Abstract
Background
H19 is one of the long non‐coding RNAs (LncRNA) that is related to the progression of many diseases including cancers. This work was carried out to study the level of the long non‐coding RNA; H19, in plasma of patients with gastric cancer (GC) and to assess its significance in their clinical management.
Methods
Sixty‐two participants were enrolled in the present study. The first group included 32 GC patients. The second group was formed of 30 age and sex matched healthy volunteers serving as a control group. Plasma samples were used to assess H19 gene expression using real‐time quantitative PCR technique.
Results
H19 expression was up‐regulated and closely related to TNM cancer stages in GC patients. Using Receiver Operating Characteristic (ROC) curve analysis, a cutoff level of 0.5 was set for H19 expression to diagnose GC cases achieving a sensitivity of 68.75%, specificity of 56.67%, positive predictive value (PPV) 62.86% and negative predictive value (NPV) 62.96% with an area under the curve (AUC) of 72.4%. Combined use of Carcinoembryonic Antigen (CEA) and H19 level in GC diagnosis was evaluated using ROC curve revealing improvement in performance with an area under the curve of 80.4%.
Conclusions
Up‐regulation of H19 is closely associated with gastric cancer displaying progressive up‐regulation in advanced stages of the disease implementing its role as a potential non‐invasive diagnostic biomarker in gastric cancer and as a novel tool in gastric cancer management with better performance achieved on using both CEA and H19 simultaneously.
Keywords: LncRNA‐H19‐gastric cancer‐biomarker
Introduction
Gastric cancer (GC) is ranked as one of the common causes of cancer‐related deaths worldwide despite the improvement in management strategies of radical surgeries and novel chemotherapies 1.
Early lymphatic, blood, and peritoneal dissemination of GC has limited the curative role of radical surgery, except in patients with early‐stage cancers who often lose the window for surgical resection due to the lack of effective tools for early diagnosis resulting in very low survival rates 2, 3.
Long non‐coding RNAs (LncRNA) are non‐coding RNAs that are longer than 200 nucleotides with many of them demonstrated to have major regulatory roles in the development and progression of many diseases including cancers 4, 5.
Production of LncRNA is regulated by mechanisms similar to these of protein‐coding genes, such as histone modifications and RNA splicing, and their expression shows tissue‐specific patterns 6.
LncRNAs have been implicated to regulate a range of biological functions and the disruption of some of these functions, such as genomic imprinting and transcriptional regulation, plays a critical role in cancer development and metastatic events 6.
One of the first LncRNA genes reported was the imprinted H19 gene 7, 8 with loss of imprinting and subsequent strong gene expression in a variety of cancers 9.
H19 gene is allocated in a region near by the telomeric area of chromosome 11p15.5 which is frequently involved in many pediatric and adult tumors. It encodes a 2.3‐kb protein which is highly expressed in fetal tissues, but is down‐regulated in most tissues shortly after birth with the exception of skeletal tissue and cartilage 10.
H19 has been implicated as having both oncogenic and tumor suppression properties being up‐regulated in a number of human cancers, including hepatocellular, bladder, and breast carcinomas, suggesting its oncogenic function, while its tumor suppressor role was proved using animal model experiments in other cancers including colorectal cancer 11.
Based on these findings, the objective of the present work was to study the level of the long non‐coding RNA; H19, in plasma of patients with GC and to assess its significance in their clinical management.
Materials and Methods
The study approval was obtained from the ethics committee of the Faculty of Medicine, Alexandria University. All patients provided signed, written informed consent to participate in this study.
A total of 62 participants were enrolled in the present study. The first group included 32 GC patients, while the second group was formed of 30 age and sex matched healthy volunteers who served as a control group. Patients were recruited at the Gastrointestinal and Clinical Oncology Units, Alexandria Main University Hospital. For all patients, diagnosis of GC was confirmed by upper gastrointestinal endoscopy and biopsy. Staging of GC was according to the TNM classification system developed by the American Joint Committee on Cancer (AJCC) 12, while grading was performed according to Laurén classification of GC 13. All blood samples were withdrawn before any surgeries or therapeutic interventions.
Liver function tests including serum aspartate aminotransferase (AST), alanine aminotransferase (ALT), and prothrombin time were assessed. Complete blood count was offered to all participants enrolled in the study. Serum Carcinoembryonic Antigen (CEA) level was assayed on Advia™ Centaur Immunoassay System.
