Abstract
In May 2016, an unusual outbreak with the Panton-Valentine leukocidin-positive human variant of meticillin-resistant Staphylococcus aureus (MRSA) clonal complex 398 occurred among mothers and infants in the maternity unit of a Danish hospital. MRSA sharing genotypic and phenotypic characteristics was confirmed in 36 cases, including 26 patients, nine household members and a healthcare worker (HCW) who had contact with all the patients. The national MRSA database contained 37 seemingly unlinked MRSA cases whose isolates shared the same genotypic and phenotypic characteristics as the outbreak strain. Whole genome sequencing showed that three of these isolates clustered together with the 36 outbreak isolates, suggesting spread outside the hospital. The HCW and 21 of 37 cases from the national MRSA database had links to south-eastern Asia, where the outbreak strain is endemic. These findings suggest that the HCW acquired the outbreak strain while travelling in south-eastern Asia and then introduced it into the hospital; from there, it spread within the patients’ households and into the community. Screening of travellers returning from countries with high levels of MRSA could be an important intervention to prevent spread of these bacteria into hospitals via patients or HCWs.
Keywords: meticillin-resistant Staphylococcus aureus, MRSA, Panton-Valentine leukocidin, PVL, hospital outbreak, surveillance, travel, epidemiology
Background
Meticillin-resistant Staphylococcus aureus (MRSA) is one of the world’s leading causes of infections in hospitals and other healthcare facilities. In 2015, MRSA accounted for an estimated 148,727 infections, 68% of which were healthcare associated, and 7,049 deaths in the European Union and European Economic Area (EU/EEA) [1].
Denmark is considered a low prevalence country for MRSA. According to the latest report from the European Antimicrobial Resistance Surveillance Network (EARS-Net), MRSA accounted for only 2.0% of S. aureus bacteraemia cases in Denmark, compared with 13.7% across Europe [2]. Nonetheless, the number of infections caused by imported, community-associated and livestock-associated (LA) MRSA clones is increasing [3]. Large hospital outbreaks with MRSA are uncommon in Denmark, with the exception of an epidemic MRSA-15 clone (EMRSA-15) outbreak that affected 440 individuals in two hospitals in Vejle County from 2002 to 2005 [4]. In 2016, MRSA only caused an estimated 30 hospital-acquired infections (HAIs), compared with 176 infections among other individuals who were recently exposed to a healthcare setting, 325 imported infections, 607 community-acquired infections and 218 infections with the LA variant of MRSA clonal complex 398 (LA-MRSA CC398) [3].
In 2006, the Danish health authorities made MRSA a notifiable organism and adopted a national multifaceted programme to prevent the introduction of MRSA into healthcare settings from expanding community and livestock reservoirs. The programme consists of several elements, including development and maintenance of a national surveillance system, a restrictive antimicrobial policy and a set of infection control guidelines. These guidelines recommend targeted screening of patients with predefined risk factors for MRSA acquisition at hospital admission, as well as decolonisation and isolation of carriers [5]. As part of the programme, the local clinical microbiology departments perform S. aureus identification and antimicrobial susceptibility testing, then submit confirmed MRSA isolates to the National Reference Laboratory for Antimicrobial Resistance (NRLAR) at Statens Serum Institut (SSI) for further genotypic and phenotypic characterisation. Within a week, the local clinical microbiology departments receive a report containing strain typing results for each submitted MRSA isolate, including the spa type and/or the clonal complex. Strain typing results and notification forms with patient information are stored in the national MRSA repository and database, which are maintained by NRLAR for surveillance purposes.
Outbreak detection
On 3 May 2016, the Department of Clinical Microbiology at Lillebælt Hospital became aware of a possible outbreak after MRSA was isolated from two women’s breast abscesses on 7 and 29 April 2016, respectively. Both had given birth in the maternity unit at Kolding Hospital in March 2016. MALDI-TOF mass spectrometry (MALDI-TOF MS) revealed that the two MRSA isolates belonged to a unique subgroup, previously termed A8.3 [6], which is related to yet distinct from the LA-MRSA CC398 MALDI-TOF MS subgroups prevalent in the hospital’s catchment area. Comparison of the antibiograms showed that the isolates had identical profiles, as they were all resistant to penicillin, cefoxitin, tetracycline, erythromycin and clindamycin. NRLAR was then contacted to obtain further genotypic and phenotypic characteristics of the two MRSA isolates, which showed that they belonged to CC398, had spa type t034 and carried mecA, scn and lukF-PV, which together with lukS-PV encodes Panton-Valentine leukocidin (PVL). The presence of lukF-PV suggested that the isolates belonged to the human variant of MRSA CC398, which was considered extremely rare in Denmark at that time [7]. This further convinced the infection control team that the two cases had acquired the strain from a common source during their stay in the maternity ward, and a formal outbreak investigation was therefore opened. Here we describe the main findings of the outbreak investigation and consider some of the many challenges associated with outbreak prevention, detection and management.
