Abstract
Background
Global reports have highlighted the increasing prevalence of Candida tropicalis infections as well as organism’s drug resistance. This study aimed at identifying azole resistance markers in clinical isolates of C. tropicalis, which will be a great resource for developing new drugs.
Methods
Two susceptible and resistant isolates of C. tropicalis were recovered from an epidemiological investigation of candidiasis in immunocompromised patients. C. tropicalis ATCC 750 was used as reference strain. Antifungal susceptibility to fluconazole and itraconazole was determined using Clinical and Laboratory Standards Institute (CLSI) method. Complementary DNA‐amplified fragment length polymorphism (cDNA‐AFLP) technology and real‐time reverse‐transcriptase (RT) PCR were used for identification of potential genes involved in azole resistance of C. tropicalis clinical isolates.
Results
Five genes encoding the following enzymes were identified as superoxide dismutase (SOD) implicated in antioxidant defense, ornithine aminotransferase (OAT), acetyl ornithine aminotransferase (ACOAT), adenosylmethionine‐8‐amino‐7‐oxononanoate aminotransferase (DAPA AT), and 4‐aminobutyrate aminotransferase (ABAT)—belonging to pyridoxal phosphate (PLP) dependent enzymes and acting in an important physiological role in many fungal‐cell cycles. Real‐time RT‐PCR confirmed mRNA level of the aforementioned genes.
Conclusion
Our findings showed that factors such as PLP‐dependent enzymes and SOD might be implicated in drug resistance in C. tropicalis clinical isolate. Therefore, further studies are required to explore the accurate biological functions of the mentioned genes that would be helpful for effective drug development.
Keywords: azole resistance, candidiasis, differential display, real‐time RT‐PCR
INTRODUCTION
Candida species are the most common cause of opportunistic fungal infections 1. Infections due to opportunistic fungal pathogens have increased mainly over the past two decades, related to increasing patient populations such as persons with AIDS, individuals undergoing solid organ and bone marrow transplantation, and immunosuppressed and cancer patients 2, 3. Candida albicans is the most significant agent of candidiasis in immunocompromised patients 4, 5.
C. tropicalis has long been proposed to be an agent of invasive fungal infections in patients with cancer, leukemia and in hematopoietic stem cell transplantation recipients 6, 7. It has been well determined that among non‐albicans Candida species C. tropicalis is the third leading cause of Candida infections as well as the second in respiratory specimens 3, 4.
There are currently three main classes of antifungal agents for treatment of candidiasis: the polyenes, azoles, and echinocandins 8. Fluconazole is the most highly used azole drug 9. Drug resistance is an important issue for treating infections caused by Candida spp. 7. Clinical isolates of C. tropicalis resistant to azoles, particularly to fluconazole, are increasingly reported 3, 5, 10. According to the literature, fluconazole resistance rate is dramatically increasing among clinical isolates of C. tropicalis, a third leading etiologic agent in candidiasis 11, 12.
The cellular and molecular foundations of resistance are strongly dependent on the method of antifungal activity. Molecular resistance may contain detection of point mutation, gene conversions, and gene amplifications causing overexpression or mitotic recombination 10, 13. Although the molecular mechanisms of drug resistance in the C. tropicalis are not completely established, azole resistance mechanisms have been studied basically in C. albicans and C. glabrata 14, 15. Lanostrol 14‐α demethylase is encoded by the ERG11 gene. Involvement of overexpression and point mutation in ERG11 have been reported in azole resistance 16, 17. The efflux pumps such as ATP‐blinding cassette (ABC) transporters and major facilitator super family (MFS) proteins including Candida drug resistance (CDR) 1 and multidrug resistance (MDR) 1 genes have been related to azole resistance in several Candida species, especially C. albicans strains 18.
Complementary DNA‐amplified fragment length polymorphism (cDNA‐AFLP) method is one of the appropriate transcriptomic methods for gene discovery that does not require prior sequence knowledge of the genes 19. This method is used for the identification of new genes in various organisms such as Candida yeast 19, 20. In this study, cDNA ‐AFLP method was utilized for identification of molecular resistance markers in C. tropicalis isolates from immunocompromised patients.
