Abstract
Background
miRNA‐181a has been implicated in autoimmunity and apoptosis. Therefore, this study was conducted to explore its possible role in pancreatic beta‐cells dysfunction.
Methods
miRNA‐181a expression was evaluated by real‐time PCR in serum of 40 type 1 diabetic children and adolescents and 40 age‐ and gender‐matched healthy controls.
Results
miRNA‐181a expression was significantly higher in diabetic children and adolescents and it was negatively correlated to fasting C‐peptide and SMAD7 levels.
Conclusion
miRNA‐181a appears to play a potential role in pancreatic beta‐cells dysfunction via SMAD7.
Keywords: C‐peptide, miRNA, miRNA‐181a, SMAD7, T1DM
INTRODUCTION
Diabetes mellitus is considered as the most common endocrine–metabolic disorder of childhood and adolescence. It is characterized by hyperglycemia that occurs when beta cells are unable to function properly or have been destroyed 1.
Type‐1 diabetes mellitus (T1DM) is a chronic autoimmune disease that results from destruction of the beta cells of islets of Langerhans 2. In most western countries, T1DM accounts for over 90% of childhood and adolescent diabetes 3, 4. In Egypt, the estimated prevalence of T1DM in children and adolescents is 0.38/1,000 and the overall incidence is 3.5/100,000 5.
Although autoimmunity is the predominant effector mechanism of T1DM, yet it may not be its primary cause 2 and it has been found that pancreatic beta cells death by apoptosis contributes significantly in T1DM 6.
In recent years, microRNAs (miRNAs), a family of short (average of 22 nucleotides long), naturally occurring, small antisense noncoding RNAs, have emerged as important posttranscriptional regulators of gene expression 3. They have since been discovered to be widely distributed, endogenous controllers of gene and protein expression by binding to the 3’‐untranslated region of specific mRNAs and interfering with protein synthesis by inducing mRNA degradation or repressing translation 7, 8. Following the discovery of the first miRNA in Caenorhabditis elegans 9, the important roles of miRNAs in a variety of biological processes, including development, differentiation, apoptosis, lipid metabolism, and cancer have been discovered 10, 11, 12, 13.
miRNA‐181a is a member of miRNA‐181 family. It has been found that miRNA‐181a induced apoptosis through downregulation of the expression of anti‐apoptotic proteins 14. Moreover, a recent study found that the downregulation of miRNA‐181a significantly inhibited the H2O2‐induced cellular apoptosis, reactive oxygen species (ROS) production, mitochondrial structure disruption, and activation of key signaling proteins in the mitochondrial apoptotic pathway 15.
Transforming growth factor beta (TGF‐β) is a pleiotropic cytokine regulating a variety of cellular processes such as apoptosis and immune response 16. It has been found that overexpression of SMAD7 facilitated cell proliferation by antagonizing TGF‐β‐mediated antiproliferative gene responses and that most members of the TGF‐β pathway are known to be targeted by one or more miRNAs 17.
Therefore, we conducted this study to evaluate the expression of miRNA‐181a and to investigate the relation between miRNA‐181a and the different clinical and laboratory parameters in type 1 diabetic children and adolescents. Additionally, the usefulness of microRNA‐181a as a biomarker of residual pancreatic beta cells function was analyzed. The circulating levels of SMAD7 were also measured and correlated to the expression of miRNA‐181a, aiming at clarifying the role of SMAD7 as a functional target of miRNA‐181a in mediating pancreatic beta cells dysfunction.
MATERIALS AND METHODS
This study was conducted on 40 children and adolescents with uncomplicated T1DM recruited from Diabetes Clinic, Children's Hospital, Ain Shams University. They were 19 males (47.5%) and 21 females (52.5%). Their age ranged from 6.5 to 16 years with a mean age of 12.38 ± 2.75 years. Duration of diabetes ranged from 5 to 13 years (8.9 ± 2.3 years).
The control group consisted of 40 healthy children and adolescents matched in age and gender to the study group. They were 16 males (40.0%) and 24 females (60.0%). Their age ranged from 6 to 16 years with a mean age of 10.88 ± 3.23 years. All participants were subjected to history taking and thorough clinical examination.