Methods
Ethics statement
This study was a collaboration between the Department of Clinical Microbiology at Lillebælt Hospital and NRLAR at SSI. Collection and use of data were approved by the Danish Data Protection Agency (protocol number 2001–14–0021).
Setting
Lillebælt Hospital serves a catchment area of 300,000 inhabitants and has about 700 beds and 200,000 occupied-bed-days per year. The trust comprises three hospitals in Kolding, Middelfart and Vejle, in the Region of Southern Denmark. The maternity unit where the outbreak occurred is located at Kolding Hospital, where an average of 3,200 births take place each year.
Case definition
On 4 May 2016, an outbreak group consisting of representatives from the maternity unit and the infection control team at the Department of Clinical Microbiology initiated a formal investigation. Possible cases were defined as mothers who had undergone caesarean delivery or other birth-related surgical procedures in the maternity unit at Kolding Hospital in early May 2016. This definition was changed on 10 May 2016 to include all mothers, infants and staff members who had worked or stayed in the maternity unit between 1 January and 30 June 2016. Probable cases were defined as all individuals who had worked or stayed in the maternity unit between 1 January and 30 June 2016 and all their household contacts who were found to be colonised or infected with MRSA CC398 of MALDI-TOF MS subgroup A8.3 between 1 January 2016 and 28 February 2017. Confirmed cases were defined as probable cases whose isolates belonged to the outbreak cluster, as determined by phylogenetic analysis of high-quality single nucleotide polymorphisms (SNPs) in the core genome.
Outbreak investigation
The outbreak investigation comprised: (i) immediate risk assessment of procedures involving the use and storage of breastfeeding equipment, as well as postoperative management of mothers who underwent caesarean delivery or other delivery-related surgical procedures; (ii) environmental sampling of common contact points in the delivery room, operating room and milk kitchen of the maternity unit, e.g. soap dispensers and handles of drawers, cabinets and doors (n = 32); (iii) interviews with hospital staff members; (iv) screening of all mothers and infants (n = 60), as well as all hospital staff members (n = 88), who were present in the maternity unit or the neonatal unit on 10 May 2016; (v) contacting all mothers who had stayed in the maternity unit between 1 April and 9 May 2016 (n = 177) to promote screening of the entire household by their general practitioner and (vi) retrospective and prospective review of the local MRSA database Laboratorieovervågning af antibiotikaresistens (LIVA) to identify individuals who had tested positive for MRSA CC398 of MALDI-TOF MS subgroup A8.3 outside the hospital between January 2016 and February 2017, including those who were contacted by the infection control team.
LIVA is a web-based database application for real-time surveillance of MRSA in the Region of Southern Denmark and contains information from the following sources: (i) demographic data from the Danish Civil Registration System (CPR); (ii) local clinical microbiology laboratory-based results of MRSA cultures from screening swabs and clinical samples, positive or negative; (iii) local and NRLAR-based strain typing results for laboratory-confirmed MRSA isolates, including MALDI-TOF MS subgroup profiles, spa types and clonal complexes; and (iv) epidemiological and clinical data for all MRSA-positive cases collected by clinical microbiologists and infection control nurses.
During the outbreak investigation, the following data were assessed for each MRSA-positive case: sex, age, country of birth, residential address, specimen source (screening swab or clinical sample), infection type, hospitalisation dates, MALDI-TOF MS subgroup profile, spa type and clonal complex.