MATERIALS AND METHODS
Yeast Isolates
Two clinical isolates of C. tropicalis composed of isolates sensitive and resistant to fluconazole and itraconazole were used in this study. These isolates were previously recovered from oropharyngeal samples of immunocompromised patients and stored in culture collection of Department of Medical Mycology and Parasitology of Tehran University of Medical Sciences.
C. tropicalis ATCC 750 was used as a reference. All clinical isolates were identified by standard mycological and biochemical methods including colony color in CHROM agar Candida (CHROMagar, Paris, France) and API20C AUX system (Bio Merieux, Lyon, France).
Culture Conditions and Drug Susceptibility Testing
Three strains of C. tropicalis including two clinical isolates and a reference strain were cultured on a yeast extract peptone dextrose (YEPD) agar plate containing 5 g/l yeast extract (Baltimore Biological Laboratory, Washington, USA), 10 g/l peptone (Merck, Frankfurt, Germany), 20 g/l dextrose (Merck, Frankfurt, Germany), and 20 g/l agar (Biolife, Milan, Italy), and then the plates were incubated for 72 h at 37°C. Later a single colony of each isolate and reference strain was transferred in YEPD broth and incubated for 24 h at 37°C.
The in vitro susceptibility of two clinical isolates to fluconazole and itraconazole was determined by broth microdilution method according to CLSI M27‐A3 standard 21. C. krusei ATCC 6258 was used for quality control. After 48 h of incubation at 35°C, minimum inhibitory concentrations (MICs) were determined visually with the aid of mirror by comparison of growth in the wells containing the drug with the drug‐free controls. The MICs of azoles were determined as lowest concentration supporting ≥50% growth inhibition compared to the growth in control wells. All tests were performed at least two times. The MICs of fluconazole and itraconazole for resistant isolate were ≥64 and ≥16 μg/ml, respectively.
RNA Extraction and cDNA Synthesis
Total RNA was extracted from the log phase using the RNeasy protect mini kit (Qiagen, Hilden, Germany), following the mechanical infraction of the yeast cells by sonication with acid‐washed glass beads (0.45–0.52 mm diameter). To avoid any genomic contamination, all RNA samples were treated with RNase‐free DNase (Qiagen, Hilden, Germany) as explained by the manufacturer. The quantity and quality of RNA were determined using nanodrop (ND‐1000, Thermo Fisher Scientific, Waltham, Massachusetts, USA) and the electrophoresis on 1.5% agarose gel.
The single‐strand cDNA (sscDNA) for each of the clinical isolates and the reference strain was synthesized using 6 μg RNA, 20 pmol/μl random hexamer (Fermentas, Burlington, Canada), 20 pmol/μl oligo‐dT (Fermentas, Burlington, Canada), and 10 mM of dNTP mix (Fermentas, Burlington, Canada) incubated at 65°C for 10 min and then cooled on ice followed by addition of 20 U Ribolock (Fermentas, Burlington, Canada), 7.5 μl of 5× reverse‐transcriptase (RT) buffer (Fermentas, Burlington, Canada) and 200 U of Moloney Murine Leukemia virus (M ‐MuLV) RT enzyme (Fermentas, Burlington, Canada) and then incubated at 72°C for 10 min followed by incubation at 42°C for 60 min. The accuracy of cDNA was checked with actin (ACT) gene primers as housekeeping (Table 2). The PCR condition was initial denaturation step for 5 min at 94°C, 30 cycles of denaturation for 30 sec at 94°C, annealing for 30 sec at 62°C, an extension for 7 min at 72°C with a final extension of 72°C.
Table 2.
