Abstract
Background
UGT1A1 is a polymorphic enzyme that has been associated with irinotecan drug metabolisms. We developed a pyrosequencing method to detect allele frequency and genotype of UGT1A1 polymorphisms (UGT1A1*28 and UGT1A1*6) in Thai colorectal cancer patients.
Method
A pyrosequencing method was designed to determine UGT1A1 genetic polymorphisms including UGT1A1*28 (A[TA]7TAA) and UGT1A1*6 (211G>A) in 91 Thai colorectal cancers.
Result
Genotyping by the pyrosequencing technique was 100% concordant with capillary electrophoresis sequencing. The allele frequencies for UGT1A1 genetic polymorphisms were *1/*1 (54.95%), *1/*6 (13.19%), *1/*28 (25.27%), *28/*6 (4.40%), and *28/*28 (2.20%). No homozygous mutation UGT1A1*6 was found in our population.
Conclusions
We developed a rapid, reliable, more cost‐effective, and simple assay to detect UGT1A1 genetic polymorphisms in routine practice before initiating irinotecan therapy. The UGT1A1*28 and UGT1A1*6 alleles were found to be similar in the Asian populations.
Keywords: UGT1A1, genetic polymorphisms, pyrosequencing, SNP, Thai
INTRODUCTION
The Uridine diphosphate (UDP)‐glucuronosyltransferase (UGT), an important class of phase II metabolic enzyme, catalyzes the glucuronidation step and facilitates the excretion of hydrophobic endobiotic and xenobiotic compounds 1, 2. The UGT enzymes are generally divided into three subfamilies, UGT1A, UGT2A, and UGT2B 3. The UGT1A1 is mediated primarily by the UGT1 family polypeptide A1, which is located on chromosome 2q37 4.
The UGT1A1*1 [A(TA)6TAA] is a wild‐type allele that includes six TA repeats in the TATA box of promoter region 5, 6, whereas UGTA1A1*28 [A(TA)7TAA] is the most common variant allele that contains seven TA repeats. UGT1A1*28 has a decreased UGT enzymatic activity accounting 60–80% of the normal level 7, 8, 9. The allele frequency of UGT1A1*28 is 33.4–36.5% in the Caucasian population and 39.0–40.4% in Africans, whereas in Asians the allele frequency is less (13.9%) 10.
The UGT1A1*6 (211G>A, G71R), polymorphism on exon 1 of the UGT1A1 gene 11, has 50% less activity than wild‐type UGT1A1 8. UGT1A1*6 is most frequent in Asians (13.0%), while it is missing or very few in Caucasians or African‐Americans 12. Previous studies have elucidated the UGT1A1 polymorphisms in Thai population, and has been reported in Gilbert syndrome, Crigler–Najjar syndrome, and HIV patients who were treated with indinavir 13, 14, 15.
Irinotecan (camptothecin‐11, CPT‐11, 7‐ethyl‐10‐[4‐(1‐(piperidino)‐1‐piperidino] carbonylcamptothecin) is a widely used anticancer drug approved for the treatment of solid tumors, especially, colorectal cancer 16, 17, 18. Irinotecan is a prodrug of topoisomerase 1 inhibitor SN‐38, which prevents the DNA strand reannealing, interrupts DNA replication, and leads to subsequent cell death. Irinotecan is metabolized by carboxylesterases to an active 7‐ethyl‐10‐hydroxycamptothecin (SN‐38; 18, 19, 20), which is 100‐ to 1,000‐fold more cytotoxic than the parent drug. SN‐38 is glucuronidated by UGTs to an inactive SN‐38 glucuronide (SN‐38G) 21, 22. The major toxicities of irinotecan are febrile neutropenia and diarrhea 23. The polymorphism of UGT1A1 has been associated with reduced enzymatic activity and SN‐38 glucuronidation, which increases toxicity of irinotecan. The polymorphism of UGT1A1*28 and UGT1A1*6 alleles has a significantly decreased SN‐38G correlated with irinotecan toxicities 12, 19, 24.