This study has complied with the principles laid down in the Declaration of Helsinki, adopted by the 18th World Medical Assembly, Helsinki, Finland, June 1964, and recently amended at the 59th World Medical Assembly, Seoul, Korea, October 2008. The entire protocol was approved by institutional ethical committee. All parents or care givers provided signed informed consent for participation in the study as required.
Blood Sample Collection and Processing
Venous blood samples (5 ml) were aseptically withdrawn from all the participants after an overnight fasting for 12–14 h and divided into two portions as follows: 2.0 ml of blood was placed in an ethylenediaminetetraacetic acid (EDTA) containing tube for complete blood picture using coulter B66, Miami, Florida, and the determination of glycated hemoglobin (HbA1c) using Helena GLYCO‐Tek affinity column method (Helena Laboratories, Beaumont, Texas). The remaining 3.0 ml of blood was used for separation of serum. The separated serum samples were kept frozen at −80°C until used in the determination of total cholesterol, HDL‐cholesterol, and triglycerides according to the manufacturers’ instructions of standard enzymatic kits (Randox Laboratories, Crumlin, UK), C‐peptide according to the manufacturer's instructions of Immuno‐Biological Laboratories (IBL) enzyme‐linked immunosorbent assay (ELISA) kit (Immuno‐Biological Laboratories, Inc., Minneapolis, MN), SMAD7 according to the manufacturer's instructions of Biocompare ELISA kit (Atlanta, GA), and miRNA‐181a expression using quantitative reverse transcription‐PCR. LDL‐cholesterol levels calculated using Friedewald equation. Urinary albumin excretion (UAE) was determined in early morning fasting urine samples as albumin‐to‐creatinine ratio by an immuno‐turbidimetric method performed on COBAS Integra 800 (Roche Diagnostics, Mannheim, Germany).
Quantitative Reverse Transcription PCR (qRT–PCR) of miRNA‐181a
Total RNA was isolated using miRNeasy Serum/Plasma Kit (Qiagen, Valencia, CA). The miRNAs were reversely transcribed into cDNAs according to the manufacturer's instructions of miScript II RT Kit (Qiagen, Valencia, CA). Each reaction mixture of qRT–PCR contained 10 μl of RT product (normalized to 500 ng), 12.5 μl of SYBR GREEN PCR Master Mix, 0.5 μM of each primer, and deionized water to a total volume of 25 μl. Reactions were run with the following thermal cycling parameters: 95°C for 5 min followed by 35 cycles of 95°C, 57°C, and 75°C for 10, 30, and 30 sec, respectively. Each sample was normalized based on its endogenous U6 RNA content. Relative expression of miRNA‐181a was presented as fold expression in relation to the control sample; the actual values were calculated using the 2−ΔΔCt equation, where ΔΔCt = [Ct miRNA‐181a – Ct U6 RNA] (diabetic sample) [Ct miRNA‐181a – Ct U6 RNA] (control sample).
STATISTICAL ANALYSIS
The analysis was done using the Statistical Package for the Social Sciences (SPSS software version 19, SPSS, Inc., Chicago, IL). Results were expressed as mean ± standard deviation (SD). Differences between continuous variables were analyzed using Student's t‐test. Correlation between different variables was performed by Pearson. Statistical significance was set at a value of P < 0.05. Receiver operating characteristic (ROC) curve was used to discriminate positive from negative results. It determined the threshold value for optimal sensitivity and specificity, which was constructed by calculating the true‐positive fraction (sensitivity percent) and false‐positive fraction (100‐specificity) of markers at several cutoff points. Sample size was calculated using the CaTS‐Power Calculator (www.sph.umich.edu/csg/abecasis/CaTS). The power of study was 80% and relative risk for power calculation was set at 2.
RESULTS
The vital signs and anthropometric measures of all participants are shown in (Table 1). N‐fold miRNA‐181a expression (P < 0.001) and HbA1C (P < 0.001) were significantly higher in type 1 diabetic children and adolescents compared to healthy controls. On the other hand, SMAD7 (P < 0.001) and fasting C‐peptide (P < 0.001) were significantly lower in type 1 diabetic children and adolescents compared to healthy controls (Table 2).
Table 1.