Primers Used in This Study
| Gene | GenBank | Product | ||
|---|---|---|---|---|
| name | Primer | Nucleotide sequence (5′–3′) | accession no. | size (bp) |
| ACT | ACT F ACT R | GAAGATCTTGTCTGAACGTGGTT GAGGTTTGCATTTCTTGTTCG | AJ389059.1 | 120 |
| SOD | SOD F SOD R | AGGTGGTGGTCAACATCCAG AAGCCCAACCAGAACCTTG | XM‐002550104 | 131 |
| OAT | OAT F OAT R | GGTATTGTTGTTCCGCCAGA TTCACCCCCTTTGAGTGTTC | XM‐002545168.1 | 146 |
| ACOAT | ACOAT F ACOAT R | TCACCTTTAATCCCAGGAG GGATTAACTCCACCTTCACC | XM‐002547867.1 | 128 |
| ABAT | ABAT F ABAT R | GTTTATCTGGTGCCGATGCT CCAAATCTGGAGAACCTGGA | XM‐002547219.1 | 147 |
| DAPA AT | DAPAAT F DAPAAT R | ACCGGTGTCTGGAAATCTCA TTCAATCACTCCCACAGCAC | XM‐002546330.1 | 129 |
Double‐strand cDNA (dscDNA) was synthesized using 40 U DNA polymerase I (Fermentas, Burlington, Canada) and 10 mM dNTP mix at 16°C for 3 h and was precipitated with ethanol. The quantity and quality of synthesized dscDNA were determined using nanodrop (ND‐1000, Thermo Fisher Scientific, Waltham, Massachusetts, USA) and the electrophoresis on 2% agarose gel.
cDNA‐AFLP
cDNA‐AFLP method was performed as explained previously with some modifications 20. Two micrograms purified dscDNA was digested with 5U EcoR1 restriction enzyme (Fermentas, Burlington, Canada) for 4 h at 37°C, and the enzyme was inactivated at 80°C for 20 min. The digested components were ligated to AFLP adaptors (Table 1), 8 μg ADEcoR1 and 4 μg adEcoR1 in a final volume of 60 μl reactions. Briefly, it was conducted 1 min at 50°C, decreasing to 10°C over 1 h (i.e., 1°C/1.5 min) and then 6 U T4 DNA Ligase (Takara, Tokyo, Japan) was added and incubated at 16°C for 16 h.
Table 1.
Adaptors Used in cDNA‐AFLP Method
| Adaptors | Sequence (5′–3′) |
|---|---|
| ADEcoR I | ACCGACGTCGACTATCCATGAAG |
| adEcoR I | AATTCTTCATGG |
| Pre amp | ACCGACGTCGACTATCCATGAAGAATTC |
| S1Ecor I | ACCGACGTCGACTATCCATGAAGAATTCC |
| S2Ecor I | ACCGACGTCGACTATCCATGAAGAATTCG |
| S3Ecor I | ACCGACGTCGACTATCCATGAAGAATTCA |
| S4Ecor I | ACCGACGTCGACTATCCATGAAGAATTCT |
The preamplification was performed with preamp primers (Table 1) on ligated fragments according to the following PCR conditions: 5 min of denaturation at 94°C; 30 cycles of 94°C for 30 sec; 63°C, 30 sec; 72°C, 30 sec; and final extension at 72°C for 5 min. The sensitive amplification was performed using sensitive primers with the same preamplification PCR conditions. These primers included adaptor sequences in addition of one new nucleotide at the 3′ end (Table 1). The PCR products resulting from this step were separated by 8% nondenaturated polyacrylamide gel electrophoresis (PAGE) and stained with silver nitrate. Then, the gels were studied for the existence of differentially expressed transcription‐derived fragments (TDFs).
Isolation, Cloning, and Sequencing of TDFs
Differentiated TDFs were extracted from the PAGE and reamplified with appropriate sensitive primers. The PCR products were checked using electrophoresis on 1.5% agarose gel. Then, they were cloned using TOPO TA cloning kit (Invitrogen, Carlsbad, California, USA) and screened with universal primers, M13 forward (−20) and M13 reverse (Table 3), according to the following PCR condition: 30 cycles of 94°C for 1 min, 55°C for 1 min, 72°C for 1 min, and 72°C for 7 min. The PCR products were checked by agarose (Merck, Frankfurt, Germany) gel electrophoresis. The recombinant plasmids containing unknown DNA were sequenced using M13 forward and reverse primers. Sequence results were analyzed in nonredundant nucleic and protein databases BLAST (www.ncbi.nlm.nih.gov/BLAST/).
Table 3.