Plasma Collection, RNA Extraction, and cDNA Synthesis
Whole‐blood EDTA samples were obtained from each participant in the study. All EDTA tubes were centrifuged at 1,200 × g for 10 min at 4°C to precipitate the blood cells. The supernatants were transferred to new plain tubes and centrifuged at 12,000 × g for 10 min at 4°C to completely remove cellular components. Plasma was then stored at −80∘C until further analysis 14. Total RNA was extracted from 1 ml plasma using miRNeasy Mini Kit (Qiagen, Hilden, Germany) according to the manufacturer's instructions.
For cDNA synthesis, High Capacity cDNA Reverse Transcription Kit (Applied Biosystems, Foster City, CA) was used. Reverse‐transcription master mix was added to RNA and incubated at 25°C for 10 min then at 37°C for 120 min, 85°C for 5 min with a final hold at 4°C.
Assessment of H19 Gene Expression Using Real‐Time Quantitative PCR
Real‐time PCR was performed on the Applied Biosystems® 7500 Real‐Time PCR Systems (Applied Biosystems) using QuantiFast Probe Assay Duplex Kit (Applied Biosystems, Cat no.243132).
Amplification was carried out in a final volume of 25 μl containing 2× QuantiFast Multiplex PCR Master Mix, 1.25 μl of each probe (with the first probe labeled with FAM and the other with MAX) and cDNA in a concentration not more than 100 ng/reaction.
Thermal cycling conditions included an initial Taq activation step at 95°C for 5 min followed by 45 cycles of 95°C for 30 sec (denaturation step) and 60°C for 30 sec (annealing/extension).
Expression of the target gene; H19 was normalized to that of GAPDH which is a house keeping gene and was calculated in plasma of gastric cancer patients' samples vs. healthy controls by the 2−ΔΔCt method.
Statistical Analysis
Data were analyzed using SPSS software package version 20.0 (SPSS, Chicago, IL). Qualitative data were described using number and percent and was compared using Chi‐square test or Fisher Exact test. Normally distributed quantitative data were expressed as mean ± SD and compared using student t‐test. Abnormally distributed data were expressed as median (Min.–Max.) and compared using Mann–Whitney test. Receiver Operating Characteristic (ROC) curve was used to assess the cutoff value for H19 expression to distinguish gastric cancer patients from controls. Sensitivity, specificity, and area under the curve (AUC) were calculated. Statistical significance was set at P < 0.05.
Based on the mean and standard deviation of plasma H19 among cases and controls and by using Epi save software, the power of the study was calculated to be 96.21%.
Results
The group of GC patients included 19 males (59.4%) and 13 females (40.6%), while the control group included 15 males (50%) and 15 females (50%).
The mean age of those with GC was 43.44 ± 6.84 years, whereas those of the control group had a mean age of 43.53 ± 7.39 years.
No significant difference was detected between both groups as regards gender (P = 0.45) or age (P = 0.95).
According to TNM‐staging system, most of GC patients were at late stages; with only 2 patients (6.2%) at stage I, 7 patients (21.9%) at stage II, 12 patients (37.5%) at stage III, and 11 patients at stage V (34.4%).
Histological grading showed 26 patients (81.3%) of intestinal type, 5 patients (15.6%) of diffuse type, and only 1 (3.1%) patient of the intermediate type.
Biochemical, hematological data, and CEA level of all those participating in the study are shown in Table 1.
Table 1.
Biochemical, hematological data, and CEA level of all those participating in the study
| Variable | GC patients (n = 32) | Control (n = 30) | P |
|---|---|---|---|
| AST (U/l) | 25.0 (16.0–76.0) | 28.0 (16.0–41.0) | 0.184 |
| ALT (U/l) | 24.50 (13.0–95.0) | 28.0 (12.0–40.0) | 0.553 |
| Prothrombin activity (%) | 98.0 (50.0–100.0) | 100.0 (95.0–100.0) | 0.018a |
| CEA (ng/ml) | 5.0 (1.20–190.0) | 3.20 (1.20–5.0) | <0.001a |
| Hemoglobin (g/dl) | 10.95 ± 1.41 | 12.87 ± 0.85 | <0.001a |
| WBCs ×109/l | 7.0 (3.90–14.50) | 5.35 (4.0–8.0) | <0.001a |
| Platelets ×109/l | 169.16 ± 30.23 | 218.33 ± 51.73 | <0.001a |
Parametric data were presented as mean ± SD and analyzed by student t‐test, while non‐parametric data were presented as median (Min.–Max.)) and compared using Mann–Whitney test.