Vital Signs and Anthropometric Measures of the Studied Groups
| Parameters | Diabetic group Mean ± SD (range) | Control group Mean ± SD (range) | P |
|---|---|---|---|
| Pulse | 75.65 ± 6.83 (60−90) | 76.25 ± 8.89 (65−95) | 0.736 |
| Systolic BP | 106.50 ± 6.62 (100−120) | 109.25 ± 7.56 (100−120) | 0.087 |
| Diastolic BP | 70.13 ± 6.45 (60−85) | 71.63 ± 5.24 (60−80) | 0.257 |
| Weight (Kg) | 33.40 ± 6.44 (22−49) | 33.58 ± 6.72 (22−49) | 0.906 |
| Height (m) | 1.44 ± 0.15 (1.18−1.68) | 1.40 ± 0.13 (1.14−1.60) | 0.189 |
| BMI (Kg/m2) | 18.14 ± 3.13 (16.4−24.08) | 17.13 ± 1.91 (15.4−22.37) | 0.083 |
| Waist circumference (cm) | 60.93 ± 4.63 (51−70) | 61.13 ± 4.59 (54−70) | 0.850 |
P > 0.05 is nonsignificant.
Table 2.
Laboratory Results of the Studied Groups
| Parameters | Diabetic group Mean ± SD (range) | Control group Mean ± SD (range) | P |
|---|---|---|---|
| Hemoglobin (g/dl) | 11.53 ± 1.80 (9.9−13.4) | 11.33 ± 1.19 (9.2−13.5) | 0.445 |
| Total leucocytic count/cmm | 8.80 ± 1.56 (4.3−12.1) | 8.08 ± 2.07 (4.3−12.5) | 0.084 |
| Platelets count/cmm | 275.03 ± 56.95 (176−400) | 251.53 ± 74.09 (155−398) | 0.116 |
| Triglycerides (mg/dl) | 77.95 ± 18.09 (65−134) | 73.08 ± 13.3 (57−98) | 0.174 |
| Total cholesterol (mg/dl) | 158.73 ± 29.55 (106−190) | 149.6 ± 22.68 (109−161) | 0.125 |
| HDL (mg/dl) | 41.28 ± 5.10 (31−53) | 40.20 ± 5.93 (32−51) | 0.387 |
| LDL (mg/dl) | 81.42 ± 17.51 (60−120) | 87.72 ± 20.06 (64−150) | 0.138 |
| UAE (mg/g urinary creatinine) | 20.82 ± 4.18 (15−29) | 19.49 ± 3.46 (15−26) | 0.123 |
| HbA1c (%) | 11.48 ± 1.79 (9.0−16.0) | 5.37 ± 0.59 (4.4−6.2) | <0.001** |
| Fasting C‐peptide (ng/ml) | 0.33 ± 0.14 (0.09−0.45) | 2.05 ± 0.87 (0.5−3.2) | <0.001** |
| SMAD7 (ng/ml) | 0.45 ± 0.09 (0.11−2.1) | 4.35 ± 1.65 (1.0−9.9) | <0.001** |
| N‐fold miRNA‐181a | 7.67 ± 2.58 (1.22−11.7) | 1.22−11.7 (0.98−5.3) | <0.001** |
**P < 0.01 is highly significant.
The correlation coefficients between N‐fold miRNA‐181a expression and the investigated laboratory parameters revealed significant positive association between N‐fold miRNA‐181a expression and HbA1c (r = 0.627, P = 0.012), triglycerides (r = 0.520, P = 0.047) and total cholesterol (r = 0.668, P = 0.007), and significant negative association with fasting C‐peptide (r = −0.391, P = 0.013) and SMAD7 (r = −0.384, P = 0.014) in type 1 diabetic children and adolescents (Table 3).
Table 3.
