Abstract
Emerging CRISPR-based nucleic acid detection shows great promise in molecular diagnosis of diseases. CRISPR-Cas12a can sensitively and specifically detect human papillomavirus (HPV) DNA in anal swabs. However, the current CRISPR-Cas12a system needs auxiliary and expensive equipment, which limit its application as a point-of-care (POC) diagnostic tool. This study aimed to develop CRISPR-Cas12a as a POC test to directly target plasma for circulating HPV DNA detection by immediately reading results with naked eyes. Cell-cultured supernatants of either HPV16- or 18-positive cancer cells were treated with lysis buffer followed by isothermal amplification without DNA isolation. Cas12a, crRNA, and fluorescent-biotin reporters were incubated with the lysates. Our data showed that integrating CRISPR-Cas12a with lateral-flow strips could directly and specifically detect HPV16 and 18 in the liquid samples with the same limit of detection (0.24 fM) as did polymerase chain reaction but requiring less time. Furthermore, the CRISPR-Cas12a system could rapidly detect presence of HPV16 and HPV18 in plasma samples of 13 of 14 and 3 of 10 the patients with histopathological diagnosis of cervical cancer, respectively. Therefore, a CRISPR-Cas12a–based POC system was developed for conveniently detecting circulating nuclei acid targets in body fluids without requiring technical expertise and ancillary machineries.
Introduction
The development of accurate and rapid nucleic acid detection for cancer and pathogen diagnoses, genotyping, and disease monitoring is clinically imperative. The current techniques, including polymerase chain reaction (PCR), have been used for nuclei acid detection [1]. However, they are costly, labor intensive, and time consuming. Furthermore, the techniques require extensive sample manipulation and expensive machineries. Rapid, reliable, and easy-to-use tests of circulating nucleic acids allowing for point-of-care (POC) without requiring special technical expertise and ancillary equipment are urgently needed.
Clustered regularly interspaced short palindromic repeats (CRISPR) are a family of DNA sequences found within the genomes of prokaryotic organisms [2]. CRISPR-associated (Cas) immune system has been applied in molecular biology to target and cleave specific nucleic acid sequences, which is commonly used in gene editing. Furthermore, upon binding to target double-stranded DNA (dsDNA) or RNA, several Cas proteins can be activated and unleash the nonspecific endoribonuclease activity to degrade the single-stranded DNA (ssDNA) and RNA and thus provide a novel diagnostic approach for nuclei acid detection [3], [4], [5], [6], [7]. For instance, based on Cas12a ssDNase activation, Chen et al. recently developed a method termed DNA endonuclease-targeted CRISPR trans reporter (DETECTR), which could detect human papillomavirus (HPV) in cell-based clinical specimens (anal swabs) [5]. However, DETECTR has some limitations in its application as a field-deployable POC tool: 1) requiring complicated process of sample and DNA preparations; 2) reliance on fluorescent detection equipment for readout; and 3) applied only to anal swabs, which are cellular specimens.
To overcome the limitations, in this study, we developed CRISPR-Cas12a as a potential POC testing for HPV by directly targeting plasma without DNA isolation and by using a visual readout with naked eyes on a paper strip.
Materials and Methods
Cell Culture
All cancer cell lines [SiHa (ATCC HTB-35), Ca Ski (ATCC CRL-1550), UPCI: SCC152 (ATCC CRL-3240), HeLa (ATCC® CCL-2), C-4 I (ATCC® CRL-1594)] were obtained from the ATCC (Manassas, VA). The cell lines were cultured in complete growth medium according to ATCC's instruction and were grown at 37°C under 5% CO2.
Clinical Specimens
From July 2006 to July 2012, we have recruited cancer-free female patients without any cancer under our protocol approved by the Institutional Review Boards of University of Maryland Baltimore. EDTA-anticoagulant blood samples were collected at the time of the interview by venipuncture from the consented subjects as previously described [8]. The samples were centrifuged at 750×g for 10 minutes, and the plasma fractions were stored at −80°C [8]. We randomly obtained plasma from 14 female subjects without diagnosis of any cancer as a control group. Fifteen plasma samples of cervical cancer patients were obtained from Tissue Solutions (Glasgow, UK) and used as a case group. The 15 cervical cancer patients underwent complete physical and gynecologic examinations, and their disease was staged according to the guidelines of the International Federation of Gynecology and Obstetrics. The demographic and clinical variables of the cases and controls are shown in Table 1.
Table 1.