Pyrosequencing is a recently developed technique to detect polymorphisms in the UGT1A1 gene and its promoter region. It involves the hybridization of a primer to a single‐stranded PCR template, and the sequencing analysis is started by adding nucleotides. This method is based on the formation of pyrophosphate and can monitor light peak in a pyrogram 25. Pyrosequencing is a high‐throughput technique with high accuracy 26. Thus, the objective of this study was to develop and evaluate a method based on pyrosequencing for detecting UGT1A1 polymorphisms (UGT1A1*28 and UGT1A1*6) in Thai colorectal cancer patients.
MATERIALS AND METHODS
Clinical Subjects
Ninety‐one Thai colorectal cancer patients were recruited from Division of Medical Oncology, Department of Medicine Ramathibodi Hospital, Mahidol University. DNA analyses of UGT1A1 polymorphisms were investigated by Division of Pharmacogenomics and Personalized Medicine, Department of Pathology Ramathibodi Hospital, Mahidol University. Written and informed consent was obtained from all participants. The study was approved by the Ethics Committee, Ramathibodi Hospital, Mahidol University, Thailand.
Human Genomic DNA Samples and Pyrosequencing Technique
Genomic DNA was extracted from 1 ml EDTA whole blood by DNA extraction automated MagNA Pure Compact (Roche Applied Science, Penzberg, Germany). The quality of genomic DNA was assessed by using NanoDrop ND‐1000. Table 1 shows the different sets of primers used for the PCR reactions and pyrosequencing, including TA insertion in TATA box of the promoter, and SNPs on the exon 1 (211G>A) of the UGT1A1. The PCR and sequencing primers were designed using PyroMark Assay Design 2.0 program (Qiagen, Singapore). The UGT1A1*28 and UGT1A1*6 were amplified using biotin tagged on 5′‐end of reverse primer.
Table 1.
Oligonucleotide Primers Used for PCR and Pyrosequencing
| Allele | PCR primers | Sequence primer | Size (bp) |
|---|---|---|---|
| UGT1A1*28 | |||
| Forward | 5′‐AGGTTCGCCTCTCCTACTTATA‐3′ | 5′‐CGCCTCTCCTACTTATATAT‐3′ | 82 |
| Reverse | B 5′‐CACGTGACAAGTCAAACATTAAC‐3′ | ||
| UGT1A1*6 | |||
| Forward | 5′‐ACCTACGCCTCGTTGTACAT‐3′ | 5′‐CTCGTGTACATCAGAGA‐3′ | 108 |
| Reverse | B 5′‐TGCCGAGACTAACAAAAGACT‐3′ | ||
B, biotinylated on 5′‐end of primer.
PCR conditions
PCRs were performed with an initial denaturation for 15 min at 95°C, followed by 42 cycles of denaturation for 20 s at 95°C, primer annealing for 30 s at 53°C, and extension for 20 s at 72°C followed by a final extension for 5 min at 72°C. All amplification reactions were performed with the Thermal cycler (Applied Biosystem, Gene Amp PCR System 9700 Foster City, California, USA) and then PCR products were checked by running on a 2% agarose gel. PCR conditions were the same for two mutations tested.
Pyrosequencing conditions
Fifteen microliters of PCR template was immobilized with 2 μL sepharose beads and 40 μL pyromark‐binding buffer by incubator (Shaker 1,400 rpm, 15 min, room temperature). For sequence primer annealing, 21 μL PyroMark annealing buffer and 4 μL sequencing primer were incorporated into each well of 24‐well pyrosequencing plate. For strand separation, all liquid was removed by the Vacuum Prep Workstation and then the bead containing immobilized PCR template was captured. The captured beads on probes were transferred to 70% ethanol for 5 s, denaturing solution for 5 s and wash buffer for 10 s, respectively. All liquid was totally drained from the filter probes. The sepharose beads with the biotinylated single‐stranded templates attached were released into 24‐well pyrosequencing plate containing sequence primer annealing. The 24‐well pyrosequencing plate was heated at 80°C for 2 min and then allowed to cool to a room temperature (25°C) for 10 min. The reaction was analyzed on PyroMark Q24 software.
