Highlights
-
•
Influenza A(H3N2) circulated in Europe in 2016–17 and 2017–18 and A(H1N1)pdm09 in 2017–18.
-
•
Changed A(H1N1)pdm09 vaccine component VE was 58% against A(H1N1)pdm09 in 2017–18.
-
•
A(H3N2) VE was 13% and 28% among all ages in 2016–17 and 2017–18, respectively.
Keywords: Influenza, Influenza vaccine, Vaccine effectiveness, Multicentre study, Case-control study, Europe
Abstract
Introduction
Influenza A(H3N2) viruses predominated in Europe in 2016–17. In 2017–18 A(H3N2) and A(H1N1)pdm09 viruses co-circulated. The A(H3N2) vaccine component was the same in both seasons; while the A(H1N1)pdm09 component changed in 2017–18. In both seasons, vaccine seed A(H3N2) viruses developed adaptations/alterations during propagation in eggs, impacting antigenicity.
Methods
We used the test-negative design in a multicentre primary care case-control study in 12 European countries to measure 2016–17 and 2017–18 influenza vaccine effectiveness (VE) against laboratory-confirmed influenza A(H1N1)pdm09 and A(H3N2) overall and by age group.
Results
During the 2017–18 season, the overall VE against influenza A(H1N1)pdm09 was 59% (95% CI: 47–69). Among those aged 0–14, 15–64 and ≥65 years, VE against A(H1N1)pdm09 was 64% (95% CI: 37–79), 50% (95% CI: 28–66) and 66% (95% CI: 42–80), respectively. Overall VE against influenza A(H3N2) was 28% (95% CI: 17–38) in 2016–17 and 13% (95% CI: −15 to 34) in 2017–18. Among 0–14-year-olds VE against A(H3N2) was 28% (95%CI: −10 to 53) and 29% (95% CI: −87 to 73), among 15–64-year-olds 34% (95% CI: 18–46) and 33% (95% CI: −3 to 56) and among those aged ≥65 years 15% (95% CI: −10 to 34) and −9% (95% CI: −74 to 32) in 2016–17 and 2017–18, respectively.
Conclusions
Our study suggests the new A(H1N1)pdm09 vaccine component conferred good protection against circulating strains, while VE against A(H3N2) was <35% in 2016–17 and 2017–18. The egg propagation derived antigenic mismatch of the vaccine seed virus with circulating strains may have contributed to this low effectiveness. A(H3N2) seed viruses for vaccines in subsequent seasons may be subject to the same adaptations; in years with lower than expected VE, recommendations of preventive measures other than vaccination should be given in a timely manner.
1. Introduction
WHO recommended the same influenza A(H3N2) Northern Hemisphere vaccine component for the 2016–17 and 2017–18 influenza seasons: A/Hong Kong/4801/2014 (H3N2)-like virus. An A/California/7/2009 (H1N1)pdm09-like virus vaccine component was recommended for the seventh consecutive year in the 2016–17 season. This was replaced with an A/Michigan/45/2015 (H1N1)pdm09-like virus vaccine component in the 2017–18 season.
Influenza A(H3N2) was the predominant circulating influenza virus in Europe in the 2016–17 season with very little A(H1N1)pdm09 and B circulating [1], [2]. While overall in Europe in the 2017–18 season influenza B/Yamagata virus lineage-mismatched to the trivalent vaccine was the main circulating strain, both A(H1N1)pdm09 and A(H3N2) circulated in varying patterns across countries [2]. Excess all-cause mortality was seen in both seasons, particularly among the elderly, in 2016–17 coinciding with a predominance of A(H3N2) [3], [4].
Since 2008–9, the I-MOVE/I-MOVE+ (Influenza Monitoring Vaccine Effectiveness in Europe) primary care multicentre case control study (MCCS) has provided vaccine effectiveness (VE) estimates by influenza virus (sub)type, age group, and target population. Since 2012–13, VE has also been estimated by vaccine type, and since 2015–16, by virus genetic clade [5], [6], [7].
We present the I-MOVE/I-MOVE+MCCS VE estimates against influenza A by subtype for the 2016–17 and 2017–18 seasons. We estimate VE by age group (including birth cohorts who may be more susceptible for infection [8]), target population, previous vaccination, vaccine type, time within the season and also estimate VE against genetic clade. The 2017–18 VE against influenza B lineage-mismatched to the trivalent vaccine will be addressed elsewhere in context of VE against influenza B in post-pandemic seasons.
2. Methods
Eleven European study sites (in Croatia, France, Germany, Ireland, Italy, Poland, Portugal, Romania, Spain, Sweden and the Netherlands) participated in our 2016–17 and 2017–18 I-MOVE/I-MOVE+MCCS, while one (in Hungary) participated in the in the 2016–17 MCCS only. Each study site used the test-negative design using the European Centre for Disease Prevention and Control (ECDC) generic case-control study protocol and the I-MOVE+protocol [9], [10]. The methods are described in detail elsewhere [5], [11].
Briefly, for each season, participating practitioners interviewed and collected nasopharyngeal or combined naso- and oro-pharyngeal specimens from a systematic sample of consenting patients seeking medical attention for influenza-like illness (ILI). Practitioners collected information including symptoms, date of onset and swabbing, current seasonal influenza vaccination status, date of influenza vaccination and vaccine product, seasonal influenza vaccination status from the previous season, sex, age and presence of chronic medical conditions in the past 12 months.
In the pooled analysis we included patients meeting the European Union ILI case definition [12], swabbed within 7 days of symptom onset, and who had not received antivirals in the 14 days prior to swabbing. Study sites with fewer than 10 influenza-positive cases by influenza subtype or with fewer than 10 vaccinated patients were excluded from the pooled analysis.
A case of confirmed influenza was an ILI patient who had been swabbed and whose test result was positive for influenza A virus using real-time, reverse-transcription polymerase chain reaction (RT-PCR). Controls were ILI patients who tested negative for any influenza virus using RT-PCR.
We defined a person as vaccinated if they had received at least one dose of current seasonal influenza vaccine more than 14 days before ILI symptom onset. Those vaccinated fewer than 15 days before ILI onset were excluded. All other patients were classified as unvaccinated.
2.1. Statistical methods
We conducted a complete case analysis, in which patients with missing values for any of the variables in the model measuring adjusted VE are excluded. We computed the pooled VE for each influenza A subtype as (1-OR) * 100 using a one-stage logistic regression model with study site as a fixed effect.
We calculated VE adjusting for a priori confounding factors: symptom onset date, age, sex, and presence of at least one chronic disease or other risk conditions such as pregnancy and obesity (where available). The continuous variables symptom onset date and age in years were modelled as restricted cubic splines with 3, 4 or 5 knots, or as categorical variables (age group, onset month or onset week), the choice of which was determined by sample size and the Akaike information criterion. We used the “one in ten” rule of covariate degrees of freedom to events to determine if we were overfitting the model [13]. If the number of events/parameters was less than 10, we conducted a sensitivity analysis using Firth’s method of penalised logistic regression [14].
To study the effect of prior seasonal influenza vaccination on the current season VE, we conducted an indicator analysis using four categories: individuals unvaccinated in both seasons (reference group), vaccinated in the previous season only, vaccinated in the current season only, and those vaccinated in both seasons. We did not measure effect of previous vaccination among children aged <9 years, due to their multi-seasonal vaccination schedule.
