all genotypes between homozygotes, homozygotes, and heterozygotes on R1628P and G2385R loci in LRRK2 gene within 1.5 h, but also identified successfully an unexpected SNP locus (rs11176013) in the present study. Ease of use, simplicity, low cost, superb sensitivity, and specificity of HRM analysis suggested that the assay should become the tool of choice to screen patients for PD susceptibility genes and pathogenic variants.
Multiplex genotyping by amplicon melting is also possible and has many important advantages. Multiple amplicons targeting different loci may be separated naturally at Tm based on their guanine–cytosine content and sequence. As an initial attempt to solution, genotyping by multiplexed amplicon melting was carried out using human platelet antigens 1 and 5 duplex assay 20. Next, a duplex amplicon genotyping assay was used for the MTHFR variants 1298 A > C and 677 C > T and successfully obtained HRM curves of PCR products 21. A developed ability to genotype up to four loci in the same reaction using amplicon melting has also been reported using rapid‐cycle PCR for amplification in one instrument and HRM in separate instruments 22. More meaningfully, the four most common mutations associated with thrombophilia were efficiently genotyped in a single homogeneous assay 23. Thus, multiplex analysis by HRM could provide an accurate and reliable method for mutation detection in known genes after the identification of particular diseases’ susceptibility loci through the powerful existing strategies for elucidating genetic variations. In the present study, we also performed a multiplexed HRM analysis on R1628P and G2385R loci of LRRK2 gene associated with PD based on conditions of genotyping two individual SNPs. As is well known, design and concentration of primers are proved to be the key step in this technique. Amplicons with no‐overlapping Tm were first obtained in each locus. The remained solution was to balance efficiency between two loci to discern and call all genotypes. By adjusting the ratio of concentrations of the two pairs of primers (R1628P:G2385R = 8:3), we were able to successfully identify all genotypes on two SNPs loci. In addition, unexpected sequence variants appeared in R1628P amplicons were clearly demonstrated under the multiplexed HRM system. Thus, with optimal primer design based on Tm's difference and minimal adjustments of oligonucleotide concentrations, duplexed loci could not only be efficiently amplified and analyzed by HRM, but also all genotypes and unexpected sequences might be accurately determined, suggesting the multiplexed HRM analysis lowers both the analysis time of each sample and cost. Further work will focus on more and more loci to make HRM analysis as a valuable tool that should be present in any laboratory studying nucleic acids.
Numerous SNPs associated with PD and distributed among different races have been reported so far. In our previous study, a significant difference between PD patients and controls was observed in G2385R locus of LRRK2 gene. At the same time, allele frequency of R1628P locus of LRRK2 gene was also investigated in the northern Han Chinese population, which was similar to the results from Japanese and Korean populations and was not shown as a genetic risk factor. To innovate HRM analysis into efficiently genotyping PD susceptibility genes, individual and multiplexed amplicons above two loci were tried to analyze automatic HRM curves. Both individual and multiplexed HRM curves could be distinguishable among all genotypes and discernable to unexpected sequences on the two PD susceptibility loci. Considering HRM's rapidity, easiness, cost, and reliability, it may replace current PCR‐RFLP method that is applied in our routine outpatient genotyping for genetic information collection. Note that the present HRM method is suitable for large‐scale study of the association between the SNPs loci on other PD‐associated genes and disease susceptibility.
In conclusion, the present HRM assay is a reliable and rapid method for detection of PD susceptibility SNPs, on R1628P and G2385R loci in LRRK2 gene, and multiplex HRM analysis results in high throughput and has the potential to facilitate a wide range of genotyping studies on PD susceptibility genes.
ACKNOWLEDGMENTS
This work was supported by a grant from the National Natural Science Foundation of China (81172713 to Dr. Hao Pang).
Grant sponsor: National Natural Science Foundation of China; Grant number: 81172713.