Sequences Identified by cDNA‐AFLP
| Code no. | Annotation | Length (bp) | Accession no. |
|---|---|---|---|
| 1 | SOD | 234 | XP 002550150.1 |
| 2 | OAT | 436 | XP 002545232.1 |
| 3 | ACOAT | 448 | XP 002547913.1 |
| 4 | DAPA AT | 433 | XP 002546376.1 |
| 5 | ABAT | 471 | XP 002547265.1 |
| 6 | Hypothetical proteins | 147 | XP 002547165.1 |
Real‐Time RT‐PCR Analysis
Real‐time RT‐PCR was carried out to detect the mRNA expression level of TDF‐derived genes between two resistant and sensitive C. tropicalis isolates by specific primers designed by primer3‐Bio Tools‐University of Massachusetts Medical (biotools. Umassmed. edu/ bioapps/ primer3‐www. cgi, Table 2). The sscDNA for each of the clinical isolates and the reference strain was synthesized from 6 μg of total RNA using Quantitect Reverse Transcription kit (Qiagen, Hilden, Germany) following the manufacturer’s instructions. RT‐PCR was performed in 25 μl reactions containing 2.5 μl cDNA target, 300 nM forward and reverse primers, and 1× Real‐Time PCR Master Mix E3 containing EvaGreen, dUTP, and ROX (500 nM) (GeneON, Ludwigshafen, Germany). Tests were performed in triplicate by StepOnePlus Real‐Time PCR system (Applied Biosystems, Foster City, California, USA).
The PCR condition was as follows: denaturation at 95°C for 60 sec and 40 cycles of denaturation at 95°C for 15 sec, annealing at 55°C for 30 sec, and extension at 72°C for 30 sec. The actin gene was used as a housekeeping gene and expression level of each gene was calculated by the 2−ΔΔCt method as previously described 22. Serial dilutions of the sscDNA were used for calculation of the efficiencies of the primer sets on real‐time PCR.
Statistical Analysis
All tests were performed at least two times and the results are explained as the mean ± SDs. The value of differences was defined by Student's t‐test and the acceptable level of significance was 95% (P < 0.05).
RESULTS
Antifungal susceptibility
Drug sensitivity of C. tropicalis isolates to fluconazole and itraconazole was determined CLSI guideline (document M27‐A3). Results showed that MIC values of fluconazole were ≥64 and 1 μg/ml and for itraconazole were ≥16 and 0.25 μg/ml in resistant and sensitive isolates, respectively. The MIC values of fluconazole and itraconazole for resistant isolate were about 64 times higher than for the sensitive isolate.
cDNA‐AFLP Analysis of Clinical Isolates
To detect the mRNA expression level related to molecular resistance, a cDNA‐AFLP technique was used on drug‐resistant and drug‐sensitive C. tropicalis isolates. To study the validity of cDNA, the actin gene was used as a housekeeping gene and RT‐PCR using specific primers related to actin (120 bp) was conducted (data not shown). cDNA‐AFLP results determined differential expression of TDFs on 8% nondenaturating PAGE using silver staining by 10 differential primer combinations (Fig. 1). Because of restriction enzyme digestion in this method, several transcripts may have the similar size. Accordingly, several bands may be one gene or one band may contain more than one gene. Fourteen TDFs were extracted from the cDNA‐AFLP on PAGE and reamplified.
Figure 1.
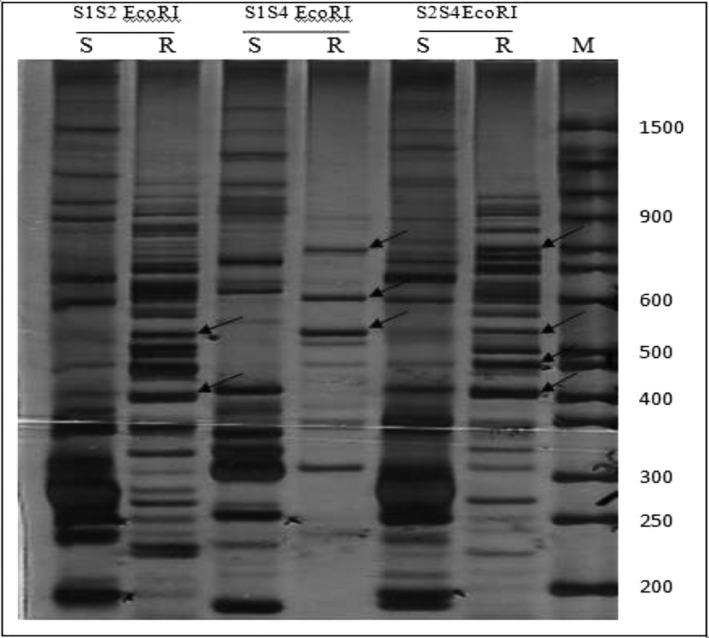
An illustration representative of cDNA‐AFLP on PAGE. Sensitive amplification of cDNA‐AFLP on a PAGE from three different primer combinations: S1S2EcoRI, S1S4EcoRI, S2S4EcoRI. The arrows show differentially expressed TDFs in resistant and sensitive C. tropicalis isolates. M, (50 bp) molecular weight marker; S, sensitive; R, resistant.