We measured VE by age group (0–14, 15–64 and 65 years and older) and in the target group for vaccination, which includes persons aged 60 or 65 years and older (depending on study site), persons with underlying medical conditions and other risk groups [15]. For the A(H1N1)pdm09 age-specific analysis among adults, we also stratified VE by the Linderman birth cohort of heightened susceptibility (1965–1979) due to influenza pre-exposure history, resulting in the age groups of 15–37, 38–52, and 53 years and over [8]. We measured VE by type of vaccine (inactivated subunit, inactivated split virion and MF59 adjuvanted), including only study sites in the analysis where vaccines of that type were available.
We measured VE by calendar time, estimating influenza A subtype-specific VE from October to December, in January and in February–April, in order to obtain early, peak and end of season estimates, respectively.
Data management and statistical analyses were carried out using Stata 15.1 [16].
2.2. Laboratory methods
Nine study sites in 2016–17 and eight in 2017–18 selected either all influenza virus-positive specimens or a random proportion of specimens for sequencing the haemagglutinin gene segment (HA) for each influenza A subtype. HA consensus sequences were uploaded by each site to GISAID and downloaded for centralised phylogenetic and amino acid substitution analysis of the HA1 coding portion in MEGA6 to determine clade distribution at the National Influenza Centre, Madrid. Amino acid composition analysis was done relative to the egg-adapted high growth reassortant X-263B vaccine strain of A/Hong Kong/4801/2014 H3N2 (GISAID accession number EPI731469). For H1N1pdm09 viruses the egg-adapted high growth reassortant X-179A vaccine strain of A/California/07/2009 (GISAID accession number EPI257201) and X-275A vaccine strain of A/Michigan/45/2015 (GISAID accession number EPI830230) were used as a reference for seasons 2016–17 and 2017–18, respectively.
3. Results
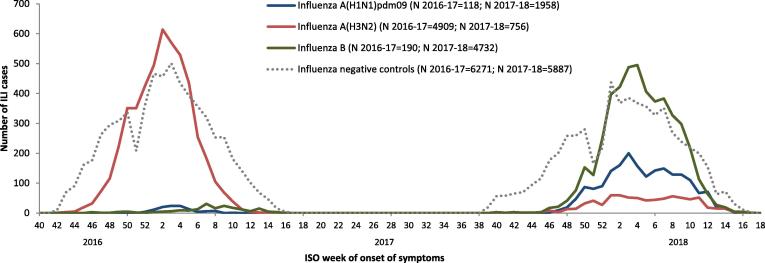
In the 2016–17 season we included 4909 influenza A(H3N2) cases and 6098 test-negative controls. Due to low numbers of influenza A(H1N1)pdm09 (N = 118) and B (N = 190) cases, no VE analysis against these was attempted (Fig. 1).
Fig. 1.
Number of ILI patients by case status (test negative controls and cases by influenza (sub)type) and week of symptom onset, I-MOVE/I-MOVE+ primary care multicenter case control study, Europe, influenza seasons 2016–17 and 2017–18. ILI: Influenza-like illness; ISO: International Organization for Standardization.
In the 2017–18 season, we included 1958 influenza A(H1N1)pdm09 cases, 756 influenza A(H3N2) cases and 5887 test-negative controls in the I-MOVE/I-MOVE+MCCS VE analyses (Fig. 1).
3.1. Participant profile and virological description
3.1.1. Influenza A(H3N2): 2016–17 and 2017–18
The median age of A(H3N2) cases was 32 years in 2016–17 and 41 years in 2017–18, compared to 31 and 32 years in controls in the same seasons, respectively (Supplementary Table 1). The vaccine coverage among cases was 11% in 2016–17 and 16% in 2017–18, compared to 12% in controls in both seasons.
In the 2016–17 and 2017–18 influenza seasons we included 1050 and 312 sequenced A(H3N2) viruses, respectively in the genetic analysis (Table 1). In 2016–17, 256 (24%) belonged to the 3C.2a clade represented by A/HongKong/4801/2014, 781 (74%) belonged to the 3C.2a1 clade represented by A/Singapore/INFIMH-16–0019/2016 (in 2016–17 the representative virus for this clade was A/Bolzano/7/2016, however we use the 2017–18 representative virus for easier comparison between seasons) and 13 (1%) belonged to the 3C.3a clade represented by A/Switzerland/9715293/2013. In 2017–18, 173 (55%) belonged to the 3C.2a clade, 134 (43%) belonged to the 3C.2a1 clade and 5 (2%) belonged to the 3C.3a clade.
Table 1.
Genetic group distribution among nine study sites participating in the random sequencing of influenza virus positive specimens. I-MOVE/I-MOVE+ primary care multicenter case control study, Europe, influenza season 2016–17 and 2017–18.
| Characterised viruses1 | Clade/subclade | DE |
ES |
FR |
HU |
IE |
NL |
PT |
RO |
SE |
Total |
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| N | % | N | % | N | % | N | % | N | % | N | % | N | % | N | % | N | % | N | % | |||
| 2016–17 |
A(H3N2) |
207 |
346 |
87 |
51 |
63 |
80 |
124 |
25 |
67 |
1050 | |||||||||||
| A/HongKong/4801/2014-like | 3C.2a | 47 | 23 | 59 | 17 | 28 | 32 | 15 | 29 | 14 | 22 | 16 | 20 | 42 | 34 | 10 | 40 | 25 | 37 | 256 | 24 | |
| * | 1 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 2 | 13 | 2 | 5 | 0 | 0 | 0 | 0 | 5 | 2 | ||
| N121K+S144K+(N122D+262N) | 3C.2a3 | 30 | 64 | 54 | 92 | 27 | 96 | 14 | 93 | 11 | 79 | 8 | 50 | 38 | 90 | 10 | 100 | 16 | 64 | 208 | 81 | |
| T131K+R142K+R261Q |
3C.2a2 |
16 |
34 |
5 |
8 |
1 |
4 |
1 |
7 |
3 |
21 |
6 |
38 |
2 |
5 |
0 |
0 |
9 |
36 |
43 |
17 |
|
| A/Bolzano/7/2016-like2 | 3C.2a1 | 159 | 77 | 280 | 81 | 59 | 68 | 36 | 71 | 45 | 71 | 63 | 79 | 82 | 66 | 15 | 60 | 42 | 63 | 781 | 74 | |
| N171K | 14 | 9 | 137 | 49 | 40 | 68 | 1 | 3 | 1 | 2 | 9 | 14 | 1 | 1 | 6 | 40 | 7 | 17 | 216 | 28 | ||
| N171K+N121K+I140M | – | 32 | 20 | 58 | 21 | 7 | 12 | 2 | 5 | 6 | 13 | 14 | 22 | 6 | 7 | 7 | 47 | 8 | 19 | 140 | 18 | |
| N171K+N121K+K92R+H311Q | 3C.2a1b | 43 | 27 | 59 | 21 | 1 | 2 | 4 | 11 | 5 | 11 | 18 | 29 | 29 | 35 | 1 | 7 | 1 | 2 | 161 | 21 | |
| N171K+N121K+T135K | 3C.2a1a | 54 | 34 | 8 | 3 | 2 | 3 | 28 | 78 | 1 | 2 | 11 | 17 | 0 | 0 | 1 | 7 | 22 | 52 | 128 | 16 | |