Correlation Between N‐fold miRNA‐181a and the Studied Parameters
| Diabetic group | Control group | |||
|---|---|---|---|---|
| Parameters | r | P | r | P |
| Age (years) | −0.107 | 0.505 | 0.201 | 0.213 |
| Weight (Kg) | −0.008 | 0.958 | 0.119 | 0.465 |
| Height (m) | −0.07 | 0.662 | 0.216 | 0.181 |
| BMI (Kg/m2) | −0.076 | 0.636 | 0.059 | 0.717 |
| Pulse | −0.099 | 0.542 | 0.129 | 0.428 |
| Systolic blood pressure | −0.152 | 0.349 | −0.404 | 0.135 |
| Diastolic blood pressure | 0.098 | 0.546 | −0.154 | 0.343 |
| Hemoglobin (g/dl) | −0.061 | 0.709 | 0.155 | 0.340 |
| Total leucocytic count/cmm | 0.259 | 0.107 | 0.045 | 0.784 |
| Platelets count/cmm | 0.306 | 0.055 | −0.140 | 0.389 |
| Triglycerides (mg/dl) | 0.520a | 0.047 | 0.139 | 0.394 |
| Total cholesterol (mg/dl) | 0.668b | 0.007 | −0.0167 | 0.302 |
| HDL (mg/dl) | 0.204 | 0.208 | −0.115 | 0.480 |
| LDL (mg/dl) | 0.080 | 0.625 | 0.090 | 0.759 |
| UAE (mg/g urinary creatinine) | 0.040 | 0.805 | −0.222 | 0.168 |
| HbA1c (%) | 0.627a | 0.012 | −0.206 | 0.202 |
| Fasting C‐peptide (ng/ml) | −0.391a | 0.013 | 0.251 | 0.146 |
| SMAD7 (ng/ml) | −0.384a | 0.014 | −0.13 | 0.456 |
Correlation is significant at the 0.05 level.
Correlation is highly significant at the 0.01 level.
The overall diagnostic performance of N‐fold miRNA‐181a expression for pancreatic beta cells dysfunction was assessed by ROC curve analysis and the best cutoff value was 2.45 (Fig. 1). Table 4 shows sensitivity, specificity, positive predictive value (PPV), negative predictive value (NPV), and accuracy of N‐fold miRNA‐181a expression in determination of pancreatic beta cells dysfunction.
Figure 1.
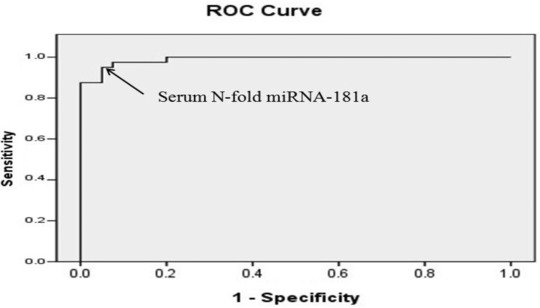
The ROC curve analysis for N‐fold miRNA‐181a expression in the diabetic group versus healthy group to calculate the best cutoff value. Area under the curve is 0.989, standard error is 0.006, and 95% confidence limits are 0.977–1.001. Arrows denote cutoff point at 2.45.
Table 4.
Sensitivity and Specificity of N‐Fold miRNA‐181a in Detection of Pancreatic Beta Cells Dysfunction
| Variables False‐negative (n, %) False‐positive (n, %) | Sensitivity | Specificity | NPV | PPV | Accuracy |
|---|---|---|---|---|---|
| N‐fold miRNA‐181a expression: False‐negative (1, 2.5%) False‐positive (4, 10%) | 97.3% | 90.7% | 97.5% | 90% | 94% |
Total number of cases (n = 80), diabetic group (n = 40), and healthy group (n = 40).
DISCUSSION
A number of miRNAs have been identified in the extracellular environment. As they may regulate a significant portion of the transcriptome and proteome, considerable attention has focused on miRNAs as mediators or biomarkers of illness 18.
Oxidative stress is involved in beta cells destruction 19. It is recognized as a mediator in the development of macrovascular or cardiovascular complications in T1DM 20.
miRNA‐181a has been implicated in apoptosis 14, 15. Researchers found that H2O2 induced upregulation of miRNA‐181a resulting in apoptosis of cardiac myocytes. They also demonstrated that the use of anti‐miRNA‐181a blocked H2O2‐induced apoptosis by regulating mitochondria‐related apoptotic pathways 15. In another study, researchers found that the transfection of primary chronic lymphocytic leukemia cells with mature mimics of miRNA‐181a increased apoptosis 14. In addition to its role in apoptosis, miRNA‐181a has been implicated in autoimmunity via regulation of a number of phosphatases known to negatively control T‐cell sensitivity 21.