Sample collection and laboratory investigations
Clinical samples were obtained from patients with active infection and were cultured directly on 5% blood agar and incubated for 24–48 hours at 35 °C. Healthy mothers, infants and hospital staff members were screened for MRSA by swabbing the nose and throat. Screening swabs and environmental swabs collected in the maternity unit were enriched for at least 24 hours at 35 °C in Contrast MRSA Broth (Oxoid, Basingstoke, United Kingdom). A 10 μl loopful of broth from positive cultures (colour change from red) was plated onto a CHROMID MRSA agar plate (bioMérieux, Marcy l'Etoile, France) and incubated for 24 hours at 35 °C, whereafter one presumptive MRSA colony (green colour) was subcultured on 5% blood agar for 24 hours at 35 °C. Colonies with morphological characteristics of S. aureus on 5% blood agar underwent antimicrobial susceptibility testing against penicillin, cefoxitin, tetracycline, erythromycin, clindamycin, gentamicin, rifampicin, fusidic acid, mupirocin and linezolid using the disk diffusion method in accordance with the European Committee on Antimicrobial Susceptibility Testing (EUCAST) guidelines [8]. Presumptive MRSA colonies growing within the inhibition zone of the cefoxitin disks were subjected to MALDI-TOF MS for species identification and subgrouping of MRSA isolates using the microflex LT/SH instrument (Bruker, Bremen, Germany), as described elsewhere [9], and investigated for the presence of PBP2a using a commercial latex agglutination kit, Slidex MRSA detection (bioMérieux).
All MRSA isolates were routinely submitted to NRLAR, where they were investigated for the presence of spa, mecA, scn, lukF-PV and the S. aureus CC398-specific sau1-hsdS1 variant using a multiplex PCR assay. Methodological details are provided in Supplement S1. spa-typing was performed as described previously [10].
Identification of other Danish cases between January 2006 and April 2017
The national MRSA database was reviewed to identify other Danish cases of PVL-positive MRSA CC398 between 1 January 2006 and 30 April 2017. The following data were assessed for each case: sex, age, country of birth, residential address, travel history, specimen source (screening swab or clinical sample), infection type and hospitalisation dates.
Whole genome sequencing and bioinformatics analyses
Whole genome sequencing and bioinformatics analyses were used to study the relationship of the PVL-positive MRSA CC398 isolates identified in this study. Methodological details are provided in Supplement S1.
Data access
The whole genome sequence data from this study have been submitted to the National Center for Biotechnology Information (NCBI) Sequence Read Archive (SRA) under BioProject PRJNA508272.
Results
Outbreak cases identified in the maternity unit
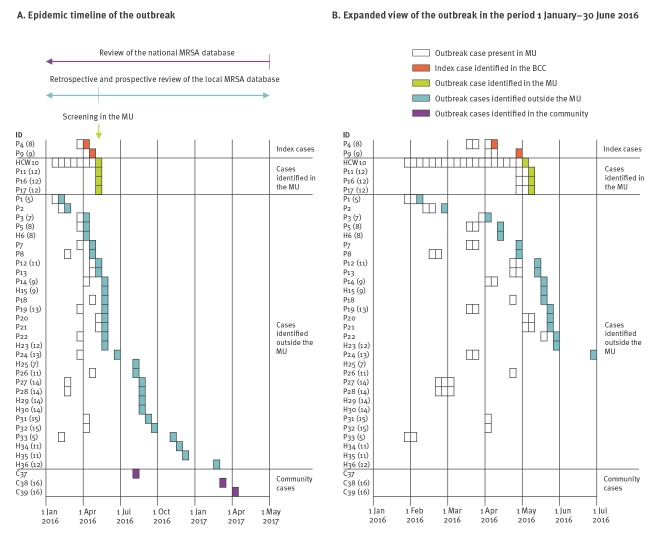
On 5 May 2016, a healthcare worker (HCW) (outbreak case 10) who had been in contact with the two index cases (outbreak cases 4 and 9) screened positive for MRSA. The HCW had also been in contact with mothers whose children were transferred to the neonatal unit at the same hospital. This prompted the infection control team to include the neonatal unit in the outbreak investigation. Five days later, on 10 May 2016, all 23 mothers and 16 infants in the maternity unit were screened for MRSA carriage, as were eight mothers and 13 infants in the neonatal unit and 88 hospital staff members. This led to the identification of three probable outbreak cases in the maternity unit, a mother and her newborn twins (outbreak cases 11, 16 and 17) (Figure 1 and Table 1). All three cases had already been discharged when the results of their MRSA status became available and, therefore, isolation precautions were not needed. The isolates from these cases and the HCW belonged to the same unique MALDI-TOF MS subgroup A8.3 and antibiogram profile as the isolates from the two index cases. All other hospital staff members, mothers and infants in the maternity and neonatal units screened negative.