| N171K+R142G |
– |
16 |
10 |
18 |
6 |
9 |
15 |
1 |
3 |
32 |
71 |
11 |
17 |
46 |
56 |
0 |
0 |
4 |
10 |
137 |
18 |
|
| A/Switzerland/9715293/2013-like |
3C.3a |
1 |
0 |
7 |
2 |
0 |
0 |
0 |
0 |
4 |
6 |
1 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
13 |
1 |
|
| Characterised viruses | Clade/subclade |
DE |
ES |
FR |
HU |
IE |
NL |
PT |
RO3 |
SE |
Total |
|||||||||||
| N | % | N | % | N | % | N | % | N | % | N | % | N | % | N | % | N | % | N | % | |||
| 2017–18 | A(H1N1)pdm09 | 43 | 10 | 64 | – | 13 | 31 | 22 | 13 | 9 | 205 | |||||||||||
| A/Michigan/45/2015-like |
6B.1 |
43 |
100 |
10 |
100 |
64 |
100 |
13 |
100 |
31 |
100 |
22 |
100 |
13 |
100 |
9 |
100 |
205 |
100 |
|||
|
A(H3N2) |
25 |
141 |
35 |
– |
48 |
26 |
13 |
– |
24 |
312 |
||||||||||||
| A/HongKong/4801/2014-like | 3C.2a | 23 | 92 | 44 | 31 | 25 | 71 | – | 41 | 85 | 18 | 69 | 7 | 54 | – | 15 | 63 | 173 | 55 | |||
| * | 0 | 0 | 0 | 0 | 0 | 0 | – | 0 | 0 | 0 | 0 | 0 | 0 | – | 0 | 0 | 0 | 0 | ||||
| N121K+S144K+(N122D+262N) | 3C.2a3 | 2 | 9 | 0 | 0 | 1 | 4 | – | 3 | 7 | 0 | 0 | 0 | 0 | – | 0 | 0 | 6 | 3 | |||
| T131K+R142K+R261Q |
3C.2a2 |
21 |
91 |
44 |
100 |
24 |
96 |
– |
38 |
93 |
18 |
100 |
7 |
100 |
– |
15 |
100 |
167 |
97 |
|||
| A/Singapore/INFIMH-16-0019/2016-like2 | 3C.2a1 | 2 | 8 | 97 | 69 | 8 | 23 | – | 6 | 13 | 8 | 0 | 6 | 46 | – | 7 | 29 | 134 | 43 | |||
| N171K+N121K | 0 | 0 | 0 | 0 | 0 | 0 | – | 0 | 0 | 0 | 0 | 0 | 0 | – | 0 | 0 | 0 | 0 | ||||
| N171K+N121K+I140M | – | 0 | 0 | 0 | 0 | 0 | 0 | – | 0 | 0 | 0 | 0 | 0 | 0 | – | 0 | 0 | 0 | 0 | |||
| N171K+N121K+K92R+H311Q | 3C.2a1b | 1 | 50 | 96 | 99 | 8 | 100 | – | 6 | 100 | 8 | 100 | 6 | 100 | – | 7 | 100 | 132 | 99 | |||
| N171K+N121K+T135K | 3C.2a1a | 1 | 50 | 1 | 1 | 0 | 0 | – | 0 | 0 | 0 | 0 | 0 | 0 | – | 0 | 0 | 2 | 1 | |||
| N171K+R142G |
– |
0 |
0 |
0 |
0 |
0 |
0 |
– |
0 |
0 |
0 |
0 |
0 |
0 |
– |
0 |
0 |
0 |
0 |
|||
| A/Switzerland/9715293/2013-like |
3C.3a |
0 |
0 |
0 |
0 |
2 |
6 |
– |
1 |
2 |
0 |
0 |
0 |
0 |
0 |
2 |
8 |
5 |
2 |
|||
| Total 2017–18 | 68 | 151 | 89 | – | 61 | 57 | 35 | 13 | 33 | 517 | ||||||||||||
Viruses with no amino acid substitutions or substitutions outside of the A–E antigenic sites.
We use for both seasons the clade/subclade denomination created in mid-season in 2017-18 (report prepared by the Francis Crick Institute for the WHO annual consultation on the composition of influenza vaccine for the Northern Hemisphere 2018-2019). Therefore some clades/subclades circulating in 2016–17, but not in 2017–18, have no specific clade/subclade denomination. All amino acid substitutions shown for the A(H3N2) overarching clade 3C.2a viruses are relative to the vaccine virus A/HongKong/4801/2014.
A/Bolzano/7/2016 and A/Singapore/INFIMH-16-0019/2016 are two representative viruses of the same clade 3C.2a1, carrying the N171K substitution in the HA gene compared to the vaccine virus A/HongKong/4801/2014. In 2016–117, A/Bolzano/7/2016 was used as the representative virus and in 2017–118 A/Singapore/INFIMH-16-0019/2016 was used being representative for clade 3C.2a1 viruses carrying N121K in addition to N171K and chosen as vaccine strain for the 2018 Southern Hemisphere and 2018/2019 Northern Hemisphere vaccines.
Romania sequenced 7 A(H3N2) viruses, but they are excluded from the pooled analysis as they had fewer than 10 cases.
In 2016–17 there was a variety of amino acid substitutions in antigenic sites within the 3C.2a and 3C.2a1 clades in most studies, the proportion of which varied between countries (Table 1).
Conversely, in 2017–18, 167/173 (97%) of the viruses of the A/HongKong/4801/2014 3C.2a clade belonged to the 3C.2a2 subclade with the T131K, R142K and R261Q amino acid substitutions relative to the vaccine strain A/HongKong/4801/2014. Among the 134 viruses in 2017–18 belonging to the A/Singapore/INFIMH-16-0019/2016 3C.2a1 clade, 132 (99%) belonged to the 3C.2a1b subclade with the N171K, N121K, K92R and H311Q amino acid substitutions relative to the vaccine strain A/HongKong/4801/2014.
3.1.2. Influenza A(H1N1)pdm09: 2017–18
The median age of influenza A(H1N1)pdm09 cases was 19 years, compared to 32 years for influenza test-negative controls (Supplementary Table 1). Among A(H1N1)pdm09 cases, 47% belonged to the 0–14 years age group, compared to 33% of controls. The vaccination coverage was 4% among A(H1N1)pdm09 cases, compared to 12% among controls.
All A(H1N1)pdm09 sequenced viruses belonged to the clade represented by the A/Michigan/45/2015 vaccine virus (clade 6B.1) (Table 1).
3.1.3. Vaccines used in 2016–17 and 2017–18
In both seasons, all vaccines used were egg-propagated. Among vaccinated controls with known vaccine brand in 2016–17 (572/685, 84%), 100 (17%) received adjuvanted vaccine, 52 with aluminium phosphate gel and 48 with MF59 adjuvant, and nine (2%) received a live attenuated influenza vaccine (LAIV) (Supplementary table 1). Among vaccinated controls with known vaccine brand in 2017–18 (556/687, 81%), 30 (5%) received adjuvanted vaccine, all with MF59 adjuvant, and eight vaccinated participants (1%) received LAIV. In 2016–17 and 2017–18, 79% and 84% of vaccines with known brand were inactivated non-adjuvanted trivalent vaccines.
3.2. 2016–17 and 2017–18 vaccine effectiveness estimates against A(H3N2)
3.2.1. Overall
The VE against influenza A(H3N2) among all ages in 2016–17 was 28% (95% CI: 17–38) and in 2017–18 13% (95% CI: −15 to 34) (Table 2). Among all ages in the target group for vaccination the VE was 22% (95% CI: 7–34) in 2016–17 and 13% (95% CI: −21 to 38) in 2017–18
Table 2.