Based on the previous results, our study evaluated the expression of miRNA‐181a in peripheral blood of children and adolescents with T1DM. Our results revealed overexpression of miRNA‐181a among diabetic children and adolescents compared to healthy controls. We found significant positive correlation between the expression of miRNA‐181a and triglycerides and total cholesterol in the diabetic group, which indicates that miRNA‐181a correlates with parameters of lipid metabolism in T1DM. Furthermore, our results revealed significant positive correlation between the expression of miRNA‐181a and the levels of HbA1c of diabetic group.
Although no enough studies were found as regards the expression of miRNA‐181a in T1DM, our findings were in agreement with 21, 22 who found an upregulation of miRNA‐181a in serum of diabetic patients and with Klöting et al. 23 who found that the expression of miRNA‐181a was positively correlated with HbA1c levels in obese subjects.
As connecting peptide (C‐peptide) is used as a marker of endogenous insulin production and reflection of pancreatic beta cells function 24. We assessed residual beta cells function through the measurement of fasting C‐peptide in our diabetics. We found that the mean level of C‐peptide was significantly decreased among diabetics compared to healthy controls. Furthermore, the expression of miRNA‐181a was negatively correlated with C‐peptide levels.
Our results revealed that overexpression of miRNA‐181a in diabetic children and adolescents was associated with significant reduction in the mean level of SMAD7. This comes in concordance with the study done by Parikh et al. 25, who demonstrated that miRNA‐181a activated TGF‐β via suppression of SMAD7. Consequently, our data confirmed SMAD7 as a functional target of miRNA‐181a and we therefore postulated that miRNA‐181a may induce its damaging effect to pancreatic beta cells via the negative regulation to SMAD7.
The ROC curve was used to determine the best cutoff value for N‐fold miRNA‐181a expression to discriminate between healthy and diabetic children and it was 2.45. Applying this cutoff value, the sensitivity and specificity of miRNA‐181a were 97.3% and 90.7%, respectively. The relatively high false‐positive results (10%) of N‐fold miRNA‐181a expression may be an indicator of its predictive role for beta cells dysfunction.
In conclusion, the control of pancreatic beta cells apoptosis is important for the prevention and treatment of T1DM. The concordance between increase expression of miRNA‐181a and decrease residual beta cells function supports the potential role of this miRNA during disease progression of T1DM. Assessment of circulating levels of miRNA‐181a may be a potential valuable future tool for treatment planning and monitoring of intervention therapies designed to preserve/regenerate beta cells function in T1DM.
CONFLICT OF INTEREST
The authors report no conflicts of interest. The authors alone are responsible for the content and writing of this article.
REFERENCES
- 1. International Society of Pediatric and Adolescent Diabetes (ISPAD) . ISPAD Clinical Practice Consensus Guidelines 2011 Compendium: Definition, epidemiology and classification of diabetes in children and adolescents. Pediatr Diabetes 2014;15:8–15. [DOI] [PubMed] [Google Scholar]
- 2. van Belle TL, Coppieters KT, von Herrath MG. Type 1 diabetes: Etiology, immunology, and therapeutic strategies. Physiol Rev 2011;91:79–118. [DOI] [PubMed] [Google Scholar]
- 3. Carthew RW, Sontheimer EJ. Origins and Mechanisms of miRNAs and siRNAs. Cell 2009;136:642–655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Lee RC, Ambros V. An extensive class of small RNAs in Caenorhabditis elegans . Science 2001;294:862–864. [DOI] [PubMed] [Google Scholar]
- 5. Salem MA, Tantawy AA, Radwan M, Mansour E, El Nekhely I. Clinico‐epidemiological study of type 1 diabetes mellitus in Egyptian children and adolescents. Pediatr Diabetes 2010;11:94–95. [Google Scholar]