Characteristics of Cervical Cancer Patients and Cancer-Free Subjects
| Cervical Cancer Cases (n = 15) | Controls (n = 14) | |
|---|---|---|
| Age | 38.28 (SD 9.35) | 43.34 (SD 9.98) |
| Sex | Female | Female |
| Race | All are Caucasian | All are Caucasian |
| Stage | ||
| Stage 0 (carcinoma in situ) | 5 | |
| Stage IA1 | 1 | |
| Stage IB1 | 4 | |
| Stage IB2 | 3 | |
| Stage IIIA | 1 | |
| Stage IIIB | 1 | |
| Histological type | All are squamous cell carcinoma | |
Crude DNA Preparation and Recombinase Polymerase Amplification (RPA)
For cultured cell supernatants, 25 μl of each supernatant sample was mixed with 25 μl of phosphate buffered saline (PBS) (Corning) containing 0.2% Triton X-100 (Promega) followed by boiled mixture at 95°C for 5 minutes. Five microliters of the preparation was used as direct input into RPA. RPA reaction was run as instructed with TwistAmp Basic (TwistDx). The 50-μl reaction mixture contained 0.48 μM forward and reverse primers (Table 2), 29.5 μl primer free rehydration buffer, 5 μl DNA from boiled liquid fluids, and 14 mM magnesium acetate (MgOAc). The PRA mixture was incubated at 37°C for 20 minutes. Five microliters of RPA reaction was used as direct input. The samples were then preceded with heating unextracted diagnostic samples to obliterate nucleases (HUDSON) [3]. For human plasma, we made a minor modification to HUDSON. Briefly, plasma was first diluted 1:3 to avoid solidification. Twenty-five microliters of plasma sample was mixed with 50 μl of PBS (Corning) containing 0.53% Triton X-100 (Promega) followed by boiled mixture at 95°C for 5 minutes.
Table 2.
Sequences of RPA and PCR Primers, Guide RNA, and Report Substrates
| Primers for RPA and PCR | |
|---|---|
| HPV16-L1_F | TTGTTGGGGTAACCAACTATTTGTTACTGTT |
| HPV16-L1_R | CCTCCCCATGTCTGAGGTACTCCTTAAAG |
| HPV18-L1_F | GCATAATCAATTATTTGTTACTGTGGTAGATACCACT |
| HPV18-L1_R | GCTATACTGCTTAAATTTGGTAGCATCATATTGC |
| Guide RNA | Sequence (5′->3′) |
| Cas12a crRNA-HPV16-L1 | UAAUUUCUACUAAGUGUAGAUUGAAGUAGAUAUGGCAGCAC |
| Cas12a crRNA-HPV18-L1 | UAAUUUCUACUAAGUGUAGAUACAAUAUGUGCUUCUACACA |
| Substrates | Sequence (5′->3′) |
| ssDNA-FQ reporter | /56-FAM/TTATT/3IABKFQ/ |
| ssDNA-FB reporter | /56-FAM/TTATT/3Bio/ |
Abbreviations: RPA, recombinase polymerase amplification; PCR, polymerase chain reaction; and FQ, FAM-quencher.
Plasmid DNA Preparation and 10-Fold Serial Dilution
Plasmid of HPV type 16 (ATCC 45113) and HPV type 18 (ATCC 45152) in Escherichia coli DH5α were obtained from ATCC and grown in LB medium with 50 μg/ml ampicillin at 37°C for 16 hours in a shaking incubator. Plasmid DNA was isolated using QIAprep spin miniprep kit (QIAGEN) according to manufacturer's instruction. Briefly, the bacterial culture was harvested and lysed. The plasmid DNA was adsorbed on a QIAprep membrane, washed, and then eluted with nuclease free water. Concentration was measured by Qubit 4 (ThermoFisher) and prepared in 10 ng/μl followed by 10-fold serial dilution into water. One microliter of the sample was used as direct input into RPA or PCR reaction.
Polymerase Chain Reaction
Each PCR was prepared in a total volume of 50 μl containing 1× TaqMan Universal PCR Master Mix, no AmpErase UNG (ThermoFisher), 1 μl plasmid with serial diluted concentrations, and HPV specific primer pairs (Table 2). Each reaction was performed using Bio-Rad CFX96 Real-time system (Bio-Rad). Thermal cycle conditions are as follow: an initial incubation at 95°C for 10 minutes followed by 50 cycles of alternating 95°C for 10 seconds, 50°C for 10 seconds and 72°C for 30 seconds. A further incubation at 72°C for 10 minutes was done to complete the extension step. Five microliters of amplified DNA was used for performing electrophoresis in 2% agarose gel (Promega).