Pooled adjusted seasonal vaccine effectiveness against influenza A(H1N1)pdm09 and A(H3N2), overall, by age groups and by vaccine type, previous vaccination and time within the season. I-MOVE/I-MOVE+ primary care multicenter study, Europe, influenza seasons 2016–17 and 2017–18.
| Influenza A(H3N2) 2016–17 | |||||
|---|---|---|---|---|---|
| Age group | Population | Na | Cases; vacc /Controls; vaccb | Adjusted VE (%) | 95% CI (%) |
| All ages | 10,591 | 4733;516/5858;680 | 28 | 17–38 | |
| 0–14 years | 3452 | 1494;46/1958;66 | 28 | −10 to 53 | |
| 15–64 years | 5840 | 2606;183/3234;286 | 34 | 18–46 | |
| ≥65 years | 1298 | 633;287/665;328 | 15 | −10 to 34 | |
| All ages | Target group for vaccination | 3181 | 1440;431/1741;543 | 22 | 7–34 |
| All ages | Subunit vaccinea | 8304 | 3832;179/4472;238 | 31 | 13–46 |
| Split virion vaccinea | 9303 | 4209;188/5094;220 | 23 | 4–39 | |
| Adjuvanted vaccinea | 4678 | 2027;32/2651;48 | 49 | 12–71 | |
| ≥9 years | Unvaccinated | 7822 | 3168/3343b | Ref | |
| 2016–17 vaccine only | 69/132b | 50 | 32–63 | ||
| 2015–16 vaccine only | 87/144b | 29 | 5–47 | ||
| 2015–16 and 2016–17 vaccines | 412/467b | 20 | 5–32 | ||
| All ages | Oct-Dec | 3541 | 1442;141/2099;206 | 38 | 16–54 |
| Jan | 4379 | 2389;260/1990;289 | 34 | 18–47 | |
| Feb-April | 2671 | 902;115/1769;185 | 1 | −34 to 27 | |
| Influenza A(H3N2) 2017–18 | |||||
| Age group | Population | N | Cases; vacc /Controls; vacc | Adjusted VE (%) | CI (%) |
| All ages | 5607 | 731;119/4876;623 | 13 | −15 to 34 | |
| 0–14 years | 1772 | 160;6/1612;74 | 29 | −87 to 73 | |
| 15–64 years | 3128 | 444;33/2684;245 | 33 | −3 to 56 | |
| ≥65 years | 673 | 127;80/546;283 | −9 | −74 to 32 | |
| All ages | Target group | 1763 | 251;106/1512;512 | 13 | −21 to 38 |
| All ages | Subunit vaccine a | 4556 | 471;56/3739;290 | −3 | −50 to 30 |
| Split virion vaccine a | 4637 | 555;36/3906;140 | 19 | −29 to 49 | |
| ≥9 years | Unvaccinated | 4051 | 491/2783b | ||
| 17–18 vaccine only | 11/105b | 49 | 1–74 | ||
| 16–17 vaccine only | 17/119b | 16 | −47 to 52 | ||
| 16–17 and 17–18 vaccines | 95/430b | 7 | −29 to 33 | ||
| All ages | Oct-Dec | 1413 | 138;19/1275;133 | 11 | −72 to 54 |
| Jan | 1849 | 230;42/1619;234 | 5 | −54 to 41 | |
| Feb-April | 2345 | 363;58/1982;256 | 18 | −24 to 45 | |
| Influenza A(H1N1)pdm09 2017–18 | |||||
| Age group | Population | N | Cases; vacc /Controls; vacc | Adjusted VE (%) | CI (%) |
| All ages | 7175 | 1875;81/5300;643 | 59 | 47–69 | |
| 0–14 years | 2680 | 888;18/1792;80 | 64 | 37–79 | |
| 15–64 years | 3806 | 893;37/2913;256 | 50 | 28–66 | |
| ≥65 years | 684 | 94;26/590;307 | 66 | 42–80 | |
| 15–37 years | 1609 | 339;4/1270;54 | 71 | 18 to 90 | |
| 38–52 years | 1311 | 369;18/942;70 | 23 | −38 to 57 | |
| 53+years | 1572 | 279;41/1293;439 | 64 | 47 to 76 | |
| All ages | Target group | 2075 | 424;65/1651;526 | 56 | 40–68 |
| All ages | Subunit vaccine a | 6268 | 1745;39/4192;292 | 59 | 40–71 |
| Split virion vaccine a | 6184 | 1745;11/4288;140 | 71 | 45–85 | |
| ≥9 years | Unvaccinated | 4784 | 981/3023b | ||
| 17–18 vaccine only | 6/110b | 79 | 51–91 | ||
| 16–17 vaccine only | 25/137b | 24 | −22 to 52 | ||
| 16–17 and 17–18 vaccines | 61/441b | 46 | 26–61 | ||
| All ages | Oct-Dec | 2009 | 326;4/1683;152 | 87 | 62–95 |
| Jan | 2277 | 673;33/1604;230 | 53 | 27–69 | |
| Feb-April | 2826 | 856;44/1970;257 | 53 | 32–67 | |
CI: confidence interval; vacc: vaccinated; VE: vaccine effectiveness.
Among study sites where this vaccine type was used. Subunit and split virion vaccine refers to egg-propagated inactivated trivalent subunit and split virion vaccines, respectively. For adjuvanted vaccine only study sites using the MF59 adjuvant included.
Cases/controls.
3.2.2. By age group
The VE against A(H3N2) in 2016–17 was 28% (95% CI: −10 to 53), 34% (95% CI: 18–46) and 15% (95% CI −10 to 34) among those aged 0–14, 15–64 and ≥65 years, respectively.
The VE against influenza A(H3N2) in 2017–18 was 29% (95% CI: −87 to 73), 33% (95% CI: −3 to 56) and −9% (-74 to 32) among those aged 0–14, 15–64 and ≥65 years, respectively.
3.2.3. VE by clade/subclade
The VE in 2016–17 against all variants of 3C.2a excluding 3C.3a1 was 31% (95% CI: −12 to 57) and the VE against the 3C.2a3 subclade was 37% (95% CI: −9 to 63) (Table 3). In 2016–17 the VE against all 3C.2a1 variants was 25% (95% CI: −1 to 44). The VE against 3C.2a1 with no mutations in A-E antigenic sites except for N171K was 56% (95% CI: 20–76). The VE against the 3C.2a1b subclade was 7% (95% CI: −56 to 45).
Table 3.