Statistically significant at P < 0.05.
H19 expression showed a statistically significant difference between both groups being up‐regulated in the GC patients (P < 0.001) (Fig. 1).
Figure 1.
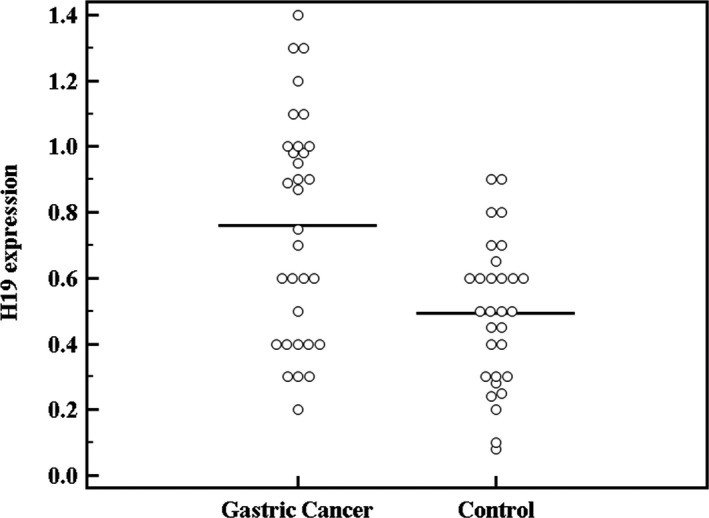
Comparison between the studied groups as regards H19 expression.
On comparing H19 level between the different TNM stages in GC patients, a statistically significant difference was detected between the four stages (P = 0.014). Applying post hoc test revealed a greater difference of H19 gene expression in advanced stages as compared to early ones (Table 2).
Table 2.
Relation between TNM stage and H19 expression
| TNM stage | KWχ | P | ||||
|---|---|---|---|---|---|---|
| I (n = 2) | II (n = 7) | III (n = 12) | IV (n = 11) | |||
| H19 expression | ||||||
| Min.–Max. | 0.20–0.50 | 0.30–1.0 | 0.30–1.10 | 0.30–1.40 | 10.574a | 0.014a |
| Mean ± SD. | 0.35 ± 0.21 | 0.53 ± 0.24 | 0.76 ± 0.27 | 0.99 ± 0.33 | ||
| P | P 1 = 0.370, P 2 = 0.100, P 3 = 0.048a, P 4 = 0.135, P 5 = 0.014a, P 6 = 0.048a | |||||
KW: Kruskal–Wallis test, comparison between groups using Mann–Whitney test.
P 1: P value for comparing between I and II group.
P 2: P value for comparing between I and III group.
P 3: P value for comparing between I and IV group.
P 4: P value for comparing between II and group III.
P 5: P value for comparing between II and IV group.
P 6: P value for comparing between III and IV group.
Statistically significant at P < 0.05.
In addition, H19 expression showed positive correlation with CEA level (r = 0.470, P = 0.007) (Fig. 2).
Figure 2.
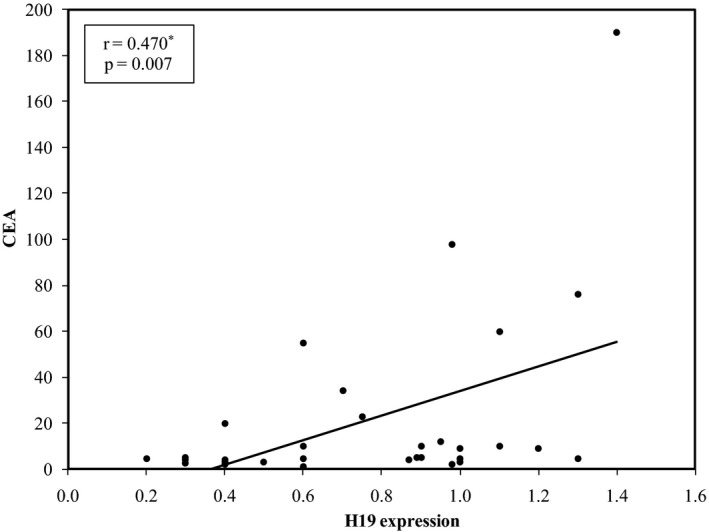
Correlation between H19 expression and CEA in Gastric Cancer patients.