Fluorescent Readout of CRISPR-Cas12a Activity Using FAM-Quencher (FQ) Reporters
LbCas12a was preassembled with an HPV16- or HPV18-targeting CRISPR RNA (crRNA) at 37°C for 30 minutes and diluted in 1× binding buffer (20 mM Tris-HCl, pH 7.5, 100 mM KCl, 5 mM MgCl2, 1 mM DTT, 5% glycerol, 50 μg/ml heparin) and custom ssDNA-FQ reporter (IDT) (Table 2) [5]. Differently diluted RPA reactions were transferred to a 384-well microplate (Corning). Different concentrations of LbCas12a, crRNA, and custom ssDNA-FQ reporter were added directly to the reactions in total 20 μl volume. Reactions were proceeded at 37°C for 2.5 hours on a fluorescence plate reader (Biotek Synergy H1) with fluorescence kinetics measured every 10 minutes.
Lateral Flow Readout of CRISPR-Cas12a Activity Using FAM-Biotin (FB) Reporters
Lateral flow readout was based on cleavage of dual-labeled ssDNA [4]. Preparation was performed with the modification using FAM-biotin labeled ssDNA reporter (IDT) as a substrate but not quenched fluorescent ssDNA reporter. After incubation at 37°C for 3 hours, 80-μl HybriDetect assay buffer was added to the 20-μl reaction mixture. Milena HybriDetect1 lateral flow dipstick (TwistDx) was dipped into the reaction mixture and incubated for 5 minutes at room temperature. Paper strip was removed, and results were interpreted immediately by photograph using a smartphone camera. A sample with signal significantly increasing on the second line (test bend) and decreasing on the first line (control line) was considered to have a positive result. A sample without incubation of CRISPR-Cas12a was used as a control and should have intense line at the control line.
Statistical Analysis
We performed statistical analysis using the Statistical Package for the Social Sciences Software version 11.5 (SPSS, Chicago, IL). Graphical presentation was plotted and analyzed by using Prism 4 software (Graphpad Inc., San Diego CA). P values less than .05 were considered as statistically significant in all the analyses. Differences in values were evaluated using Student t test. We used Bland-Altman plots to analyze the correlation of two different techniques for quantification of HPV DNA.
Results
The CRISPR-Cas12a System Can Directly and Specifically Detect HPV DNA in Liquid Fluids
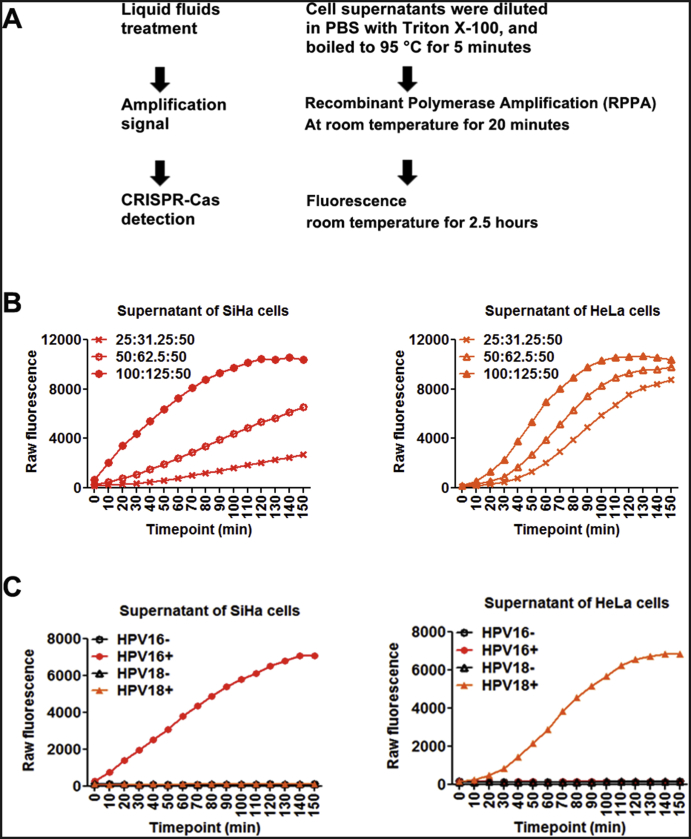
To evaluate whether CRISPR-Cas12a could directly detect HPV in liquid fluids without DNA isolation, we collected cell-cultured supernatants of three HPV16-positive cell lines (SiHa, Ca Ski, and SCC152) and two HPV18-positive cell lines (HeLa and C-4I). The cell supernatants were diluted 1:2 in PBS containing 0.2% Triton X-100 and boiled at 95°C for 5 minutes. Five microliters of boiled liquid fluids was directly applied in RPA. RPA is a regular temperature isothermal amplification method to exponentially amplify DNA amount without requiring an additional temperature step [4]. LbCas12a with its crRNAs and ssDNA FQ reporter was directly added in the RPA reactions without any purification step. The results were determined by using a fluorescence plate reader with fluorescence kinetics (Figure 1A). Several reaction parameters, including the concentrations of LbCas12a, crRNA, ssDNAFQ reporter, and RPA reactions, were investigated in the cell supernatants. When 1:10 diluted RPA reactions were used, the optimized concentrations of LbCas12a, crRNA, and ssDNA FAM-biotin reporter were 100 nM, 125 nM, and 50 nM, respectively. At the optimum reaction condition, LbCas12a with either HPV16 or 18 crRNA unambiguously and rapidly identified HPV16 or HPV18 in the cell-supernatants of HPV16- or 18-postive cells, respectively (Figure 1, B-C). Therefore, the CRISPR-Cas12a system could be directly applied to liquid fluids for sensitivity detecting and specifically classifying HPV DNA.