Statistical Analysis
Genetic equilibrium was tested by Haploview 4.2 according to the formula of Hardy–Weinberg. The frequencies of all genotypes in this study were observed within the 95% confidence interval. Chi‐squared test or Fisher's exact test was used to compare genotype frequencies in different population. A P value of less than 0.05 was considered to be statistically significant. All analyses were performed using SPSS version 18.0 (SPSS Inc., Chicago, IL).
RESULTS
Characteristics of Patients
We recruited 91 Thai colorectal cancer patients include 52 males (57.14%) and 39 females (42.86%). The median age was 63 years (range, 27–94 years). Clinical characteristics are described in Table 2. The most common sites of disease were rectum (44.66%) and sigmoid (34.95%). The predominant metastatic organs were the liver (57.57%) and lung (34.85%).
Table 2.
Characteristics of Patients
| Clinical characteristics | No. of patients | Percentage or range |
|---|---|---|
| Age (years), mean | 63 | 27–94 |
| Sex | ||
| Male | 52 | 57.14 |
| Female | 39 | 42.86 |
| Site of disease | ||
| Ascending | 11 | 10.68 |
| Transverse | 4 | 3.88 |
| Descending | 6 | 5.83 |
| Sigmoid | 36 | 34.95 |
| Rectum | 46 | 44.66 |
| Metastatic focus | ||
| Liver | 38 | 57.57 |
| Lung | 23 | 34.85 |
| Other | 5 | 7.58 |
| Histopathology type | ||
| Well differentiated | 34 | 37.36 |
| Moderately differentiated | 52 | 57.14 |
| Poorly differentiated | 5 | 5.49 |
Sensitivity and Specificity of Pyrosequencing
To analyze the frequencies of the UGT1A1 polymorphisms (UGT1A1*28 and UGT1A1*6) included in the research, we developed genotyping assays based on the pyrosequencing method. A representative pyrogram for each genotype is shown in Fig. 1. The assay was designed to generate unique peak for UGT1A1 polymorphisms by setting a suitable nucleotide additional order. The sequence data identified from pyrosequencing were validated by capillary electrophoresis sequencing. The result showed that genotypes were 100% concordant between pyrosequencing and capillary electrophoresis sequencing method (Table 3).
Figure 1.
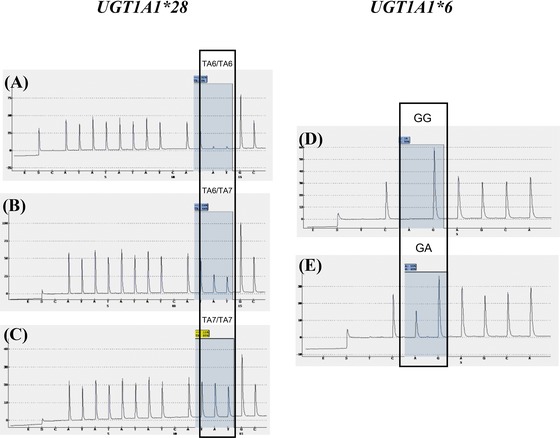
Pyrosequencing histogram for UGT1A1 by pyrosequencing software. (A) Wild‐type TA6/TA6, (B) heterozygous TA6/TA7, (C) homozygous TA7/TA7, (D) wild‐type G/G, (E) heterozygous G/A.
Table 3.