On the other hand, no statistically significant relation was detected between H19 level and histological grade (P = 0.505).
Using ROC curve analysis, a cutoff level of 0.5 was set for H19 expression to diagnose GC cases achieving a sensitivity of 68.75%, specificity of 56.67%, positive predictive value (PPV) 62.86%, and negative predictive value (NPV) 62.96% with an area under the curve (AUC) of 72.4%.
Combined use of CEA and H19 level in GC diagnosis was evaluated using ROC curve revealing improvement in performance with an AUC of 80.4% (Fig. 3).
Figure 3.
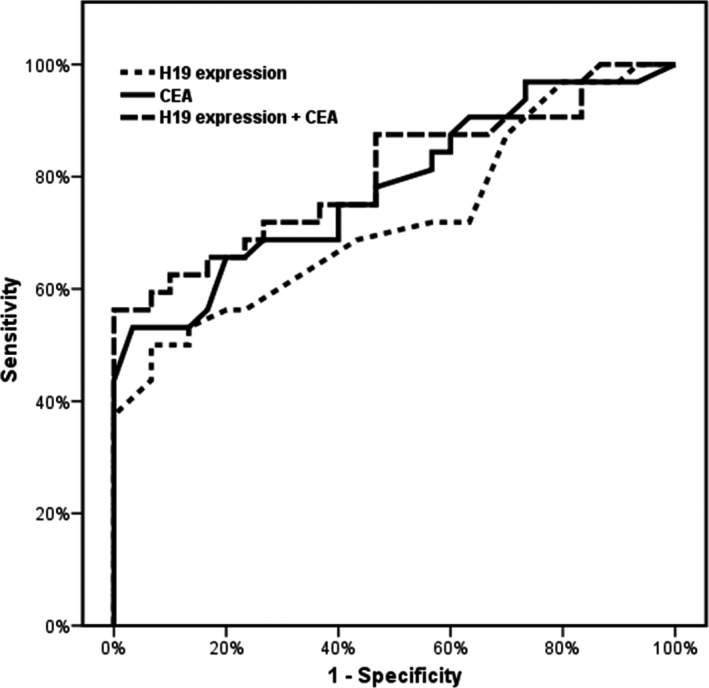
ROC curve for H19 expression to diagnose gastric cancer patients.
Discussion
Recent advances in the molecular techniques had led researchers to investigate LncRNAs as novel biomarkers suggested especially for their potential role in cancer progression 15 and as GC is one of the common malignancies worldwide, the urge to discover new biomarkers for GC prediction, disease monitoring, and prognosis assessment is of utmost importance for clinical management of GC cases 16.
In the present study, most of GC patients (71.8%) were of late stages (stage III and IV) implying late detection of cancer which is known to be a relatively hidden malignancy with most cases presented at advanced, metastatic stages at first diagnosis 17.
This explains the efforts targeting the detection of a biomarker that can efficiently screen for early GC giving those patients the benefits of early detection.
Although CEA is one of the most frequently used serological biomarkers in cases of GC, yet its use in screening and diagnosis of gastrointestinal tract cancers is hindered by its limited sensitivity and specificity particularly in early cases 18.
In the current study, H19 was up‐regulated in plasma of GC patients (P < 0.001). Many studies associated H19 with many pediatric 19 and adult digestive system tumors 20.
Up‐regulation of H19 in the group of GC patients mostly supports its oncogenic role which is attributed in many studies to loss of imprinting ending in altered gene expression 21, 22.
A study by Yang et al. 23 assayed H19 levels in GC tissues and investigated its biological effect in carcinogenesis. They demonstrated that H19 expression is increased in GC cells compared to normal control cells and that association between H19 and p53 results in p53 inactivation proving the oncogenic role of H19 in gastric cancerous tissues and its potential application in GC therapy.
Association of H19 with factors involved in tumor growth and proliferation as angiopoietin and fibroblast growth factor‐18 is also suggested as a factor behind its oncogenic role 24.
Researchers assayed H19 in post‐operative tissue samples 25, plasma samples 26, and in bone marrow mononuclear cells 27.
Quantification of LncRNAs is performed by real‐time PCR technique as a simple effective procedure in many studies 23, 26 with some researchers 25 using microarray technique to assess multiple genes' level simultaneously.