Figure 1.
Specific distinguishing HPV16 and HPV18 DNA in liquid fluids with the CRISPR-Cas12a system.
(A) Schematic of HPV detection by using fluorescence plate reader from liquid fluids without extraction of DNA. (B) Time courses of fluorescence detection with different concentrations (nM) of LbCas12a:crRNA complexes and fixed 50 nM ssDNAFQ reporter to target HPV16 in supernatant of SiHa cells (left) and target HPV18 in supernatant of HeLa cells (right). (C) Detection of HPV16 (red circle) or HPV18 (orange triangle) in supernatant of SiHa cells containing HPV16 genomic fragment (left) and supernatant of Hela cells containing HPV18 genomic fragment. + or −: incubating with or without LbCas12a-crRNA that targets either HPV16 or HPV18.
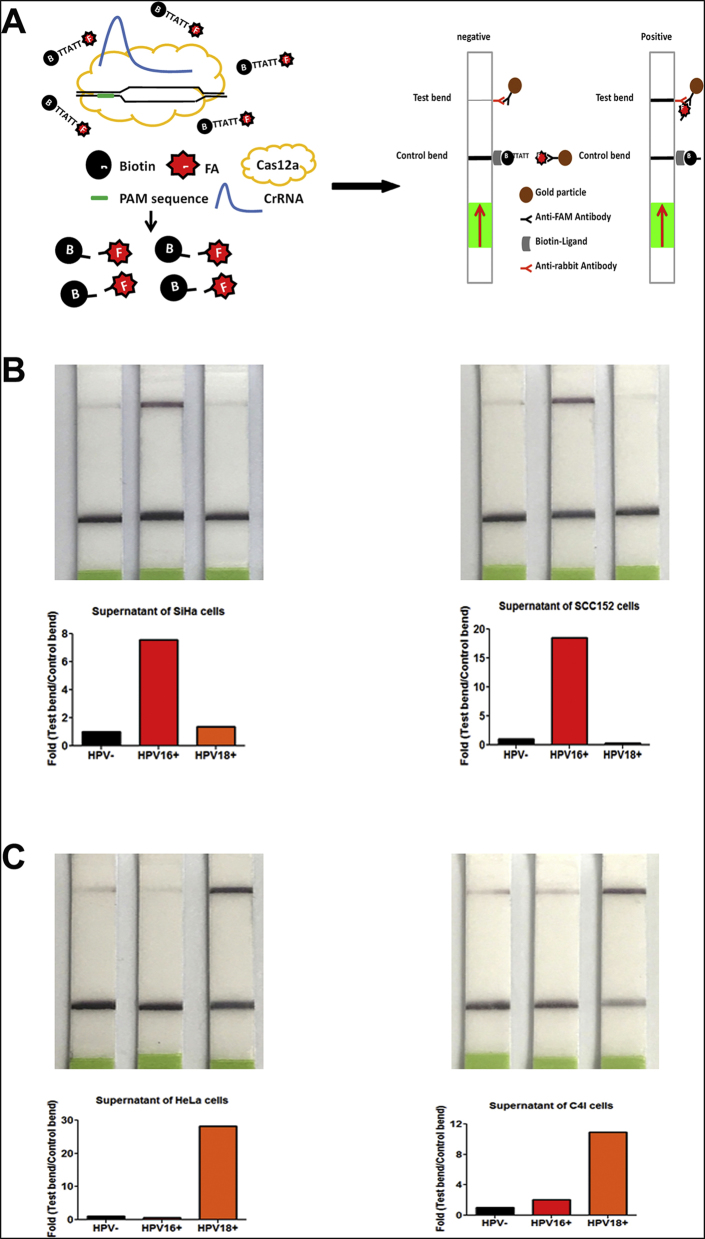
The CRISPR-Cas12a System Can Rapidly and Directly Read the Results on Paper Strips
To determine whether the results of CRISPR-Cas12a could be directly visualized with the naked eye, we used lateral flow dipsticks to read out the results (Figure 2A). The lateral-flow readout is based on the destruction of a FAM-biotin reporter, by which plentiful reporter gathers anti-FAM antibody-gold nanoparticle conjugates at the first line on the strip, hindering binding of the antibody-gold conjugates to protein A on the second line. In our experiments, when CRISPR-Cas12a cleaved ssDNA FAM-biotin reporter, the signal significantly increased on the second line as the test bend and indicated a positive result on the paper strip (Figure 2A). Using the optimized protocol established above, we incubated RPA reactions of the cell supernatants with 100 nM LbCas12a: 125 nM crRNA: 50 nM FAM-biotin reporter at room temperature. CRISPR-Cas12a intensely increased the test bend and therefore clearly identified HPV16 and 18 in the cell supernatants of HPV16-positive (Figure 2B) or 18-positive cells (Figure 2C), respectively. Furthermore, the total turnaround time for combing CRISPR-Cas12a with lateral-flow readout was about 3 hours, implying that the CRISPR-Cas12a system could be used as a rapid and portable test for directly reading HPV DNA.