The Comparison of Result UGT1A1 Polymorphisms Between 2 Methods
| Capillary electrophoresis sequencing (gold standard) | Pyrosequencing | |
|---|---|---|
| Genotypes | n = 91 (%) | n = 91 (%) |
| *1/*1 | 50 (54.95) | 50 (54.95) |
| *1/*6 | 12 (13.19) | 12 (13.19) |
| *1/*28 | 23 (25.27) | 23 (25.27) |
| *6/*28 | 4 (4.40) | 4 (4.40) |
| *28/*28 | 2 (2.20) | 2 (2.20) |
Frequencies of UGT1A1*28 and UGT1A1*6 in Thai Colorectal Cancer
The frequencies of UGT1A1*28 (A(TA)7TAA) and UGT1A1*6 (211G>A) were determined in individual samples (n = 91) by PSQ 24 in Thai colorectal cancer patients and all were in Hardy–Weinberg equilibrium.
The results showed the allele frequencies of *1/*1 (50/91, 54.95%), *1/*6 (12/91, 13.19%), *1/*28 (23/91, 25.27%), *6/*28 (4/91, 4.40%),*28/*28 (2/91, 2.20%). No homozygous UGT1A1*6 was found in our population. The allele frequencies of UGT1A1*1, UGT1A1*6, and UGT1A1*28 were 0.74, 0.09, and 0.17, respectively (Table 4). Each pairwise comparison of Thai population with Chinese, Japanese, and Caucasian populations is shown in Table 5. The allele frequencies of UGT1A1*28 variant are similar among Thai, Chinese, and Japanese, whereas the homozygous UGT1A1*28 (TA7/TA7) was found lower in our population (3.0%) than Caucasian population (11.50%; P = 0.006). On the other hand, the homozygous UGT1A1*6 variant is absent in Caucasian population, while UGT1A1*6 variant is more commonly found in Thai, Chinese, and Japanese populations 27, 28.
Table 4.
Genotype and Allele Frequencies of UGT1A1 in Thai Colorectal Cancer
| UGT1A1 | Frequency | 95% CI |
|---|---|---|
| Alleles | n = 182 (%) | |
| *1 | 135 (74.18) | 67.63–80.37 |
| *6 | 16 (8.79) | 4.84–13.16 |
| *28 | 31 (17.03) | 11.54–22.46 |
| Genotypes | n = 91 (%) | |
| *1/*1 | 50 (54.95) | 44.78–65.22 |
| *1/*6 | 12 (13.19) | 6.09–19.91 |
| *1/*28 | 23 (25.27) | 16.37–34.23 |
| *6/*28 | 4 (4.40) | 0.19–8.61 |
| *28/*28 | 2 (2.20) | 0–5.21 |
Table 5.
Genotype Frequencies of UGT1A1*28 and UGT1A1*6 in Four Populations
| Gene | Thaia | Chineseb | Japaneseb | Caucasianc | P value | ||
|---|---|---|---|---|---|---|---|
| UGT1A1*28 | (A, %) | (B, %) | (C, %) | (D, %) | A vs. B | A vs. C | A vs. D |
| TA6/TA6 | 68.2 | 70.6 | 82.7 | 37.2 | 0.645 | 0.014 | 0.000 |
| TA6/TA7 | 28.8 | 24.5 | 15.3 | 51.4 | 0.871 | 0.054 | 0.000 |
| TA7/TA7 | 3.0 | 4.9 | 2.0 | 11.5 | 0.470 | 1.00 | 0.006 |
| UGT1A1*6 | |||||||
| G/G | 81.3 | 74 | 73.7 | 100 | 0.236 | 0.236 | 0.000 |
| G/A | 18.7 | 18 | 22.4 | 0 | 0.856 | 0.599 | 0.000 |
| A/A | 0 | 3 | 3.8 | 0 | 0.081 | 0.043 | ‐ |
Present study.
(27).
(28).
DISCUSSION
The designed pyrosequencing was validated and confirmed by capillary electrophoresis sequencing (Gold standard) 29. The results of pyrograms (patterns of peaks) showed clear distinction between mutation genotypes, and each genotype exhibited a unique peak. Therefore, interpretation will be easier and accurate. The throughput of the pyrosequencing instrument was 24 genotypes (24‐well plate) in 15 min.