REFERENCES
- 1. Anand VS, Braithwaite SP. LRRK2 in Parkinson's disease: Biochemical functions. FEBS J 2009;276:6428–6435. [DOI] [PubMed] [Google Scholar]
- 2. Shin N, Jeong H, Kwon J, et al. LRRK2 regulates synaptic vesicle endocytosis. Exp Cell Res 2008;314:2055–2065. [DOI] [PubMed] [Google Scholar]
- 3. Piccoli G, Condliffe SB, Bauer M, et al. LRRK2 controls synaptic vesicle storage and mobilization within the recycling pool. J Neurosci 2011;31:2225–2237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Imai Y, Gehrke S, Wang HQ, et al. Phosphorylation of 4E‐BP by LRRK2 affects the maintenance of dopaminergic neurons in Drosophila. EMBO J 2008;27:2432–2443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Liu Z, Lee J, Krummey S, et al. The kinase LRRK2 is a regulator of the transcription factor NFAT that modulates the severity of inflammatory bowel disease. Nat Immunol 2011;12:1063–1070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Gillardon F, Schmid R, Draheim H. Parkinson's disease‐linked leucine‐rich repeat kinase 2 (R1441G) mutation increases proinflammatory cytokine release from activated primary microglial cells and resultant neurotoxicity. Neuroscience 2012;208:41–48. [DOI] [PubMed] [Google Scholar]
- 7. Parisiadou L, Cai H. LRRK2 function on actin and microtubule dynamics in Parkinson disease. Commun Integr Biol 2010;3:396–400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Zhang FR, Huang W, Chen SM, et al. Genomewide association study of leprosy. N Engl J Med 2009;361:2609–2618. [DOI] [PubMed] [Google Scholar]
- 9. Franke A, McGovern DP, Barrett JC, et al. Genome‐wide meta‐analysis increases to 71 the number of confirmed Crohn's disease susceptibility loci. Nat Genet 2010;42:1118–1125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Hassin‐Baer S, Laitman Y, Azizi E, et al. The leucine rich repeat kinase 2 (LRRK2) G2019S substitution mutation. Association with Parkinson disease, malignant melanoma and prevalence in ethnic groups in Israel. J Neurol 2009;256:483–487. [DOI] [PubMed] [Google Scholar]
- 11. Lewis PA, Manzoni C. LRRK2 and human disease: A complicated question or a question of complexes? Sci Signal 2012;5:pe2. [DOI] [PubMed] [Google Scholar]
- 12. Macedo MG, Verbaan D, Fang Y, et al. Genotypic and phenotypic characteristics of Dutch patients with early onset Parkinson's disease. Mov Disord 2009;24:196–203. [DOI] [PubMed] [Google Scholar]
- 13. Tsika E, Moore DJ. Mechanisms of LRRK2‐mediated neurodegeneration. Curr Neurol Neurosci Rep 2012;12:251–260. [DOI] [PubMed] [Google Scholar]
- 14. Jaleel M, Nichols RJ, Deak M, et al. LRRK2 phosphorylates moesin at threonine‐558: characterization of how Parkinson's disease mutants affect kinase activity. Biochem J 2007;405:307–317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Cookson MR. The role of leucine‐rich repeat kinase 2 (LRRK2) in Parkinson's disease. Nat Rev Neurosci 2010;11:791–797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Peeraully T, Tan EK. Genetic variants in sporadic Parkinson's disease: East vs West. Parkinsonism Relat Disord 2012;18:S63–S65. [DOI] [PubMed] [Google Scholar]
- 17. Zhou Y, Luo X, Li F, et al. Association of Parkinson's disease with six single nucleotide polymorphisms located in four PARK genes in the northern Han Chinese population. J Clin Neurosci 2012;19:1011–1015. [DOI] [PubMed] [Google Scholar]
- 18. Cao HC, Lin J, Qian J, et al. Detection of the JAK2 mutation in myeloproliferative neoplasms by asymmetric PCR with unlabeled probe and high‐resolution melt analysis. J Clin Lab Anal 2011;25:300–304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Vossen RH, Aten E, Roos A, et al. High‐resolution melting analysis (HRMA): More than just sequence variant screening. Hum Mutat 2009;30:860–866. [DOI] [PubMed] [Google Scholar]
- 20. Liew M, Nelson L, Margraf R, et al. Genotyping of human platelet antigens 1 to 6 and 15 by high‐resolution amplicon melting and conventional hybridization probes. J Mol Diagn 2006;8:97–104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Seipp MT, Durtschi JD, Liew MA, et al. Unlabeled oligonucleotides as internal temperature controls for genotyping by amplicon melting. J Mol Diagn 2007;9:284–289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Seipp MT, Pattison D, Durtschi JD, et al. Quadruplex genotyping of F5, F2, and MTHFR variants in a single closed tube by high‐resolution amplicon melting. Clin Chem 2008;54:108–115. [DOI] [PubMed] [Google Scholar]
- 23. Seipp MT, Durtschi JD, Voelkerding KV, et al. Multiplex amplicon genotyping by high‐resolution melting. J Biomol Tech 2009;20:160–164. [PMC free article] [PubMed] [Google Scholar]