Finally, each favorable band was identified by cloning and DNA sequencing. The BLAST analysis indicated that six sequences had remarkable similarities with genome database (E‐value ≤ 10−5). Five target genes containing superoxide dismutase (SOD), ornithine aminotransferase (OAT), acetyl ornithine aminotransferase (ACOAT), adenosylmethionine‐8‐amino‐7‐oxononanoate aminotransferase (DAPA AT), 4‐aminobutyrate aminotransferase (ABAT) and one unknown gene coding hypothetical proteins were detected with different mRNA expression levels in resistant C. tropicalis isolate compared to sensitive isolate and sensitive reference strain. Some characteristics of these genes are presented in Table 3.
Real‐time RT‐PCR was used to investigate the different mRNA expression of target genes in the resistant and sensitive C. tropicalis clinical isolates (Fig. 2). Data show a very considerable upregulation of SOD (32.221‐fold), OAT (88.208‐fold), ACOAT (69.277‐fold), DAPA AT (59.933‐fold), and ABAT (59.933‐fold) in resistant clinical isolate compared to sensitive clinical isolate (P < 0.05).
Figure 2.
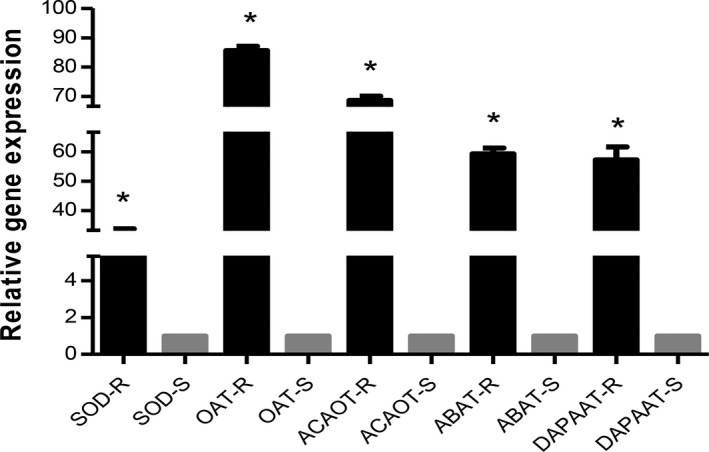
Relative gene expression pattern of target genes in resistant and sensitive Candida tropicalis isolates by real‐time RTPCR. The expression of actin gene was used to normalize the data. R, resistant; S, sensitive. Values are mean ± SD. *P < 0.05.
DISCCUSION
Among non‐albicans Candida species, C. tropicalis represents the third most common species but the second in respiratory specimens 3, 4. Resistance to azoles, especially to fluconazole, in clinical isolates of C. tropicalis is increasingly reported 11, 12. Antifungal resistance of Candida species is a great problem in treating infections caused by these agents 7.
This study describes the role of five target genes including SOD, OAT, ACOAT, DAPA AT, and ABAT on the azole resistance of C. tropicalis, which were identified using cDNA‐AFLP. Our results demonstrate upregulation of these five genes in resistant C. tropicalis clinical isolate for the first time. According to this study, these five genes seem to be potential new markers in molecular resistance that confirmed upregulation of expression level of these genes by real‐time RT‐PCR.
SOD catalyzes the conversion of superoxide radical to hydrogen peroxide and dioxygen 23. According to the metal cofactor, four classes of SOD exist: there are iron (Fe SOD), manganese (Mn SOD), copper and zinc (CuZn SOD), and nickel (Ni SOD) types of enzymes 23, 24. Most antifungal drugs were shown to induce cell death in yeast with apoptotic mechanism by the generation of high intracellular reactive oxygen species (ROS) levels 25, 26.