Figure 2.
Applying the CRISPR-Cas12a system with paper strips to rapidly and directly visualize the result with naked eyes.
(A) Schematic of CRISPR-Cas12a system with lateral flow technology by paper strips to read out the negative or positive results. (B) Lateral flow detection of HPV16 in supernatants of SiHa cells (left) and SCC152 cells (right). (C) Lateral flow detection of HPV18 in supernatants of HeLa cells (left) and C4I cells (right). Quantification of band intensity from detection is shown below. Black bar: incubating without LbCas12a-crRNA as control. Red/orange bar: incubating with LbCas12a-crRNA that targeted HPV16 or HPV18, respectively.
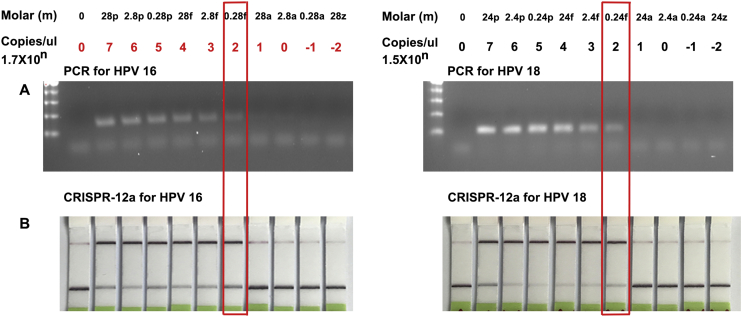
The CRISPR-Cas12a System Has a Similar Sensitivity for Detecting HPV DNA in Liquid Fluids Compared with PCR-Based Assay
To define analytic performance of the CRISPR-Cas12a system, the plasmids containing the complete genomic DNA of HPV genotype 16 and 18 were serially (10-fold) diluted into water. Each diluted sample was processed with the CRISPR-Cas12a-paper test and PCR analysis. CRISPR-Cas12a and PCR had the same limit of detection of 170.6 copies/μl of input or 0.28 fM and 151.8 copies/μL or 0.24 fM in quantification of HPV-16 and 18, respectively (Figure 3). Furthermore, the CRISPR-Cas12a-paper test and PCR analyses exhibited excellent linearity between the target input and measured values in a dynamic range of six orders of magnitude of input. The two techniques displayed an estimated slope coefficient that was in the range of 0.89-0.94 and R2 of 0.90-0.92 for HPV detection, suggesting an excellent correlation between the CRISPR-Cas12a techniques and PCR assays.
Figure 3.
The limit of detection of PCR assay and CRISPR-Cas12a-paper test.
Ten-fold serial dilutions of HPV16 plasmid (left) and HPV18 plasmid (right) in nuclease-free water were tested by PCR (A) and CRISPR-Cas12a system with lateral flow detection (B), respectively.