Figure 1.
Timeline of the Staphylococcus aureus clonal complex 398 hospital outbreak, Kolding, Denmark, 1 January 2016–30 April 2017 (n = 39)
BCC: breast care centre; C: community case; H: household contact; HCW: healthcare worker; P: patient; MU: maternity unit.
Cases were categorised as healthcare workers, patients, household contacts or community cases, and were assigned case numbers according to the sampling date.
Household cluster IDs are indicated in parentheses.
Table 1. Description of MRSA cases and isolates identified during hospital outbreak investigation, Kolding, Denmark, 2016–2017 (n = 36).
| Outbreak case numbera | Isolate ID | Case description | Household cluster | Hospitalisation dates in the maternity unitb
(month and year) |
Sampling date | Sampling place | Specimen source |
|---|---|---|---|---|---|---|---|
| Index cases | |||||||
| 4 | 216065 | Mother | 8 | Mar 2016 | Apr 2016 | BCC | Breast abscess |
| 9 | 216913 | Mother | 9 | Apr 2016 | Apr 2016 | BCC | Breast abscess |
| Cases identified in the MU | |||||||
| 10 | 217161 | HCW in MU | NA | NA | May 2016 | MU | Screeningc |
| 11 | 217479 | Mother | 12 | May 2016 | May 2016 | MU | Screeningd |
| 16 | 217637 | Infant | 12 | May 2016 | May 2016 | MU | Conjunctivitis, scalp pustules |
| 17 | 217639 | Infant | 12 | May 2016 | May 2016 | MU | Scalp pustules |
| Cases identified outside the MU | |||||||
| 1 | 213707 | Mother | 5 | Feb 2016 | Feb 2016 | GP | Postoperative wound |
| 2 | 214441 | Infant | NA | Feb 2016 | Feb 2016 | HCF | Scalp pustules |
| 3 | 216051 | Infant | 7 | Mar 2016 | Apr 2016 | HCF | Conjunctivitis |
| 5 | 216247 | Infant | 8 | Mar 2016 | Apr 2016 | GP | Screening |
| 6 | 216245 | Father | 8 | Not hospitalised | Apr 2016 | GP | Screening |
| 7 | 216863 | Mother | NA | Mar 2016 | Apr 2016 | GP | Axillary abscess |
| 8 | 216867 | Infant | NA | Feb 2016 | Apr 2016 | GP | Conjunctivitis |
| 12 | 217389 | Infant | 11 | Apr 2016 | May 2016 | GP | Screening |
| 13 | 217491 | Mother | NA | Apr 2016 | May 2016 | GP | Breast abscess, postoperative wound |
| 14 | 217573 | Infant | 9 | Apr 2016 | May 2016 | GP | Screening |
| 15 | 217681 | Grandparent | 9 | Not hospitalised | May 2016 | GP | Screening |
| 18 | 217683 | Infant | NA | Apr 2016 | May 2016 | HCF | Screening |
| 19 | 217817 | Mother | 13 | Mar 2016 | May 2016 | GP | Breast abscess |
| 20 | 217831 | Infant | NA | May 2016 | May 2016 | GP | Screening |
| 21 | 218157 | Infant | NA | May 2016 | May 2016 | GP | Bulla (gluteal region) |
| 22 | 218159 | Mother | NA | Mar 2016 | May 2016 | GP | Axillary abscess |
| 23 | 219585 | Sibling | 12 | Not hospitalised | May 2016 | GP | Wound (leg) |
| 24 | 219599 | Infant | 13 | Mar 2016 | Jun 2016 | GP | Screening |
| 25 | 220909 | Father | 7 | Not hospitalised | Aug 2016 | GP | Wound (leg) |
| 26 | 220981 | Mother | 11 | Apr 2016 | Aug 2016 | BCC | Breast abscess |
| 27 | 221423 | Infant | 14 | Feb/Mar 2016 | Aug 2016 | GP | Eczema |
| 28 | 221793 | Mother | 14 | Feb/Mar 2016 | Aug 2016 | GP | Screening |
| 29 | 221789 | Sibling | 14 | Not hospitalised | Aug 2016 | GP | Screening |
| 30 | 221735 | Father | 14 | Not hospitalised | Aug 2016 | GP | Screening |
| 31 | 222407 | Mother | 15 | Apr 2016 | Sep 2016 | GP | Skin abscess (mosquito bite) |
| 32 | 223053 | Infant | 15 | Apr 2016 | Sep 2016 | GP | Screening |
| 33 | 224287 | Infant | 5 | Jan/Feb 2016 | Nov 2016 | GP | Screening |
| 34 | 225425 | Grandparent | 11 | Not hospitalised | Nov 2016 | GP | Screening |
| 35 | 225423 | Father | 11 | Not hospitalised | Dec 2016 | GP | Screening |
| 36 | 229989 | Sibling | 12 | Not hospitalised | Feb 2017 | GP | Screening |
BCC: breast care centre; GP: general practitioner; HCF: other healthcare facility; HCW: healthcare worker; MRSA: meticillin-resistant Staphylococcus aureus; MU: maternity unit; NA: not applicable.