ROS is one of the major causes of damage to DNA, proteins and lipids, and mutations 27, 28. In this study we report the upregulation of SOD in resistant C. tropicalis clinical isolate. SODs play a significant role in protecting cells from the superoxide ion toxicity 23, 29. Several studies have described the effects of Cu/Zn SOD on the morphological phenotype, oxygen metabolism, and virulence of C. albicans 23, 24.
SOD5 gene, which seems to be an important cofactor in the virulence, switches from yeast to hyphal forms and protects C. albicans cell against intracellularly generated superoxide radicals 24. SOD5 expression levels were increased when cells grew in the presence of high salt concentrations. Martchenko et al. reported there is a correlation between generation and detoxification of superoxide in respiratory of C. albicans 24.
It has been suggested that different factors such as antioxidant defense systems of C. albicans play critical roles for resistance against host immune response 23. Hwang et al. showed that the Cu/Zn SOD deficient strain revealed an increased susceptibility to fungicidal damage by macrophages and attenuated virulence in a mouse model for systemic candidiasis. Our results are in accordance with the above‐mentioned studies that indicated upregulation of SOD in resistant C. tropicalis clinical isolate.
OAT, ACOAT, DAPA AT, and ABAT belong to pyridoxal phosphate (PLP) dependent aspartate aminotransferase superfamily. Aspartate aminotransferase is a key enzyme in the nitrogen metabolism of all organisms 30, 31. Various studies determined that all proteins belonging to this family are involved in different biological processes including transamination (movement of amino groups), decarboxylation (removing COOH groups), racemization (redistribution of enantiomers), and various side‐chain reactions depending on the enzyme involved 30, 31, 32.
These enzymes have been considered to play important physiological role of fungal‐cell cycle 33, 34. ABAT metabolic machinery is interfaced with some of the major facets of fungal‐cell cycle. These include nitrogen and energy metabolism, sporulation, differentiation, and development; and the most important facets include nitrogen and carbon source, energy metabolism, sporulation, differentiation, and development 33. It has been understood that some biomolecules including putrescine, spermidine, and spermine produced by the OAT and ABAT enzymes known as compounds implicated in developmental processes and stress tolerance 33, 35, 36, 37. Fungi have an unusually high requirement of polyamines as essential components for cell growth 33, 35.
It has been reported that overexpression and genetic alterations of various genes such as ERG11 depend on azole resistance 16, 17. The efflux pumps such as ABC transporters and MFS proteins including CDR1 and MDR1 genes have been related to azole resistance in several Candida species, especially in C. albicans strains 18.
The most ubiquitous mechanism resistant to drug toxicity is the elimination of drugs out of the cell using ABC membrane transporters. These molecular pumps actively translocate drugs through the cell membrane by using the energy obtained from ATP hydrolysis 38. The OAT, ACOAT, DAPA AT, and ABAT enzymes play a main role in the production of tricarboxylic acid cycle (TCA cycle) precursors. Therefore, overexpression of these genes leads to the production of high amounts of ATP and energy in fungal cells, so that a part of the ATPs may be used by multidrug efflux pumps for translocation of Azole drugs out of the yeast cells.
We used cDNA‐AFLP method for the identification of target genes involved in azole resistance of C. tropicalis clinical isolate. We discovered five genes including SOD, OAT, ACOAT, ABAT, and DAPA AT as well as one hypothetical protein. This is the first report of indication of the above‐mentioned genes in azole resistance of C. tropicalis clinical isolate. Our findings showed that factors such as PLP‐dependent enzymes and SOD might be implicated in drug resistance in C. tropicalis clinical isolate. Therefore further studies are required to explore the accurate biological functions of the mentioned genes, which would be helpful for effective drug development.
ACKNOWLEDGMENT
The authors thank Dr. Hossein Mirhendi and Pegah Ardi for their kind cooperation.
Grant sponsor: Tehran University of Medical Sciences (TUMS); Grant number: 17694.