Pooled adjusted seasonal vaccine effectiveness against influenza A(H3N2) clades and subclades, I-MOVE/I-MOVE+ primary care multicenter study, Europe, influenza seasons 2016–17 and 2017–18.
| Season | Clade/subclade | N | Cases; vacc /Controls; vacc | Adjusted VE (%) | CI (%) |
|---|---|---|---|---|---|
| 2016–17 | 3C.2a (all variants excluding 3C.2a1) | 4492 | 252;28/4240;496 | 31 | −12 to 57 |
| 3C.2a3 N121K+S144K+(N122D+262 N) |
4445 | 205;20/4240;496 | 37 | −9 to 63 | |
| 3C.2a1 (all variants) | 5007 | 767;99/4240;496 | 25 | −1 to 44 | |
| 3C.2a1 (no mutations in A-E antigenic sites other than N171K) | 4454 | 214;19/4240;496 | 56 | 20 to 76 | |
| 3C.2a1b N171K+N121K+K92R+H311Q |
4397 | 157;30/4240;496 | 7 | −56 to 45 | |
| 2017–18 | 3C.2a (all variants) | 3732 | 166;40/3566;445 | −29 | −109 to 21 |
| 3C.2a2 T131K+R142K+R261Q |
3726 | 160;38/3566;445 | −19 | −95 to 27 | |
| 3C.2a1 (all variants) | 3698 | 132;17/3566;445 | 41 | −19 to 71 | |
| 3C.2a1b N171K+N121K+K92R+H311Q |
3697 | 131;17/3566;445 | 42 | −18 to 71 | |
CI: confidence interval; vacc: vaccinated; VE: vaccine effectiveness.
In 2017–18 the VE against all variants of 3C.2a excluding 3C.3a1 was −29% (−109 to 21). The vast majority (160/166) of 3C.2a viruses belonged to the subclade 3C.2a2, against which the VE was −19 (95% CI: −95 to 27). The VE against 3C.2a1b was 43% (95% CI: −16 to 72).
3.2.4. By vaccine group
In 2016–17, among all ages, the VE of the trivalent subunit vaccine against A(H3N2) was 31% (95% CI: 13–46) and of the trivalent split virion vaccine was 23% (95% CI: 4–39). The VE of the MF59 adjuvanted trivalent subunit vaccine, used in three study sites, against A(H3N2) was 49% (95% CI: 12–71).
In 2017–18, the VE of the trivalent subunit vaccine against A(H3N2) was −3% (95% CI: −50 to 30) and the VE of the trivalent split virion vaccine against A(H3N2) was 19 (95% CI: −29 to 49).
3.2.5. By previous vaccination
In 2016–17, using those unvaccinated in current and prior season as a reference group, the VE among those aged ≥9 years was 50% (95% CI: 32–63) in those vaccinated in the current season only, 20% (95% CI: 5–32) in those vaccinated in both seasons, and 29% (95% CI: 5–47) in those vaccinated in previous season only.
In 2017–18, using those unvaccinated in current and prior season as a reference group, the VE among those aged ≥9 years was 49% (95% CI: 1–74) in those vaccinated in the current season only, 7% (95% CI: −29 to 33) in those vaccinated in both seasons and 16% (95% CI: −47 to 52) in those vaccinated in previous season only.
3.2.6. By calendar time
In 2016–17 the VE was 38% (95% CI: 16–54) in October to December, 34% (95% CI: 18–47) in January and 1% (95% CI: −34 to 27) in February to April among all ages.
In 2017–18 the VE was 11% (95% CI: −72 to 54), 5% (95% CI: −54 to 41) and 18% (95% CI: −24 to 45) in October to December, January and in February to April among all ages, respectively.
3.3. 2017–18 vaccine effectiveness estimates against A(H1N1)pdm09
3.3.1. Overall and by age group
In the 2017–18, 1875 A(H1N1)pdm09 cases and 5300 test-negative controls were included in the complete case VE analysis. The overall VE was 59% (95% CI: 47–69). Among the target group for vaccination (N = 2075) the VE was 56% (95% CI: 42–68) (Table 2).
The VE was 64% (95% CI: 37–79), 50% (95% CI: 28–66%) and 66% (95% CI 42–80) among patients aged 0–14, 15–64 and ≥64 years, respectively.
The VE among those aged 15–37 years was 71% (95% CI: 18–90), among those aged 38–52 years (corresponding to the Linderman birth cohort of susceptibility of 1965–79) 23% (95% CI: −38 to 57) and among those aged 53 and over 64% (95% CI: 47–76).
3.3.2. By vaccine type
VE was 59% (95%CI: 40–71) for the trivalent inactivated subunit vaccine and 71% (95% CI: 45–85) for the trivalent inactivated split virion vaccine. Numbers were too low to estimate VE for the MF59 adjuvanted vaccine.
3.3.3. By previous vaccination
In the ≥9 years olds (N = 4784), compared to those receiving no vaccine in current or prior season, the VE was 79% (95% CI: 51–91) among those receiving the 2017–18 vaccine only and 46% (95% CI: 26–61) among those receiving both 2016–17 and 2017–18 vaccine.
3.3.4. By calendar time
VE was 87% (95% CI: 62–95) in October to December, 53% (95% CI: 27–69) in January and 53% (95% CI: 32–67%) in February to April (see Table 2).
4. Discussion
In 2017–18 the influenza VE against primary care medically attended ILI due to influenza A(H1N1)pmd09 in the I-MOVE/I-MOVE+MCCS in Europe was 56 and 59% in the total and target population, respectively. The VE against A(H3N2) in 2016–17 was 22% and 28% in the total and target populations, respectively, and in 2017–18 it was 13% in both populations.
4.1. Influenza A(H1N1)pdm09
The 2017–18 season was the first season of the new A/Michigan/45/2015-like virus vaccine component after seven consecutive seasons of the A/California/7/2009 (H1N1)-like virus vaccine component. The A/California/7/2009 (H1N1)-like virus was maintained from 2010-11 to 2016–17 in the Northern Hemisphere vaccine. Despite evolution of the A(H1N1)pdm09 viruses into distinct genetic clades there were was no significant antigenic drift, making a vaccine strain switch unnecessary until recently. Clades 6B and 6C started to circulate in Europe in 2012–13. From 2014 to 15 onwards, all genetically characterised viruses reported to ECDC belonged to genetic subgroup 6B, characterised by a K163Q substitution in HA1 [17], [18]. In 2015–16, 90% (652/723) of all systematically selected A(H1N1)pdm09 viruses characterised in the I-MOVE/I-MOVE+MCCS belonged to the 6B.1 subgroup, defined by the S84N, I216T and S162N substitutions. This latter substitution may result in a potential new glycosylation site [6]. These recent genetic clades were antigenically distinct when using human antisera following vaccination [1], [18]. This was also reflected in the 2015–16 VE against A(H1N1)pdm09 of 33% (95% CI: −4 to 57) among all ages in the I-MOVE/I-MOVE+MCCS, which was lower than previous seasons. This decline in VE across seasons was also seen in Canada, but not in the UK [6], [7], [15], [19], [20], [21]. The I-MOVE/I-MOVE+MCCS 2017–18 season VE of 59% (95% CI: 47–69) indicates that the vaccine component change was successful, underlining the importance of complementation of antigenic characterisation of viruses using naïve ferret antisera with antisera derived from vaccinated humans from different age groups [22].
The process of “original antigenic sin” is where human antibody responses cross-react to influenza strains experienced in early childhood when challenged by a newer viral strain [23]. In 2014, Linderman et al. proposed that this process explained a heightened susceptibility to the circulating A(H1N1)pdm09 6B clade in some birth cohorts. These people may have experienced childhood exposure to A(H1N1) viruses with the K163 epitope, not the case in younger adults whose childhood exposure was to A(H1N1) viruses with this epitope masked [8]. Linderman et al. reported that particular birth cohorts 1965 to 1979 had K163-specific antibodies in their sera. They therefore recommended a change in vaccine from the A/California/7/2009 (H1N1)-like virus, whilst indicating nevertheless the uncertainty that a vaccine change will break the “original antigenic sin”. The VE against A(H1N1)pdm09 among the 1965–1979 birth cohort (38–52 years) in the 2017–18 I-MOVE/I-MOVE+MCCS was 22%, lower than among other age groups. This VE against A(H1N1)pdm09 is similar to the VE reported in 2015–16 in the US among similar birth cohorts (VE = 23%) [24]. The dip in VE was seen in slightly older age groups in Canada and was not apparent in the 2015–16 I-MOVE/I-MOVE+MCCS, where VE was low overall [6], [20]. The age-specific VE we observe in the our study in the 2017–18 season may be due to random variation, or due to a more complex interplay of influenza virus exposure and vaccinations [20]. Monitoring age-specific effects is warranted to better understand susceptible populations in future influenza seasons.