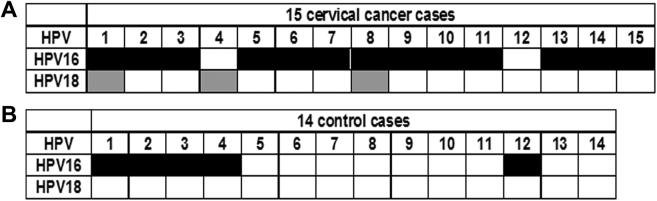
The CRISPR-Cas12a System Can Be Used as a POC Diagnostic Test for Detecting HPV DNA in Clinical Plasma Specimens
We obtained plasma of 15 cervical cancer patients and 14 cancer-free subjects (Table 1). The final diagnosis of the cancer cases and control subjects was made by clinical standard procedures, including HPV testing, Pap testing, and HPV/Pap contesting. Clinically, of the 15 cervical cancer patients, 5 are positive for HPV16, 1 for HPV18, and 9 for both HPV16 and 18.
Among the five HPV 16-positive cervical cancer patients, all had the evidence of HPV16 in plasma determined by the CRISPR-Cas12a system. The one HPV18-positive cancer patient also had the evidence of HPV-18 in plasma. Of cervical cancer cases that were both HPV16 and 18 positive, eight had positive HPV16, while 2 had positive HPV 18 results in the plasma samples (Figure 4A). Furthermore, both of the HPV DNA 16 and 18 statuses in plasma had no special relationship with histology and stage of cervical cancer and age of the patients (all P > .05). Of the 14 control individuals, 5 had the evidence of HPV16 in plasma (Figure 4B) (Supplementary Figure 1A). Therefore, the CRISPR-Cas12a system could rapidly detect circulating nuclei acid targets in plasma without requiring technical expertise and ancillary machineries.
Figure 4.
The CRISPR-Cas12a system for detecting HPV DNA in clinical plasma specimens.
Using CRISPR-Cas12a-paper to detect HPV DNA in plasma of 15 cervical cancer patients (A) and 14 cancer-free female individuals without any cancer (B). Black cells indicate positive HPV16 results, while gray cells indicate positive HPV18 results in plasma.
Discussion
CRISPR-Cas biology has revolutionized the field of molecular diagnostics for infectious and noninfectious diseases, including cancer [7]. Based on RNA-guided Cas13a-mediated collateral cleavage of RNA reporter, Gootenberg et al. recently developed specific high-sensitivity enzymatic reporter UnLOCKing (SHERLOCK) [9]. SHERLOCK could detect multiple synthetic ssRNA targets (RNA1, ZIKV, and DENV) and one synthetic dsDNA target in the same reaction [3], [4], [9]. However, since Cas13a targets RNA, SHERLOCK requires reverse transcription (RT)-RPA coupled with T7 RNA polymerase to transcribe amplified DNA to RNA for subsequent detection [3], [4], [9]. The multiple procedures may lead to the associated analytical variations and cost. Furthermore, since RNA is less stable than DNA in human specimens, SHERLOCK might generate unreliable results, presenting challenges to be used in the clinics.
In contrast to SHERLOCK, the Cas12a-based DETECTR could detect HPV DNA without RT reaction and thus is more practically useful [5]. However, some obstacles exist in the use of DETECTR as a field-deployable POC tool, including requiring complex procedure of sample and DNA preparations and needing expensive fluorescent detection equipment for readout. Furthermore, many pathogens, such as viruses, are shed in body fluids (e.g., plasma, urine, or saliva), which provide noninvasively and easily obtained surrogate materials. A field-deployable, rapid diagnostic test that can directly detect the circulating nucleic acids in the body fluids would be clinically useful. However, the DETECTR system is only applicable to cell-based specimens rather than circulating body fluids.
In this study, we demonstrate that CRISPR-Cas12a can directly target body fluids for sensitivity detecting and specifically classifying circulating HPV DNA without extraction step. Furthermore, we use lateral flow dipstick to rapidly visualize the results of CRISPR-Cas12a without requiring special technical expertise and ancillary equipment. In addition, the CRISPR-Cas12a system has a similar sensitivity for detecting HPV DNA in liquid fluids as does PCR. Moreover, combining CRISPR-Cas12a with a lateral-flow paper strip could be a rapid and portable test for directly reading HPV DNA. Therefore, our further development of CRISPR-Cas12a for detecting HPV DNA in plasma might overcome the obstacles of DETECTR and hence provide a potential field-deployable POC tool.
HPV16 and HPV18 are considered as the highest-risk viruses in the development of cervical cancer, and almost all cervical cancer patients bear HPV DNA in their affected tissues [10]. PCR-based assays have been used for the detection of HPV DNA in tissue samples. Furthermore, the presence of circulating HPV DNA in body fluids can act as a determining factor for identifying the patients at risk for the development and progression of cervical cancer [11], [12]. However, PCR is costly, labor intensive, and time consuming, leading to difficulties in the detection of HPV infection in plasma [13], [14]. Intriguingly, the CRISPR-Cas12a system could rapidly and cost-effectively detect presence of HPV16 and HPV18 in the plasma of 13 of 14 and 3 of 10 the patients with histopathological diagnosis of cervical cancer, respectively.