a Outbreak cases were assigned case numbers according to the sampling date.
b This includes all mothers, infants and staff members who had worked or stayed in the maternity unit between 1 January and 30 June 2016.
c MRSA was isolated 3 months after travel to Thailand.
d MRSA was subsequently isolated from a postoperative wound on 12 May 2016.
Outbreak cases identified outside the maternity unit
Retrospective and prospective review of the LIVA database revealed that 30 other individuals outside the hospital had tested positive for MRSA CC398 of MALDI-TOF MS subgroup A8.3 between January 2016 and February 2017. All 30 cases had links to the maternity unit, including eight mothers and 13 infants who had stayed in the unit between 1 January and 9 May 2016 and nine household contacts (Figure 1 and Table 1). Notably, the LIVA database does not indicate whether mothers, infants and household contacts were tested because they were contacted by the local infection control team or if they were tested for other reasons.
Outbreak description
In total, 36 MRSA cases with epidemiological links to the outbreak in the maternity unit were identified between 10 February 2016 and 22 February 2017. Of these, 27 (15 infants, 11 mothers and one HCW) had stayed in the maternity unit between 31 January and 10 May 2016 (Figure 1 and Table 1). The mean length of stay was 5.1 days (range: 2–11 days). Four of these infants (outbreak cases 3, 12, 14 and 18) had subsequently stayed in the neonatal unit, while one infant (outbreak case 2) had been visiting the neonatal unit’s outpatient clinic (Figure 1 and Table 1), but this did not lead to any secondary cases. Eighteen of the 36 possible outbreak cases (seven infants, nine mothers, one father and one sibling) were infected at the time of diagnosis (Figure 1 and Table 1). The majority of mothers had abscesses in the breast or axillary area, while pustulosis of the scalp and face predominated among infants. All MRSA isolates from the 36 outbreak cases had the same unique MALDI-TOF MS subgroup A8.3, belonged to CC398, had spa type t034 and carried mecA, scn and lukF-PV. The HCW was identified as the most likely source of the outbreak and was therefore relieved from duty for 3 days, during which the HCW began MRSA decolonisation therapy. By the end of May 2016, no more cases were detected in the maternity unit and the outbreak was declared over.
Identification of cases from the national MRSA database
Review of the national MRSA database identified 37 cases that had no epidemiological links to the outbreak in the maternity unit, but whose isolates also belonged to CC398, had spa type t034 and carried mecA, scn and lukF-PV (Table 2). Twenty-nine of these were infected at the time of diagnosis, most of which presented in primary care with skin abscesses.
Table 2. Description of MRSA cases and isolates in the national MRSA database, Denmark, 2006–2017 (n = 37).