In this study, the designed pyrosequencing method has analyzed UGT1A1*28 and UGT1A1*6. The allele frequency of UGT1A1*28 was higher than UGT1A1*6 in our population. Among Thai, Chinese, and Japanese populations, the genotype distributions of the UGT1A1*28 variant are not different, whereas among the Caucasian population these are much higher. On the other hand, the UGT1A1*6 variant is absent in Caucasian population, while UGT1A1*6 variant is more commonly found in Thai, Chinese, and Japanese populations.
Several genetic polymorphisms of UGT1A1 gene have been associated with the occurrence of severe neutropenia and diarrhea toxicities during treatment with irinotecan 30. Severe neutropenia or diarrhea is associated with UGT1A1*28 31. In 2005, the US Food and Drug Administration (FDA) recommended for UGT1A1*28 testing in patients who receive irinotecan 22. In Asian population, UGT1A1*6 is the most common allele correlated with reduced SN‐38 glucuronidation activity and drug toxicity 32. Therefore, UGT1A1 genotyping must be performed before initiating treatment with irinotecan. Moreover, the UGT1A1 polymorphisms are currently known to affect enzyme activity, leading to constitutional unconjugated jaundice (Crigler–Najjar types I and II or Gilbert's syndromes) 9. Therefore, we can study Crigler–Najjar and Gilbert's syndromes associated with UGT1A1 polymorphisms in Thai populations. However, this is the first study in Thai colorectal cancer associated with UGT1A1 polymorphisms.
CONCLUSION
In conclusion, the pyrosequencing method for detecting UGT1A1*28 and UGT1A1*6 seems to be rapid, reliable, and more cost‐effective. Thus, we developed a pyrosequencing method to detect UGT1A1*28 and UGT1A1*6 for a routine clinical practice. Our results suggested that genotyping of UGT1A1*28 and UGT1A1*6 should be considered before colorectal cancer patients are prescribed with irinotecan. Pyrosequencing technique can be used for further additional study of association between UGT1A1 genetic polymorphisms and pharmacokinetics drug plasma concentration and clinical outcome in Thai colorectal cancer patients in large sample population.
ACKNOWLEDGMENTS
The authors thank (a) Ramathibodi Hospital Cancer Center for the support of reagents, chemicals, clinical data, and specimens; (b) Suwannee Sirilerttrakul and Somthawin Lukerak for colorectal cancer collections; (c) Department of Pathology, Faculty of Medicine Ramathibodi Hospital, Mahidol University for the support of facilities; and (d) Yupin Wisetpanit for colorectal cancer samples bank.
REFERENCES
- 1. Lankisch TO, Vogel A, Eilermann S, et al. Identification and characterization of a functional TATA box polymorphism of the UDP glucuronosyltransferase 1A7 gene. Mol Pharmacol 2005;67(5):1732–1739. [DOI] [PubMed] [Google Scholar]
- 2. Fang JL, Lazarus P. Correlation between the UDP‐glucuronosyltransferase (UGT1A1) TATAA box polymorphism and carcinogen detoxification phenotype: Significantly decreased glucuronidating activity against benzo(a)pyrene‐7,8‐dihydrodiol(‐) in liver microsomes from subjects with the UGT1A1*28 variant. Cancer Epidemiol Biomarkers Prev 2004;13(1):102–109. [DOI] [PubMed] [Google Scholar]