In the present study, H19 level was successfully and easily assayed in plasma samples then quantified using a real‐time PCR technique. Arita et al. 26 investigated the stability of a number of plasma LncRNAs including, H19, HOX antisense intergenic RNA (HOTAIR), and metastasis associated lung adenocarcinoma transcript‐1 (MALAT1), in patients with gastric carcinoma and proved that LncRNAs can be easily and efficiently extracted out of plasma and that they only exhibited minimal instability under severe conditions.
According to TNM staging system, H19 in the present study, showed progressive up‐regulation in advanced stages of GC.
In other studies, H19 as well as other LncRNAs were related to metastatic events, poor patients' prognosis and diminished survival in many malignancies 28. HOX Antisense Intergenic RNA (HOTAIR) showed strong correlation with liver metastasis and poor patient outcome in colorectal carcinoma 28.
In addition, HOTAIR was considered a significant predictor of disease recurrence and was closely related to patients' aggressive phenotype in cases of cancer breast 29 and hepatocellular carcinoma 30. A study by Li et al. 31 used cell lines and knockdown models to prove the importance of H19 in tumorigenesis and metastasis in GC cells.
As the present work implies, limited sensitivity (68.75%) and specificity (56.67%) of H19 associated with its tendency to up‐regulation in late cancer stages limits its use as a potential screening tool for GC.
Using both markers together showed increased AUC reflecting improved diagnostic performance.
Researchers as Arita et al. 26 suggested an optimal cutoff of 0.32 for H19 plasma level to distinguish between GC cases and normal healthy controls at a sensitivity of 74% and a specificity of 58%. Song et al. (25) estimated an AUC of 61.3% for H19 when used alone and of 76.1% when H19 was used in combination with another LncRNA (uc001lsz).
In conclusion, up‐regulation of H19 is closely associated with gastric cancer displaying progressive up‐regulation in advanced stages of the disease. Convenient, efficient quantification of H19 in plasma using real‐time PCR technique implements its role as a potential non‐invasive diagnostic biomarker in gastric cancer and as a novel tool in gastric cancer management with better performance achieved on using both CEA and H19 simultaneously.
References
- 1. Jemal A, Bray F, Center M, Ferlay J, Ward E, Forman D. Global cancer statistics. CA Cancer J Clin 2011;61:69. [DOI] [PubMed] [Google Scholar]
- 2. Wu C, Hsiung C, Lo S, et al. Nodal dissection for patients with gastric cancer: A randomised controlled trial. Lancet Oncol 2006;7:309–315. [DOI] [PubMed] [Google Scholar]
- 3. Dassen A, Lemmens VE, van de Poll‐Franse LV, et al. Trends in incidence, treatment and survival of gastric adenocarcinoma between 1990 and 2007: A population‐based study in the Netherlands. Eur J Cancer 2010;46:1101–1110. [DOI] [PubMed] [Google Scholar]
- 4. Malone C, Hannon G. Small RNAs as Guardians of the Genome. Cell 2009;136:656–668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Plath K, Fang J, Mlynarczyk‐Evans SK, et al. Role of histone H3 lysine 27 methylation in X inactivation. Science 2003;300:131–135. [DOI] [PubMed] [Google Scholar]
- 6. Derrien T, Johnson R, Bussotti G, et al. The GENCODE v7 catalog of human long noncoding RNAs: analysis of their gene structure, evolution, and expression. Genome Res 2012;22:1775–1789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Gabory A, Jammes H, Dandolo L. The H19 locus: Role of an imprinted non‐coding RNA in growth and development. BioEssays 2010;32:473–480. [DOI] [PubMed] [Google Scholar]
- 8. Brannan C, Dees E, Ingram R, Tilghman S. The product of the H19 gene may function as an RNA. Mol Cell Biol 1990;10:28–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Fellig Y, Ariel I, Ohana P, et al. H19 expression in hepatic metastases from a range of human carcinomas. J Clin Pathol 2005;58:1064–1068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Tabano S, Colapietro P, Cetin I, et al. Epigenetic modulation of the IGF2/H19 imprinted domain in human embryonic and extra‐embryonic compartments and its possible role in fetal growth restriction. Epigenetics 2010;5:313–324. [DOI] [PubMed] [Google Scholar]