| Isolate/case ID | Case description | Household cluster | Sampling date | Sampling place | Specimen source | Relation to south-eastern Asia | Lineage | Phylogenetic cluster |
|---|---|---|---|---|---|---|---|---|
| 50148 | Adult | 1 | Jan 2006 | HCF | Skin abscess | Yes | L1 | A |
| 51306 | Child | 1 | May 2006 | HCF | Screening | Yes | L1 | A |
| 51726 | Child | NA | Jun 2006 | HCF | Skin abscess | Yes | L1 | NA |
| 55600 | Child | 1 | Mar 2007 | HCF | Wound infection | Yes | L1 | A |
| 55730 | Child | 1 | Mar 2007 | HCF | Screening | Yes | L1 | A |
| 160793 | Adult | NA | Jan 2012 | Unknown | Skin abscess | Yes | L2 | NA |
| 173861 | Adult | 2 | May 2013 | GP | Skin abscess | Yes | L2 | B |
| 174399 | Adult | 2 | May 2013 | Unknown | Screening | Yes | L2 | B |
| 176761 | Adult | 3 | Aug 2013 | HCF | Skin abscess | No | L2 | C |
| 176987 | Adult | 3 | Sep 2013 | GP | Screening | Unknown | L2 | C |
| 185025 | Adult | NA | Mar 2014 | GP | Skin abscess | Yes | L2 | D |
| 187597 | Adult | NA | Jun 2014 | GP | Screening | No | L2 | D |
| 194365 | Adult | NA | Dec 2014 | HCF | Skin abscess | Yes | L2 | NA |
| 203065 | Child | NA | May 2015 | GP | Wound infection | Unknown | L2 | E |
| 203343 | Adult | NA | Jun 2015 | GP | Skin abscess | Yes | L2 | NA |
| 205059 | Adult | NA | Jul 2015 | HCF | Skin abscess | Yes | L2 | F |
| 206889 | Adult | 4 | Sep 2015 | Unknown | Wound infection | No | L2 | E |
| 208403 | Adult | 4 | Oct 2015 | GP | Screening | No | L2 | E |
| 212591 | Adult | NA | Jan 2016 | GP | Wound infection | Yes | L2 | NA |
| 214119 | Adult | 6 | Feb 2016 | GP | Paronychia | No | L2 | G |
| 214133 | Adult | NA | Jan 2016 | GP | Skin abscess | No | L2 | G |
| 214137 | Adult | 6 | Feb 2016 | HCF | Skin abscess | No | L2 | G |
| 217191 | Child | 10 | May 2016 | GP | Screening | Yes | L2 | H |
| 218093 | Adult | NA | May 2016 | HCF | Skin abscess | Yes | L2 | I |
| 219061 | Adult | NA | Jun 2016 | GP | Wound infection | Yes | L2 | I |
| 219077 | Adult | NA | Jun 2016 | GP | Skin abscess | No | L2 | I |
| 219969 | Adult | NA | Jun 2016 | GP | Wound infection | No | L2 | NA |
| 220875 | Adult | NA | Aug 2016 | HCF | Wound infection | Yes | L2 | NA |
| 223021 | Adult | NA | Sep 2016 | GP | Wound infection | Yes | L2 | NA |
| 224479 | Adult | 10 | Nov 2016 | HCF | Skin abscess | Yes | L2 | H |
| 225753 | Adult | NA | Aug 2016 | HCF | Wound infection | Yes | L2 outbreakb | J |
| 230011 | Adult | NA | Feb 2017 | GP | Skin abscess | No | L2 | NA |
| 230183 | Adult | NA | Feb 2017 | GP | Skin abscess | Yes | L2 | NA |
| 230425 | Adult | 16 | Mar 2017 | GP | Skin abscess | No | L2 outbreakc | J |
| 231383 | Child | NA | Mar 2017 | GP | Skin abscess | No | L2 | F |
| 231391 | Adult | NA | Apr 2017 | Unknown | Skin abscess | No | L2 | E |
| 231535 | Adult | 16 | Apr 2017 | GP | Screening | No | L2 outbreakd | J |
GP: general practitioner; HCF: other healthcare facility; MRSA: meticillin-resistant Staphylococcus aureus; NA: not applicable.
a Relation to south-eastern Asia comprises cases who were either born in or had recently visited south-eastern Asia or had known contact with such individuals.
b Outbreak case 37.
c Outbreak case 38.
d Outbreak case 39.