- 3. Zhou J, Tracy TS, Remmel RP. Correlation between bilirubin glucuronidation and estradiol‐3‐gluronidation in the presence of model UDP‐glucuronosyltransferase 1A1 substrates/inhibitors. Drug Metab Dispos 2011;39(2):322–329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Levesque E, Girard H, Journault K, Lepine J, Guillemette C. Regulation of the UGT1A1 bilirubin‐conjugating pathway: Role of a new splicing event at the UGT1A locus. Hepatology 2007;45(1):128–138. [DOI] [PubMed] [Google Scholar]
- 5. Guillemette C, De Vivo I, Hankinson SE, et al. Association of genetic polymorphisms in UGT1A1 with breast cancer and plasma hormone levels. Cancer Epidemiol Biomarkers Prev 2001;10(6):711–714. [PubMed] [Google Scholar]
- 6. Hirose K, Kozu C, Yamashita K, et al. Correlation between plasma concentration ratios of SN‐38 glucuronide and SN‐38 and neutropenia induction in patients with colorectal cancer and wild‐type UGT1A1 gene. Oncol Lett 2012;3(3):694–698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Rodriguez‐Novoa S, Martin‐Carbonero L, Barreiro P, et al. Genetic factors influencing atazanavir plasma concentrations and the risk of severe hyperbilirubinemia. AIDS 2007;21(1):41–46. [DOI] [PubMed] [Google Scholar]
- 8. Bae JW, Choi CI, Lee JH, Jang CG, Chung MW, Lee SY. Effects of UDP‐glucuronosyltransferase polymorphisms on the pharmacokinetics of ezetimibe in healthy subjects. Eur J Clin Pharmacol 2011;67(1):39–45. [DOI] [PubMed] [Google Scholar]
- 9. Marcuello E, Altes A, Menoyo A, Del Rio E, Gomez‐Pardo M, Baiget M. UGT1A1 gene variations and irinotecan treatment in patients with metastatic colorectal cancer. Br J Cancer 2004;91(4):678–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Kobayashi E, Satoh N. Clinical applications of UGT1A1 polymorphisms for irinotecan therapy. Pharmacogenom Pharmacoproteom 2012;3(2):e117. [Google Scholar]
- 11. Shimoyama S. Pharmacogenetics of irinotecan: An ethnicity‐based prediction of irinotecan adverse events. World J Gastrointest Surg 2010;2(1):14–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Takano M, Kato M, Yoshikawa T, et al. Clinical significance of UDP‐glucuronosyltransferase 1A1*6 for toxicities of combination chemotherapy with irinotecan and cisplatin in gynecologic cancers: A prospective multi‐institutional study. Oncology 2009;76(5):315–321. [DOI] [PubMed] [Google Scholar]
- 13. Boyd MA, Srasuebkul P, Ruxrungtham K, et al. Relationship between hyperbilirubinaemia and UDP‐glucuronosyltransferase 1A1 (UGT1A1) polymorphism in adult HIV‐infected Thai patients treated with indinavir. Pharmacogenet Genomics 2006;16(5):321–329. [DOI] [PubMed] [Google Scholar]
- 14. Amornvipas P, Suphapeetiporn K, Shotelersuk V, Chongsrisawat V, Desudchit T, Poovorawan Y. Crigler–Najjar syndrome: A case report. Asian Biomed 2009;3(2):165–170. [Google Scholar]
- 15. Sutomo R, Laosombat V, Sadewa AH, et al. Novel missense mutation of the UGT1A1 gene in Thai siblings with Gilbert's syndrome. Pediatr Int 2002;44(4):427–432. [PubMed] [Google Scholar]
- 16. Hu ZY, Yu Q, Pei Q, Guo C. Dose‐dependent association between UGT1A1*28 genotype and irinotecan‐induced neutropenia: Low doses also increase risk. Clin Cancer Res 2010;16(15):3832–3842. [DOI] [PubMed] [Google Scholar]
- 17. Iyer L, Das S, Janisch L, et al. UGT1A1*28 polymorphism as a determinant of irinotecan disposition and toxicity. Pharmacogenomics J 2002;2(1):43–47. [DOI] [PubMed] [Google Scholar]