- 6. Johnson JD, Luciani DS. Mechanisms of pancreatic beta‐cell apoptosis in diabetes and its therapies. Adv Exp Med Biol 2010;654:447–462. [DOI] [PubMed] [Google Scholar]
- 7. Bartel DP. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell 2004;116:281–297. [DOI] [PubMed] [Google Scholar]
- 8. Guo H, Ingolia NT, Weissman JS, Bartel DP. Mammalian microRNAs predominantly act to decrease target mRNA levels. Nature 2010;466:835–840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Lee RC, Feinbaum RL, Ambros V. The C. elegans heterochronic gene lin‐4 encodes small RNAs with antisense complementarity to lin‐14. Cell 1993;75:843–854. [DOI] [PubMed] [Google Scholar]
- 10. Lu J, Getz G, Miska EA, et al. MicroRNA expression profiles classify human cancers. Nature 2005;435:834–838. [DOI] [PubMed] [Google Scholar]
- 11. Thum T, Galuppo P, Wolf C, et al. MicroRNAs in the human heart: A clue to fetal gene reprogramming in heart failure. Circulation 2007;116:258–267. [DOI] [PubMed] [Google Scholar]
- 12. Zhu H, Shyh‐Chang N, AV Segrè, et al. The Lin28/let‐7 axis regulates glucose metabolism. Cell 2011;147:81–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Malumbres M. miRNAs and cancer: An epigenetics view. Mol Aspects Med 2013;34:863–874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Zhu DX, Zhu W, Fang C, et al. miR‐181a/b significantly enhances drug sensitivity in chronic lymphocytic leukemia cells via targeting multiple anti‐apoptosis genes. Carcinogenesis 2012;33:1294–1301. [DOI] [PubMed] [Google Scholar]
- 15. Wang L, Huang, H , Fan, Y , et al. Effects of downregulation of microRNA‐181a on H2O2‐induced H9c2 cell apoptosis via the mitochondrial apoptotic pathway. Oxid Med Cell Longev 2014;2014. Article ID 960362. doi: 10.1155/2014/960362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Yan X, Liu Z, Chen Y. Regulation of TGF‐beta signaling by SMAD7. Acta Biochim Biophys Sin 2009;41:263–272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Huo YY, Zhang KT, Li BY et al. Effect of overexpression of SMAD7 gene on cell proliferation. Zhonghua Zhong Liu Za Zhi 2004;26(9):521–524. [PubMed] [Google Scholar]
- 18. Argyropoulos C, Wang K, McClarty S, et al. Urinary microRNA profiling in the nephropathy of type 1 diabetes. PLoS ONE 2013;8(1):e54662. doi: 10.1371/journal.pone.0054662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Kaneto H, Katakami N, Kawamori D, et al. Involvement of oxidative stress in the pathogenesis of diabetes. Antioxid Redox Signal 2007;9(3):355–366. [DOI] [PubMed] [Google Scholar]
- 20. Wegner M, Pioruńska‐Stolzmann M, Araszkiewicz A, Zozulińska‐Ziółkiewicz D, Wierusz‐Wysocka B. Evaluation of paraoxonase 1 arylesterase activity and lipid peroxide levels in patients with type 1 diabetes. Pol Arch Med Wewn 2011;121(12):448–455. [PubMed] [Google Scholar]
- 21. Li QJ, Chau J, Ebert PJ, et al. miR‐181a is an intrinsic modulator of T cell sensitivity and selection. Cell 2007;129(1):147–161. [DOI] [PubMed] [Google Scholar]
- 22. Nielsen LB, Wang C, Sørensen K, Bang‐Berthelsen CH, Hansen L, Andersen MM, Hougaard P, Juul A, Zhang C, Pociot F, Mortensen HB. Circulating levels of microRNA from children with newly diagnosed type 1 diabetes and healthy controls: Evidence that miR‐25 associates to residual beta‐cell function and glycaemic control during disease progression. Exp Diabetes Res 2012;2012. Article ID 896362. doi: 10.1155/2012/896362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Klöting N, Berthold S, Kovacs P, et al. MicroRNA expression in human omental and subcutaneous adipose tissue. PLOS ONE 2009;4(3):e4699. doi: 10.1371/journal.pone.0004699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. VanBuecken DE, Greenbaum CJ. Residual C‐peptide in type 1 diabetes: What do we really know? Pediatr Diabetes 2014;15:84–90. [DOI] [PubMed] [Google Scholar]
- 25. Parikh A, Lee C, Joseph P, et al. DiFeo A microRNA‐181a has a critical role in ovarian cancer progression through the regulation of the epithelial‐mesenchymal transition. Nat Commun 2014;5. Article number 2977. doi: 10.1038/ncomms3977. [DOI] [PMC free article] [PubMed] [Google Scholar]