Our interim VE results against A(H1N1)pdm09 were higher at 68%, with data from the start of the 2017–18 season up to the 26th of January 2018 than these end of season results (VE = 59%) [25]. Indeed, the VE in October to December (87%) was higher than the VE in January and February to April (54% and 53%). A decline in VE against A(H1N1)pdm09 by time since vaccination was also seen in the I-MOVE/I-MOVE+MCCS in 2015-16 and a decline in VE against A(H1N1)pdm09 by calendar time was seen in Canada in 2015-16 [20], but not in the I-MOVE MCCS between 2010 and 11 and 2014-15 [26]. While monitoring declining vaccine effect against the 6B.1 clade warrants further research, the decline by calendar time could possibly be a statistical artefact of the study. However, the decline was not seen in VE against A(H3N2) in 2017–18, indicating that this may be at least in part a real rather than an artefactual phenomenon for influenza A(H1N1)pdm09.
Vaccine coverage was low and numbers were small for estimates of vaccine type specific VE and indicated no real difference between VE of inactivated trivalent subunit and split virion vaccine among all ages, similar to the I-MOVE/I-MOVE+MCCS results from the 2013-14, 2014-15 and 2015-16 seasons [6], [15], [19]. Numbers for quadrivalent and adjuvanted vaccine types were too low to estimate vaccine type-specific VE.
VE among those receiving both current and prior season influenza vaccine was lower than among those receiving current season only vaccination (46% vs.79%), compared to a reference group of those not receiving vaccine in either season. A similar pattern for A(H1N1)pdm09, albeit with low precision, was seen in the 2015-16 season in the I-MOVE/I-MOVE+MCCS study and also in Canada, but not in the US [6], [20], [27]. The findings in the 2017–18 season may be explained by negative interference of the prior vaccine strain, but should be seen also in context of different profiles among unvaccinated, single season vaccinees, vaccinated and repeat vaccinated participants, as well as in the context of the influence of different antibody landscapes among individuals.
4.2. Influenza A(H3N2)
Similar clades of A(H3N2) were circulating in the 2016–17 and 2017–18 influenza seasons, albeit in different proportions and with some differing subclades, with the same vaccine component in both seasons. VE was approximately 30% among those aged <65 years in both seasons, and lower among those aged 65 and older.
A(H3N2) vaccine seed viruses can develop adaptations/alterations during propagation in eggs, impacting antigenicity, which may negatively affect VE [28], [29], [30]. Egg-propagated A/HongKong/4801/2014 (H3N2)-like seed vaccine viruses have the egg adaptation induced T160K substitution, whereas 160T is present in the original A/HongKong/4801/2014 virus and current circulating 3C.2a viruses. Viruses with 160T grow poorly in eggs, and the T160K egg adaptation results in the loss of a putative N-glycosylation site in the HA antigenic site B [31]. In the I-MOVE/I-MOVE+MCCS 98-99% of all systematically sequenced A(H3N2) viruses had 160T. These changes due to egg-adaptation may contribute to the low VE observed in the 2016–17 and 2017–18 seasons in our study. The A/Singapore/INFIMH-16-0019/2016 (H3N2)-like vaccine component for the Southern Hemisphere 2018 and the Northern Hemisphere 2018-19 influenza season, is also subject to the same problems during egg adaptation in the vaccine manufacturing process in the same critical antigenic site: vaccine seed strains having 160 K instead of 160T in the original virus. If similar genetic variants of A(H3N2) with 160T are circulating in the future we may expect VE to be similar to the 2016–17 and 2017–18 seasons, making recommendations for other preventive measures already at start of the season vital. This may be particularly crucial for older age groups, where A(H3N2) can also cause severe disease, as younger age groups had a higher VE compared to those 65 years of age and older. That said, WHO reported that A(H3N2) strains circulating in the 2017–18 season were more similar in HA assays to the egg-propagated A/Singapore/INFIMH-16-0019/2016 (H3N2)-like virus (the recommended 2018-19 Northern hemisphere A(H3N2) vaccine virus component) than the 2017–18 egg-propagated A/HongKong/4801/2014 (H3N2)-like virus vaccine strain [32]; however early results from Europe indicate that 2018-19 season VE against A(H3N2) may be low [33].
Low VE against influenza A(H3N2) may also be explained by the genetic variation among circulating strains. In 2016–17 the VE against 3C.2a variants, excluding 3C.2a1, was similar to the VE against 3C.2a1 (31% vs 25%). In 2017–18, the VE against 3C.2a, excluding 3C.2a1, of which almost all belonged to the 3C.2a2 subclade, was lower than the VE against 3C.2a1 (−29 vs 41%). The same pattern of difference in 2017–18 between 3C.2a (with 3C.2a1 excluded) and 3C.2a1 VE was seen in other study sites [34] [R Pebody, PHE, UK, Personal communication]. Sample size was low, however, in particular in the 2017–18 season, and it was not possible to disentangle age-specific and strain-specific effects on VE. Increased sequencing of A(H3N2) viruses in seasons with A(H3N2) circulating would help with interpretation of VE in light of genetic changes.
There was no statistical difference between VE of inactivated trivalent subunit and split virion vaccine in any of the age groups in 2016–17 or in 2017–18, as in the previous A(H3N2) season in 2014-15 [19].
The 2015-16 influenza A(H3N2) vaccine component was A/Switzerland/9715293/2013, which has 160 K in the original and egg-adapted strains. In 2016–17 the VE among those receiving both current and prior vaccine was lower than the VE among those receiving current vaccine and no prior season vaccine. The pattern was the same in 2017–18 when those receiving both current and prior vaccine had a VE of 7% compared to the VE of 49% among those receiving current vaccine only. The results suggest a lower VE with combined use of current and prior vaccination, as per the antigenic distance hypothesis [35], [36]. It is possible that this is a true phenomenon in the study, as due to passaging-related adaptions in the vaccine virus, the antigenic distance between vaccine and circulating strains may be large and the vaccine viruses are either identical (2017–18) or similar (2016–17), compared to previous seasons. Other potential explanations include confounding due to different participant profiles of repeat and single season vaccinees, recall bias and random variation. Prospective longitudinal studies are needed to better understand the phenomenon of effects of previous vaccination.
The interim VE against A(H3N2) in 2016–17 was 38% (95% CI: 21–51)[37], higher than the end-of-season estimate of 28% (95% CI: 17–38). When grouped into calendar months (Oct-Dec, January and Feb-April), the 2016–17 season VE declined along the season resulting in no effectiveness in February to April. This could be due to virological changes in the A(H3N2) virus across the season, waning of immunity within the individual against the vaccine, be a statistical artefact, or a combination of all. In 2016–17, 6% (14/216) of clade 3C.2a1 viruses with no mutations in A-E antigenic sites other than the N171K occurred in February to April, compared to 18% (99/565) of clade 3C.2a1 viruses with mutations in the same period (data not shown).