- 18. Gagne JF, Montminy V, Belanger P, Journault K, Gaucher G, Guillemette C. Common human UGT1A polymorphisms and the altered metabolism of irinotecan active metabolite 7‐ethyl‐10‐hydroxycamptothecin (SN‐38). Mol Pharmacol 2002;62(3):608–617. [DOI] [PubMed] [Google Scholar]
- 19. Inoue K, Sonobe M, Kawamura Y, et al. Polymorphisms of the UDP‐glucuronosyl transferase 1A genes are associated with adverse events in cancer patients receiving irinotecan‐based chemotherapy. Tohoku J Exp Med 2013;229(2):107–114. [DOI] [PubMed] [Google Scholar]
- 20. Yu J, Shannon WD, Watson MA, McLeod HL. Gene expression profiling of the irinotecan pathway in colorectal cancer. Clin Cancer Res 2005;11(5):2053–2062. [DOI] [PubMed] [Google Scholar]
- 21. Bosma PJ. Inherited disorders of bilirubin metabolism. J Hepatol 2003;38(1):107–117. [DOI] [PubMed] [Google Scholar]
- 22. Hoskins JM, Goldberg RM, Qu P, Ibrahim JG, McLeod HL. UGT1A1*28 genotype and irinotecan‐induced neutropenia: Dose matters. J Natl Cancer Inst 2007;99(17):1290–1295. [DOI] [PubMed] [Google Scholar]
- 23. Huang SH, Chao Y, Wu YY, et al. Concurrence of UGT1A polymorphism and end‐stage renal disease leads to severe toxicities of irinotecan in a patient with metastatic colon cancer. Tumori 2011;97(2):243–247. [DOI] [PubMed] [Google Scholar]
- 24. Minami H, Sai K, Saeki M, et al. Irinotecan pharmacokinetics/pharmacodynamics and UGT1A genetic polymorphisms in Japanese: Roles of UGT1A1*6 and *28. Pharmacogenet Genomics 2007;17(7):497–504. [DOI] [PubMed] [Google Scholar]
- 25. Rouits E, Boisdron‐Celle M, Dumont A, Guerin O, Morel A, Gamelin E. Relevance of different UGT1A1 polymorphisms in irinotecan‐induced toxicity: A molecular and clinical study of 75 patients. Clin Cancer Res 2004;10(15):5151–5159. [DOI] [PubMed] [Google Scholar]
- 26. Marsh S. (ed.). Chapter 8 Pyrosequencing. Canada: Molecular Diagnostics; 2010. [Google Scholar]
- 27. Teh LK, Hashim H, Zakaria ZA, Salleh MZ. Polymorphisms of UGT1A1*6, UGT1A1*27 & UGT1A1*28 in three major ethnic groups from Malaysia. Indian J Med Res 2012;136(2):249–259. [PMC free article] [PubMed] [Google Scholar]
- 28. Sung C, Lee PL, Tan LL, Toh DS. Pharmacogenetic risk for adverse reactions to irinotecan in the major ethnic populations of Singapore: Regulatory evaluation by the health sciences authority. Drug Saf 2011;34(12):1167–1175. [DOI] [PubMed] [Google Scholar]
- 29. Davidson CJ, Zeringer E, Champion KJ, et al. Improving the limit of detection for Sanger sequencing: A comparison of methodologies for KRAS variant detection. Biotechniques 2012;53(3):182–188. [DOI] [PubMed] [Google Scholar]
- 30. Xu JM, Wang Y, Ge FJ, Lin L, Liu ZY, Sharma MR. Severe irinotecan‐induced toxicity in a patient with UGT1A1 28 and UGT1A1 6 polymorphisms. World J Gastroenterol 2013;19(24):3899–3903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Han JY, Lim HS, Shin ES, et al. Comprehensive analysis of UGT1A polymorphisms predictive for pharmacokinetics and treatment outcome in patients with non‐small‐cell lung cancer treated with irinotecan and cisplatin. J Clin Oncol 2006;24(15):2237–2244. [DOI] [PubMed] [Google Scholar]
- 32. Akiyama Y, Fujita K, Ishida H, et al. Association of ABCC2 genotype with efficacy of first‐line FOLFIRI in Japanese patients with advanced colorectal cancer. Drug Metab Pharmacokinet 2012;27(3):325–335. [DOI] [PubMed] [Google Scholar]