REFERENCES
- 1. Pfaller M, Diekema D. Epidemiology of invasive candidiasis: A persistent public health problem. Clin Microbiol Rev 2007;20:133–163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Myoken Y, Kyo TI, Fujihara M, Sugata T, Mikami Y. Clinical significance of breakthrough fungemia caused by azole‐resistant Candida tropicalis in patients with hematologic malignancies. Haematologica 2004;89:378–380. [PubMed] [Google Scholar]
- 3. Vandeputte P, Larcher G, Berges T, Renier G, Chabasse D, Bouchara JP. Mechanisms of azole resistance in a clinical isolate of Candida tropicalis . Antimicrob Agents Chemother 2005;49:4608–4615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Yong AZ, Juan D, Quan BZ, Yun XM. Molecular mechanisms of fluconazole resistance in clinical isolates of Candida tropicalis . Chinese J Clin 2011;5:6329–6335. [Google Scholar]
- 5. Tortorano AM, Biraghi E, Astolfi A, et al., Group FCS . European Confederation of Medical Mycology (ECMM) prospective survey of candidaemia: Report from one Italian region. J Hosp Infect 2002;51:297–304. [DOI] [PubMed] [Google Scholar]
- 6. Bouza E, Munoz P. Epidemiology of candidemia in intensive care units. Int J Antimicrob Agents 2008;32(2):87–91. [DOI] [PubMed] [Google Scholar]
- 7. Jiang C, Dong D, Yu B, et al. Mechanisms of azole resistance in 52 clinical isolates of Candida tropicalis in China. J Antimicrob Chemother 2013;68:778–785. [DOI] [PubMed] [Google Scholar]
- 8. Pappas PG, Kauffman CA, Andes D, et al. Infectious Diseases Society of America. Clinical practice guidelines for the management of candidiasis: 2009 update by the Infectious Diseases Society of America. Clin Infect Dis 2009;48:503–535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Cornely OA, Bassetti M, Calandra T, et al.; Group EFIS . ESCMID guideline for the diagnosis and management of Candida diseases 2012: Non‐neutropenic adult patients. Clin Microbiol Infect 2012;18(7):19–37. [DOI] [PubMed] [Google Scholar]
- 10. Sanglard D, Odds FC. Resistance of Candida species to antifungal agents: Molecular mechanisms and clinical consequences. Lancet Infect Dis 2002;2:73–85. [DOI] [PubMed] [Google Scholar]
- 11. Yang YL, Ho YA, Cheng HH, Ho M, Lo HJ. Susceptibilities of Candida species to amphotericin B and fluconazole: The emergence of fluconazole resistance in Candida tropicalis . Infect Control Hosp Epidemiol 2004;25:60–64. [DOI] [PubMed] [Google Scholar]
- 12. Krcmery V, Barnes AJ. Non‐albicans Candida spp. causing fungaemia: Pathogenicity and antifungal resistance. J Hosp Infect 2002;50:243–260. [DOI] [PubMed] [Google Scholar]
- 13. Lupetti A, Danesi R, Campa M, Del Tacca M, Kelly S. Molecular basis of resistance to azole antifungals. Trends Mol Med 2002;8:76–81. [DOI] [PubMed] [Google Scholar]
- 14. Brun S, Berges T, Poupard P, et al. Mechanisms of azole resistance in petite mutants of Candida glabrata . Antimicrob Agents Chemother 2004;48:1788–1796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Sanguinetti M, Posteraro B, Fiori B, Ranno S, Torelli R, Fadda G. Mechanisms of azole resistance in clinical isolates of Candida glabrata collected during a hospital survey of antifungal resistance. Antimicrob Agents Chemother 2005;49:668–679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Sanglard D. Resistance of human fungal pathogens to antifungal drugs. Curr Opin Microbiol 2002;5:379–385. [DOI] [PubMed] [Google Scholar]
- 17. Sanglard D. Resistance and tolerance mechanisms to antifungal drugs in fungal pathogens. Mycologist 2003;17:74–78. [Google Scholar]
- 18. Morschhauser J. The genetic basis of fluconazole resistance development in Candida albicans . Biochim Biophys Acta 2002;1587:240–248. [DOI] [PubMed] [Google Scholar]