Whole genome sequencing
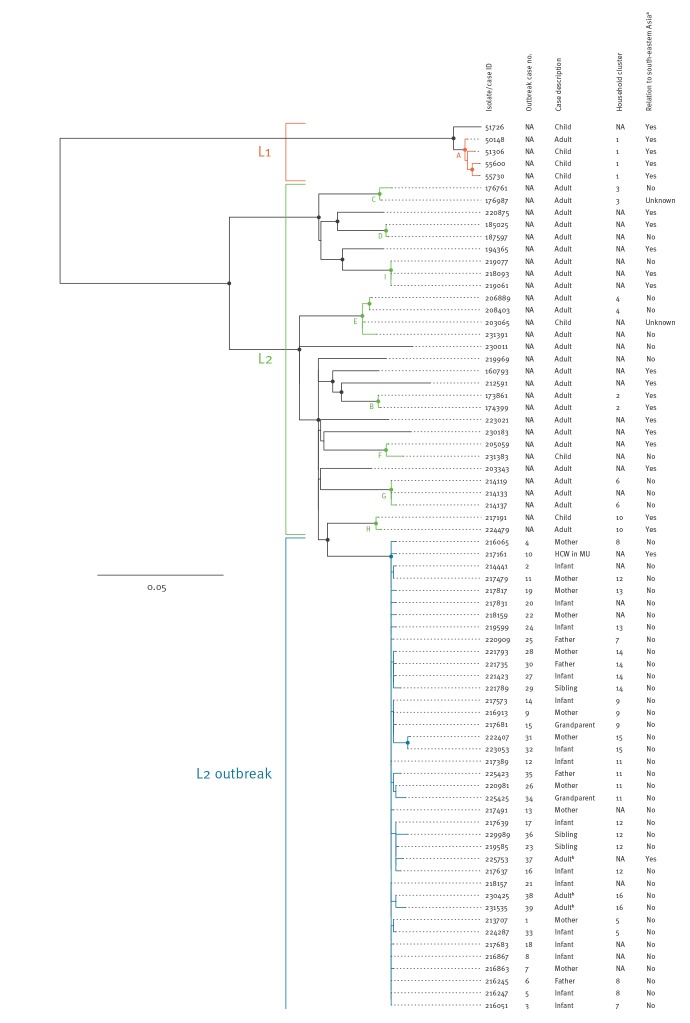
Analysis of whole genome sequence data from the 73 PVL-positive MRSA CC398 isolates, including 36 isolates from the outbreak investigation and 37 additional isolates from the national MRSA database, showed that they all contained the core SNPs used to differentiate the human variant of MRSA CC398 from the LA variant [11]. Mapping sequence reads for each isolate against the S. aureus CC398 reference isolate 71193 identified 1,071 high-quality core SNPs among the 73 isolates after recombination was removed from the SNP alignment. Phylogenetic analysis revealed two distinct lineages (L1 and L2) separated by 254–284 SNPs (Figure 2). L1 contained five isolates that differed by 5–22 SNPs, belonged to ST398, carried spa type t034, harboured a composite type Vb (5C2 and 5) SCCmec element and contained erm(C) conferring resistance to macrolides, lincosamides and streptogramin B (MLSB), as well as chromosomal mutations associated with resistance to quinolones (Ser84Leu in gyrA and Ser80Phe in grlA). L2 contained 68 isolates that differed by 0–148 SNPs, belonged to ST1232 and carried either spa type t034 (n = 66), t1255 (n = 1) or t1928 (n = 1). All L2 isolates carried a non-subtypable SCCmec V (5C2) element and contained erm(A), spc and tet(K) encoding resistance to MLSB, aminoglycosides (spectinomycin) and tetracyclines, respectively. In addition, four L2 isolates contained aac(6')-aph(2”) encoding resistance to aminoglycosides, one of which also carried erm(B) encoding resistance to MLSB. The 36 isolates considered part of the outbreak belonged to L2 and formed a well-supported cluster together with three isolates from community cases living in close proximity to Kolding Hospital. They are therefore referred to as outbreak cases 37, 38 and 39, respectively (Figure 2 and Table 2). The 39 L2 outbreak isolates differed from the L2 non-outbreak isolates by 47–140 SNPs and were more tightly clustered (0–15 SNPs vs 0–148 SNPs). Besides erm(A), spc and tet(K), which were present in all L2 isolates, the outbreak isolates also contained dfrG conferring resistance to trimethoprim.
Figure 2.
Maximum-likelihood phylogeny of Panton-Valentine leukocidin-positive meticillin-resistant Staphylococcus aureus clonal complex 398 isolates, Denmark, 2006–2017 (n = 73)
HCW: healthcare worker; MU: maternity unit; NA: not applicable.
a Relation to south-eastern Asia comprises cases who were either born in or had recently visited south-eastern Asia or had known contact with such individuals.
b The three community cases identified through review of the national MRSA database.