HPV16 was also detected in the plasma samples of 5 of 14 women with no sign of malignancy. There are two possible explanations for the observation. First, although these women had no symptoms of cervical cancer, some had signs of inflammation or bacterial infection in their Pap smear test. It would be possible that such conditions facilitate the spread of the existing virus into the blood circulation even in the absence of malignancies. Nevertheless, long-term follow-up of the subjects who have no cervical cancer with positive HPV in plasma is required. Second, CRISPR-Cas12a needs a crRNA as a guide RNA for the activities to genomic targets [15]. The specificity of the CRISPR-Cas12a system is mainly determined by how specific the crRNA sequence is for the genomic target. However, a crRNA targeting sequence might occasionally have additional sites throughout the genome where partial homology exists, causing off-target effect [16]. When used in the detection of a gene of interest, the off-target effect of the crRNA may cause a false-positive result. To reduce the possible off-target–related false-positive results, we are using new programs [17], [18], [19] that particularly take consideration of on-target cutting efficiency to design crRNAs for specifically detecting HPV16 and 18, respectively.
A major limitation of this study is that the size of the cohort (15 cases of cervical cancer and 14 healthy control individuals) is small. With the small sample size, the diagnostic performance of the CRISPR-Cas12a system in the clinical specimens could not be reliably evaluated. We are performing a large and prospective study to determine its diagnostic value for detection of circulating HPV DNA.
Conclusions
The CRISPR-Cas12a system might be a promising POC test for noninvasively monitoring and tracking other nucleic acid–related infectious and noninfectious diseases in body fluids. However, the CRISPR-Cas12a system needs further optimization for improved detecting efficiency.
The following are the supplementary data related to this article.
CRISPR-Cas12a-paper to detect HPV DNA in plasma specimens of 15 cervical cancer patients (A) and 14 cancer-free female individuals without any cancer (B). The red arrow indicates positive results of either HPV16 or 18.
Declarations
Ethical Approval
All procedures performed in studies involving human participants were in accordance with the ethical standards of the institutional and/or national research committee and with the 1964 Helsinki declaration and its later amendments or comparable ethical standards. This article does not contain any studies with animals performed by any of the authors.
The Institutional Review Boards of University of Maryland Baltimore.
Competing Interests
The authors declare no conflict of interest.
Authors' Contributions
J. T., Q. L., and F. J. conducted the experiments and participated in study design, coordination, and data interpretation, and preparing the manuscript. All authors read and approved the final manuscript.
Availability of Data and Materials
Not applicable.
Disclosure of Potential Conflicts of Interest
The authors declare no conflict of interest.
Acknowledgements
This work was supported in part by; National Institutes of Health/National Cancer Institute (USA) grant CA240556, Department of Defense (USA) grant LC160422, the Geaton and JoAnn DeCesaris Family Foundation (USA), and University of Maryland Marlene & Stewart Greenebaum Comprehensive Cancer Center Pilot Grant (USA) (All to F.J.).
References
- 1.Persing DH. Nucleic acid-based pathogen discovery techniques: potential application to xenozoonoses. Mol Diagn. 1996;1:243–254. doi: 10.1016/s1084-8592(96)70006-1. [DOI] [PubMed] [Google Scholar]
- 2.Barrangou R. The roles of CRISPR-Cas systems in adaptive immunity and beyond. Curr Opin Immunol. 2015;32:36–41. doi: 10.1016/j.coi.2014.12.008. [DOI] [PubMed] [Google Scholar]