- 19. Yang D, Ma P, Liang X, et al. Metabolic profiles and cDNA‐AFLP analysis of Salvia miltiorrhiza and Salvia castanea Diel f. tomentosa Stib. Plos One 2012;7:e29678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Farahyar S, Zaini F, Kordbacheh P, et al. Overexpression of aldo‐keto‐reductase in azole‐resistant clinical isolates of Candida glabrata determined by cDNA‐AFLP. Daru 2013;21:1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Wayne P. Clinical and Laboratory Standards Institute: Reference method for broth dilution antifungal susceptibility testing of yeasts; approved standard‐third edition; CLSI document M27‐A3. CLSI 2008a;28:6–12. [Google Scholar]
- 22. Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real‐time quantitative PCR and the 2−ΔΔct method. Methods 2001;25:402–408. [DOI] [PubMed] [Google Scholar]
- 23. Hwang CS, Rhie Ge, Oh JH, Huh WK, Yim HS, Kang SO. Copper‐and zinc‐containing superoxide dismutase (Cu/ZnSOD) is required for the protection of Candida albicans against oxidative stresses and the expression of its full virulence. Microbiology 2002;148:3705–3713. [DOI] [PubMed] [Google Scholar]
- 24. Martchenko M, Alarco AM, Harcus D, Whiteway M. Superoxide dismutases in Candida albicans: Transcriptional regulation and functional characterization of the hyphal‐induced SOD5 gene. Mol Biol Cell 2004;15:456–467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Almeida B, Silva A, Mesquita A, Sampaio‐Marques B, Rodrigues F, Ludovico P. Drug‐induced apoptosis in yeast. Biochim Biophys Acta 2008;1783:1436–1448. [DOI] [PubMed] [Google Scholar]
- 26. Perrone GG, Tan SX, Dawes IW. Reactive oxygen species and yeast apoptosis. Biochim Biophys Acta 2008;1783:1354–1368. [DOI] [PubMed] [Google Scholar]
- 27. Ikner A, Shiozaki K. Yeast signaling pathways in the oxidative stress response. Mutat Res 2005;569:13–27. [DOI] [PubMed] [Google Scholar]
- 28. Brown AJ, Haynes K, Quinn J. Nitrosative and oxidative stress responses in fungal pathogenicity. Curr Opin Microbiol 2009;12:384–391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Gunasekaran U, Yang R, Gunasekaran M. Regulation of superoxide dismutase synthesis in Candida albicans . Mycopathologia 1998;141:59–63. [DOI] [PubMed] [Google Scholar]
- 30. Cubellis MV, Rozzo C, Nitti G, Arnone MI, Marino G, Sannia G. Cloning and sequencing of the gene coding for aspartate aminotransferase from the thermoacidophilic archaebacterium Sulfolobus solfataricus . Eur J Biochem 1989;186:375–381. [DOI] [PubMed] [Google Scholar]
- 31. Kappes B, Tews I, Binter A, Macheroux P. PLP‐dependent enzymes as potential drug targets for protozoan diseases. Biochim Biophys Acta 2011;1814:1567–1576. [DOI] [PubMed] [Google Scholar]
- 32. Jensen RA, Gu W. Evolutionary recruitment of biochemically specialized subdivisions of family I within the protein superfamily of aminotransferases. J Bacteriol 1996;178:2161–2171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Kumar S, Punekar NS. The metabolism of 4‐aminobutyrate (GABA) in fungi. Mycol Res 1997;101:403–409. [Google Scholar]
- 34. Pireaux J‐C, Hayani‐Obeidou W, Chalot M, Botton B, Dizengremel P. Mitochondria in the white‐rot fungus Phanerochaete chrysosporium: Purification and evidence for a mitochondrial isoform of aspartate aminotransferase. Exp Mycol 1995;19:91–100. [Google Scholar]
- 35. Stranska J, Kopecny D, Tylichova M, Snegaroff J, Sebela M. Ornithine δ‐aminotransferase. Plant Signal Behav 2008;3:929–935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Jobbagy AJ, Wagner RP. Changes in enzyme activity of germinating conidia of Neurospora crassa . Dev Biol 1973;31:264–274. [DOI] [PubMed] [Google Scholar]
- 37. Reyes‐Garcia MG, Garcia‐Tamayo F, Hernandez‐Hernandez F. Gamma‐aminobutyric acid (GABA) increases in vitro germ‐tube formation and phospholipase B1 mRNA expression in Candida albicans . Mycoscience 2012;53:36–39. [Google Scholar]
- 38. Martinez L, Falson P. Multidrug resistance ATP‐binding cassette membrane transporters as targets for improving oropharyngeal candidiasis treatment. Adv Cell Mol Otolaryngol 2014;2:1–8. [Google Scholar]