In 2017–18, the interim VE against A(H3N2) was −16 (95% CI: −96 to 13) compared to the 13% (-15 to 34) end-of-season estimate [25]. Differences in point estimates were likely due to random variation due to sample size and there was no evidence of change in VE by calendar time. However, there was also less diversity of subclades, so that within clade 3C.2a (excluding clade 3C.2a1) and clade 3C.2a1, respectively, there was no consistent change in variants over time. Additionally, we may not expect any measurable waning of immunity, with such a low overall VE (13%).
This study uses the test-negative design that has its strengths in controlling for health care-seeking behaviour and ease of study implementation [38]. However as with all observational studies, we cannot rule out bias due to unmeasured confounding and selection bias. We believe these are minimised by the study design, as the practitioners are blinded to the case status of their patients, and to the adherence to the protocols, in which practitioners are asked to systematically or exhaustively select patients to swab and interview [10].
5. Conclusions
Our study indicates that the new vaccine component A/Michigan/45/2015 conferred greater protection to the circulating 6B.1 clade than the A/California/7/2009 vaccine strain. Receiving influenza vaccine in the 2017–18 season reduced the risk of being an A(H1N1)pdm09 case by more than half among all ages in our study. All viruses sequenced in our study belonged to the 6B.1 clade, so we expect a similar protection of the Northern Hemisphere vaccine, which contains the same A(H1N1)pdm09 strain, against A(H1N1)pdm09, should this virus circulate in the future, without additional common amino acid changes. To monitor this, representative genetic and amino acid composition characterisation of circulating A(H1N1)pdm09 should complement VE studies in coming seasons.
The VE of the A/HongKong/4801/2014 vaccine component against A(H3N2) was <30% in the 2016–17 and 2017–18 seasons. The circulating subclades in both seasons may have contributed towards the low VE, as well as the amino acid substitutions within the vaccine virus during the egg-adaption process of manufacturing. No vaccines based on cell-propagated viruses were used by participants of the 2016–17 and 2017–18 I-MOVE/I-MOVE+MCCS. The A(H3N2) component A/Singapore/INFIMH-16-0019/2016 recommended for the 2018-19 Northern Hemisphere influenza vaccine is subject to the same issues of amino acid substitutions during the egg-adaption process. As influenza A(H3N2) and A(H1N1)pdm09 co-circulated strain in the 2018-19 season, it is possible that influenza VE against A(H3N2) will be low, particularly among older adults, based on the results of our study. Excess influenza-associated mortality and morbidity has been reported among the elderly during previous seasons with A(H3N2) circulation, when the H3N2 vaccine component showed poor or moderate VE. Therefore, national authorities need to be prepared in such a situation for rapid actions in reallocation of health care resources, ensuring surge capacity and ensuring timely prescriptions and administration of influenza antivirals.
All authors attest they meet the ICMJE criteria for authorship
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
Acknowledgements
The I-MOVE/I-MOVE+ study team is very grateful to all patients, general practitioners, paediatricians, hospital teams, laboratory teams, regional epidemiologists who have contributed to the study.
We acknowledge the authors, originating and submitting laboratories of the sequences from GISAID’s EpiFlu Database used for this study. All submitters of data may be contacted directly via the GISAID website www.gisaid.org.
Funding
This project has received funding from the European Union's Horizon 2020 research and innovation programme under grant agreement no. 634446 to conduct the study in individuals aged 65 years or more.
ECDC has contributed funds for the coordination and some study sites under the Framework contract no. ECDC/2014/026 for the individuals aged less than 65 years.
The WHO Regional office for Europe has contributed funds for the Romanian study site.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.jvacx.2019.100042.
Appendix A. Supplementary material
The following are the Supplementary data to this article:
References
- 1.ECDC, ‘Influenza virus characterisation - Summary Europe, June 2016’, Jul. 2016.
- 2.C. Adlhoch, R. Snacken, A. Melidou, S. Ionescu, P. Penttinen, and The European Influenza Surveillance Network, ‘Dominant influenza A(H3N2) and B/Yamagata virus circulation in EU/EEA, 2016/17 and 2017/18 seasons, respectively’, Euro Surveill. Bull. Eur. Sur Mal. Transm. Eur. Commun. Dis. Bull., vol. 23, no. 13, 2018. [DOI] [PMC free article] [PubMed]
- 3.Vestergaard L.S. Excess all-cause and influenza-attributable mortality in Europe, December 2016 to February 2017. Euro. Surveill. Bull. Eur. Sur Mal. Transm. Eur. Commun. Dis. Bull. 2017;22(14) doi: 10.2807/1560-7917.ES.2017.22.14.30506. Apr. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.EuroMOMO, ‘European monitoring of excess mortality for public health action - EuroMOMO’. [Online]. Available: [Accessed: 14-Aug-2018]. http://www.euromomo.eu/index.html.
- 5.Valenciano M., Ciancio B., I-MOVE study team I-MOVE: a European network to measure the effectiveness of influenza vaccines. Euro Surveill. Bull. Eur. Sur Mal. Transm. Eur. Commun. Dis. Bull. 2012;17(39) doi: 10.2807/ese.17.39.20281-en. Sep. [DOI] [PubMed] [Google Scholar]
- 6.Kissling E. 2015/16 I-MOVE/I-MOVE+ multicentre case-control study in Europe: moderate vaccine effectiveness estimates against influenza A(H1N1)pdm09 and low estimates against lineage-mismatched influenza B among children. Influenza Other Respir. Viruses. 2018;12(4):423–437. doi: 10.1111/irv.12520. Jul. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kissling E. ‘Influenza vaccine effectiveness estimates in Europe in a season with three influenza type/subtypes circulating: the I-MOVE multicentre case-control study, influenza season 2012/13. Euro Surveill Bull. Eur. Sur Mal. Transm. Eur. Commun. Dis. Bull. 2014;19(6) doi: 10.2807/1560-7917.es2014.19.6.20701. Feb. [DOI] [PubMed] [Google Scholar]
- 8.Linderman S.L. Potential antigenic explanation for atypical H1N1 infections among middle-aged adults during the 2013–2014 influenza season. Proc. Natl. Acad. Sci. U. S. A. 2014;111(44):15798–15803. doi: 10.1073/pnas.1409171111. Nov. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.ECDC, ‘European Centre for Disease Prevention and Control (ECDC). Protocol for case control studies to measure pandemic and seasonal vaccine effectiveness in the European Union and European Economic Area’, European Centre for Disease Prevention and Control, Stockholm, Sweden, 2010.
- 10.‘Generic protocol for the test negative design case control studies to measure pandemic and seasonal influenza vaccine effectiveness in the European Union and European Economic Area Member States’, European Union, Jul. 2015.
- 11.Valenciano M. Estimates of pandemic influenza vaccine effectiveness in Europe, 2009–2010: results of Influenza Monitoring Vaccine Effectiveness in Europe (I-MOVE) multicentre case-control study. PLoS Med. 2011;8(1) doi: 10.1371/journal.pmed.1000388. Jan. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.ECDC, ‘European Commission. Commission Decision 2009/363/EC of 30 April 2009 amending Decision 2002/253/EC laying down case definitions for reporting communicable diseases to the Community network under Decision No 2119/98/EC of the European Parliament and of the Council.’, Jan. 2009.