- 3.Myhrvold C, Freije CA, Gootenberg JS, Abudayyeh OO, Metsky HC, Durbin AF, Kellner MJ, Tan AL, Paul LM, Parham LA. Field-deployable viral diagnostics using CRISPR-Cas13. Science. 2018;360:444–448. doi: 10.1126/science.aas8836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gootenberg JS, Abudayyeh OO, Kellner MJ, Joung J, Collins JJ, Zhang F. Multiplexed and portable nucleic acid detection platform with Cas13, Cas12a, and Csm6. Science. 2018;360:439–444. doi: 10.1126/science.aaq0179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chen JS, Ma E, Harrington LB, Da Costa M, Tian X, Palefsky JM, Doudna JA. CRISPR-Cas12a target binding unleashes indiscriminate single-stranded DNase activity. Science. 2018;360:436–439. doi: 10.1126/science.aar6245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Abudayyeh OO, Gootenberg JS, Essletzbichler P, Han S, Joung J, Belanto JJ, Verdine V, Cox DBT, Kellner MJ, Regev A. RNA targeting with CRISPR-Cas13. Nature. 2017;550:280–284. doi: 10.1038/nature24049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chertow DS. Next-generation diagnostics with CRISPR. Science. 2018;360(6387):381–382. doi: 10.1126/science.aat4982. [DOI] [PubMed] [Google Scholar]
- 8.Ma J, Li N, Guarnera M, Jiang F. Quantification of plasma miRNAs by digital PCR for cancer diagnosis. Biomark Insights. 2013;8:127–136. doi: 10.4137/BMI.S13154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gootenberg JS, Abudayyeh OO, Lee JW, Essletzbichler P, Dy AJ, Joung J, Verdine V, Donghia N, Daringer NM, Freije CA. Nucleic acid detection with CRISPR-Cas13a/C2c2. Science. 2017;356:438–442. doi: 10.1126/science.aam9321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ferlay J, Colombet M, Soerjomataram I, Mathers C, Parkin DM, Pineros M, Znaor A, Bray F. Estimating the global cancer incidence and mortality in 2018: GLOBOCAN sources and methods. Int J Cancer. 2019;144:1941–1953. doi: 10.1002/ijc.31937. [DOI] [PubMed] [Google Scholar]
- 11.Dong SM, Pai SI, Rha SH, Hildesheim A, Kurman RJ, Schwartz PE, Mortel R, McGowan L, Greenberg MD, Barnes WA. Detection and quantitation of human papillomavirus DNA in the plasma of patients with cervical carcinoma. Cancer Epidemiol Biomarkers Prev. 2002;11:3–6. [PubMed] [Google Scholar]
- 12.Ho CM, Yang SS, Chien TY, Huang SH, Jeng CJ, Chang SF. Detection and quantitation of human papillomavirus type 16, 18 and 52 DNA in the peripheral blood of cervical cancer patients. Gynecol Oncol. 2005;99:615–621. doi: 10.1016/j.ygyno.2005.07.004. [DOI] [PubMed] [Google Scholar]
- 13.Gnanamony M, Peedicayil A, Subhashini J, Ram TS, Rajasekar A, Gravitt P, Abraham P. Detection and quantitation of HPV 16 and 18 in plasma of Indian women with cervical cancer. Gynecol Oncol. 2010;116:447–451. doi: 10.1016/j.ygyno.2009.10.081. [DOI] [PubMed] [Google Scholar]
- 14.Jaberipour M, Samsami A, Sahraiian F, Kazerooni T, Hashemi M, Ghaderi A, Habibagahi M. Elevation of HPV-18 and HPV-16 DNA in the plasma of patients with advanced cervical cancer. Asian Pac J Cancer Prev. 2011;12:163–167. [PubMed] [Google Scholar]
- 15.Lee K, Zhang Y, Kleinstiver BP, Guo JA, Aryee MJ, Miller J, Malzahn A, Zarecor S, Lawrence-Dill CJ, Joung JK. Activities and specificities of CRISPR/Cas9 and Cas12a nucleases for targeted mutagenesis in maize. Plant Biotechnol J. 2019;17:362–372. doi: 10.1111/pbi.12982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chen X, Chen Y, Huang HM, Li HD, Bu FT, Pan XY, Yang Y, Li WX, Li XF, Huang C. SUN2: a potential therapeutic target in cancer. Oncol Lett. 2019;17:1401–1408. doi: 10.3892/ol.2018.9764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang M, Tang C, Chen J. Drug-target interaction prediction via dual Laplacian graph regularized matrix completion. Biomed Res Int. 2018;18:560–568. doi: 10.1155/2018/1425608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chen Y, Zhao B, Liu M, Wang J, Qiu X, Zhu C, Wu X. MicroRNAs profiling identifies miR-125a and its target gene Wnt2 in skins of different haired rabbits. Front Genet. 2018;9:628–632. doi: 10.3389/fgene.2018.00628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang L, Zhang H, Xie Y, Chen H, Ren C, Chen X. Target-mediated hyperbranched amplification for sensitive detection of human alkyladenine DNA glycosylase from HeLa cells. Talanta. 2019;194:846–851. doi: 10.1016/j.talanta.2018.10.083. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
CRISPR-Cas12a-paper to detect HPV DNA in plasma specimens of 15 cervical cancer patients (A) and 14 cancer-free female individuals without any cancer (B). The red arrow indicates positive results of either HPV16 or 18.
Data Availability Statement
Not applicable.