- 11. Matouk IJ, DeGroot N, Mezan S, et al. The H19 non‐coding RNA is essential for human tumor growth. PLoS ONE 2007;2:e845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Edge S, Byrd D, Compton C, et al. 2010. AJCC cancer staging manuel, 7th edn New York: Springer. [Google Scholar]
- 13. Lauren P. The two histological main types of gastric carcinoma: Diffuse and so‐called intestinal‐type carcinoma. An attempt at a histo‐clinical classification. Acta Pathol Microbiol Scand 1965;64:31–49. [DOI] [PubMed] [Google Scholar]
- 14. Xie H, Ma H, Zhou D. Plasma HULC as a Promising Novel Biomarker for the Detection of Hepatocellular Carcinoma. Biomed Res Int 2013;2013:136106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Esteller M. Non‐coding RNAs in human disease. Nat Rev Genet 2011;12:861–874. [DOI] [PubMed] [Google Scholar]
- 16. Hajjari M, Behmanesh M, Sadeghizadeh M, Zeinoddini M. Junctional adhesion molecules 2 and 3 may potentially be involved in progression of gastric adenocarcinoma tumors. Med Oncol 2013;30:380. [DOI] [PubMed] [Google Scholar]
- 17. Wu H, Lin W, Tsai K. Advances in molecular biomarkers for gastric cancer: miRNAs as emerging novel cancer markers. Expert Rev Mol Med 2014;16:1–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Li Y, Yang Y, Lu M, Shen L. Predictive value of serum CEA, CA19‐9 and CA72.4 in early diagnosis of recurrence after radical resection of gastric cancer. Hepatogastroenterology 2011;58:2166–2170. [DOI] [PubMed] [Google Scholar]
- 19. DeBaun M, Niemitz E, McNeil D, Brandenburg S, Lee M, Feinberg A. Epigenetic alterations of H19 and LIT1 distinguish patients with Beckwith‐Wiedemann syndrome with cancer and birth defects. Am J Hum Genet 2002;70:604–611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Oliva J, Bardag‐Gorce F, French BA, Li J, French SW. The regulation of non‐coding RNA expression in the liver of mice fed DDC. Exp Mol Pathol 2009;87:12–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Hajjari M, Khoshnevisan A. Potential long non coding RNAs to be considered as biomarkers or therapeutic targets in gastric cancer. Front Genet 2013;4:1–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Lim D, Maher E. Genomic imprinting syndromes and cancer. Adv Genet 2010;70:145–175. [DOI] [PubMed] [Google Scholar]
- 23. Yang F, Bi J, Xue X, et al. Up‐regulated long non‐coding RNA H19 contributes to proliferation of gastric cancer cells. FEBS 2012;279:3159–3165. [DOI] [PubMed] [Google Scholar]
- 24. Matouk IJ, DeGroot N, Mezan S, et al. The H19 non‐coding RNA is essential for human tumor growth. PLoS ONE 2007;2:e84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Song H, Sun W, Ye G, et al. Long non‐coding RNA expression profile in human gastric cancer and its clinical significances. J Transl Med 2013;11:225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Arita T, Ichikawa D, Konishi H, et al. Circulating Long Non‐coding RNAs in Plasma of Patients with Gastric Cancer. Anticancer Res 2013;33:3185–3194. [PubMed] [Google Scholar]
- 27. Cho S, Chang Y, Chang C. MALAT1 long non coding RNA is over expressed in multiple myeloma and may serve as a marker to predict disese progressiom. BMC Cancer 2014;14:809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Kogo R, Shimamura T, Mimori K, et al. Long non coding RNA HOTAIR regulates polycomb‐dependent chromatin modification and is associated with poor prognosis in colorectal cancers. Cancer Res 2011;71:6320–6326. [DOI] [PubMed] [Google Scholar]
- 29. Gupta R, Shah N, Wang K, et al. Long non‐coding RNA HOTAIR reprograms chromatin state to promote cancer metastasis. Nature 2010;46:1071–1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Yang Z, Zhou L, Wu L, et al. Over expression of long non‐coding RNA HOTAIR predicts tumor recurrence in hepatocellular carcinoma patients following liver transplantation. Ann Surg Oncol 2011;18:1243–1250. [DOI] [PubMed] [Google Scholar]
- 31. Li H, Yu B, Jianfang L, et al. Over‐expression of lncRNA H19 enhances carcinogenesis and metastasis of gastric cancer. Oncotarget 2014;5:2318–2329. [DOI] [PMC free article] [PubMed] [Google Scholar]