- 13.Peduzzi P., Concato J., Kemper E., Holford T.R., Feinstein A.R. A simulation study of the number of events per variable in logistic regression analysis. J Clin Epidemiol. 1996;49(12):1373–1379. doi: 10.1016/s0895-4356(96)00236-3. Dec. [DOI] [PubMed] [Google Scholar]
- 14.Firth D. Bias reduction of maximum likelihood estimates. Biometrika. 1993;80(1):27–38. Mar. [Google Scholar]
- 15.Valenciano M. The European I-MOVE multicentre 2013–2014 case-control study. homogeneous moderate influenza vaccine effectiveness against A(H1N1)pdm09 and heterogenous results by country against A(H3N2) Vaccine. 2015;33(24):2813–2822. doi: 10.1016/j.vaccine.2015.04.012. Jun. [DOI] [PubMed] [Google Scholar]
- 16.StataCorp, Stata statistical software. College Station, TX: StataCorp LLC.
- 17.European Centre for Disease Prevention and Control, ‘Influenza virus characterisation, Summary Europe, June 2015’, European Centre for Disease Prevention and Control, Stockholm, Sweden.
- 18.WHO, ‘Recommended composition of influenza virus vaccines for use in the 2017 southern hemisphere influenza season’, WHO, Sep. 2016.
- 19.Valenciano M. Vaccine effectiveness in preventing laboratory-confirmed influenza in primary care patients in a season of co-circulation of influenza A(H1N1)pdm09, B and drifted A(H3N2), I-MOVE Multicentre Case-Control Study, Europe 2014/15. Euro Surveill. Bull. Eur. Sur Mal. Transm. Eur. Commun. Dis. Bull. 2016;21(7) doi: 10.2807/1560-7917.ES.2016.21.7.30139. p. pii=30139. [DOI] [PubMed] [Google Scholar]
- 20.Skowronski D.M. Beyond antigenic match: possible agent-host and immuno-epidemiological influences on influenza vaccine effectiveness during the 2015–2016 season in Canada. J. Infect. Dis. 2017;216(12):1487–1500. doi: 10.1093/infdis/jix526. Dec. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pebody R. Effectiveness of seasonal influenza vaccine for adults and children in preventing laboratory-confirmed influenza in primary care in the United Kingdom: 2015/16 end-of-season results. Euro Surveill. Bull. Eur. Sur Mal. Transm. Eur. Commun. Dis. Bull. Sep. 2016;21(38) doi: 10.2807/1560-7917.ES.2016.21.38.30348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hensley S.E. Challenges of selecting seasonal influenza vaccine strains for humans with diverse pre-exposure histories. Curr. Opin. Virol. 2014;8:85–89. doi: 10.1016/j.coviro.2014.07.007. Oct. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Davenport F.M., Hennessy A.V., Francis T. Epidemiologic and immunologic significance of age distribution of antibody to antigenic variants of influenza virus. J. Exp. Med. 1953;98(6):641–656. doi: 10.1084/jem.98.6.641. Dec. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Flannery B. Influence of Birth Cohort on Effectiveness of 2015–2016 Influenza Vaccine Against Medically Attended Illness Due to 2009 Pandemic Influenza A(H1N1) Virus in the United States. J. Infect. Dis. 2018;218(2):189–196. doi: 10.1093/infdis/jix634. Jun. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rondy M. Interim 2017/18 influenza seasonal vaccine effectiveness: combined results from five European studies. Euro Surveill. Bull. Eur. Sur Mal. Transm. Eur. Commun. Dis. Bull. 2018;23(9) doi: 10.2807/1560-7917.ES.2018.23.9.18-00086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kissling E. I-MOVE multicentre case-control study 2010/11 to 2014/15: Is there within-season waning of influenza type/subtype vaccine effectiveness with increasing time since vaccination? Euro Surveill Bull. Eur. Sur Mal. Transm. Eur. Commun. Dis. Bull. 2016;21(16) doi: 10.2807/1560-7917.ES.2016.21.16.30201. Apr. [DOI] [PubMed] [Google Scholar]
- 27.Jackson M.L. Influenza Vaccine Effectiveness in the United States during the 2015-2016 Season. N. Engl. J. Med. 2017;377(6):534–543. doi: 10.1056/NEJMoa1700153. 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chen Z., Zhou H., Jin H. The impact of key amino acid substitutions in the hemagglutinin of influenza A (H3N2) viruses on vaccine production and antibody response. Vaccine. May 2010;28(24):4079–4085. doi: 10.1016/j.vaccine.2010.03.078. [DOI] [PubMed] [Google Scholar]
- 29.Skowronski D.M., De Serres G. Role of egg-adaptation mutations in low influenza A(H3N2) vaccine effectiveness during the 2012–13 season. Clin. Infect. Dis. Off. Publ. Infect. Dis. Soc. Am. 2018 doi: 10.1093/cid/ciy350. Apr. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Parker L. Effects of egg-adaptation on receptor-binding and antigenic properties of recent influenza A (H3N2) vaccine viruses. J. Gen. Virol. 2016;97(6):1333–1344. doi: 10.1099/jgv.0.000457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zost S.J. Contemporary H3N2 influenza viruses have a glycosylation site that alters binding of antibodies elicited by egg-adapted vaccine strains. Proc. Natl. Acad. Sci. U. S. A. 2017;114(47):12578–12583. doi: 10.1073/pnas.1712377114. 21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Worldwide Influenza Centre, ‘Report prepared for the WHO annual consulation on the composition of influenza vaccine for the Northern Hemisphere 2018-19’, The Francis Crick Institute, Feb. 2018
- 33.Kissling E. Interim 2018/19 influenza vaccine effectiveness: six European studies, October 2018 to January 2019. Eurosurveillance. 2019;24(8) doi: 10.2807/1560-7917.ES.2019.24.1900121. Feb. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Skowronski, Danuta M., ‘Viral genomic variation and vaccine effectiveness across consecutive influenza A(H3N2) epidemics in Canada, 2016-17 and 2017-18’, presented at the Canadian Immunization Conference, Ottawa, Canada, 04-Dec-2018.
- 35.Skowronski D.M. Serial vaccination and the antigenic distance hypothesis: effects on influenza vaccine effectiveness during A(H3N2) epidemics in Canada, 2010–2011 to 2014–2015. J. Infect. Dis. 2017;215(7):1059–1099. doi: 10.1093/infdis/jix074. Apr. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Smith D.J., Forrest S., Ackley D.H., Perelson A.S. Variable efficacy of repeated annual influenza vaccination. Proc. Natl. Acad. Sci. U. S. A. 1999;96(24):14001–14006. doi: 10.1073/pnas.96.24.14001. Nov. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kissling E., Rondy M., I-MOVE/I-MOVE+ study team Early 2016/17 vaccine effectiveness estimates against influenza A(H3N2): I-MOVE multicentre case control studies at primary care and hospital levels in Europe. Euro Surveill. Bull. Eur. Sur Mal. Transm. Eur. Commun. Dis. Bull. 2017;22(7) doi: 10.2807/1560-7917.ES.2017.22.7.30464. 16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fukushima W., Hirota Y. Basic principles of test-negative design in evaluating influenza vaccine effectiveness. Vaccine. 2017;35(36):4796–4800. doi: 10.1016/j.vaccine.2017.07.003. 24. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.