The phylogeny was estimated from 1,071 high-quality core single nucleotide polymorphisms (SNPs) among the 73 isolates after recombination was removed from the SNP alignment, by using a general time reversible (GTR) model of nt substitution. Coloured branches indicate phylogenetic clusters (A-I) containing L1 and L2 isolates that differed by no more than 15 SNPs, which corresponds to the maximum number of pairwise SNP differences among the L2 outbreak isolates. Bootstrap values above 90% are illustrated by filled circles at the nodes. The scale bar represents the number of nt substitutions per variable site.
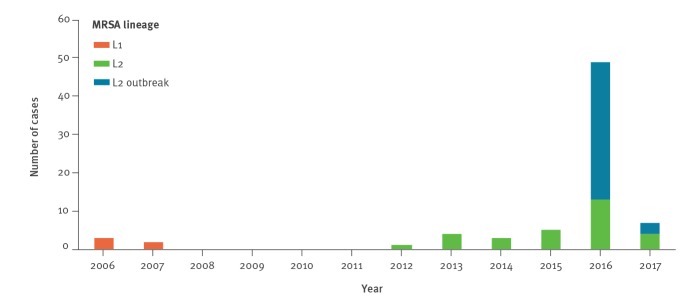
The temporal distribution of L1, L2 non-outbreak and L2 outbreak isolates is illustrated in Figure 3. The L1 isolates were detected in five cases from two households between 18 January 2006 and 15 March 2007, including four children adopted from south-eastern Asia and one of their parents (Figure 2 and Table 2). The L2 non-outbreak isolates were detected in 29 cases from 24 households between 16 January 2012 and 3 March 2017; 15 cases were either born in or had recently visited south-eastern Asia (i.e. Vietnam, Thailand or Cambodia) or had known contact with such individuals (Figure 2 and Table 2). We then divided the L1 and L2 non-outbreak isolates into clusters containing isolates that differed by no more than 15 SNPs, which corresponds to the maximum number of pairwise SNP differences among the L2 outbreak isolates. This led to the identification of nine phylogenetic clusters (A-I) involving four of five L1 isolates and 20 of 29 L2 non-outbreak isolates (Figure 2 and Table 2). L1 and L2 non-outbreak isolates from the same households always clustered together, supporting that transmission had occurred in at least six households. In addition, five clusters (D, E, F, G and I) contained L2 non-outbreak isolates from more than one household. Four of these clusters involved individuals with established or putative epidemiological links, including known community contacts (D and I), living in the same town (G) or living in the same municipality (E), while one cluster (F) involved people with no apparent epidemiological links. This indicates spread of L2 non-outbreak isolates from the households into the local community.
Figure 3.
Temporal trend of Panton-Valentine leukocidin-positive meticillin-resistant Staphylococcus aureus clonal complex 398, Denmark, 2006–2017 (n = 73)
MRSA: meticillin-resistant Staphylococcus aureus.
The L2 outbreak strain was detected in 39 cases between 10 February 2016 and 5 April 2017, including 15 infants, 11 mothers, nine household contacts, one HCW and three community cases (Figure 1, Figure 2, Table 1, Table 2). Two of 39 outbreak cases had links to south-eastern Asia, including the HCW who had been on a 2-week holiday in Thailand some weeks before the outbreak and one community case who was Vietnamese. The community case lived ca 500 m from five of the outbreak cases. The isolate formed a well-supported cluster together with the outbreak isolates from the children, but not with that from the mother (Figure 2).
Outbreak control measures
The HCW who screened positive for MRSA was promptly relieved from clinical duties for 3 days, during which the HCW began MRSA decolonisation therapy. Since the HCW had probably carried the outbreak strain since January 2016, a special team composed of members of the local infection control team and NRLAR epidemiologists was deployed to identify probable cases among mothers and infants who stayed in the maternity unit between 1 January and 9 May 2016 (i.e. before the outbreak investigation in the maternity unit was initiated) through retrospective and prospective review of local and national MRSA databases. In addition, all mothers who stayed in the maternity unit between 1 April and 9 May 2016 were contacted by letter and encouraged to undergo screening, along with their infants and household contacts. Decolonisation therapy was offered to all MRSA-positive individuals and their household contacts, in line with the Danish guidelines for the management of MRSA in healthcare settings [5].
Discussion
This study describes an unusual MRSA outbreak in the maternity unit at Kolding Hospital. The outbreak strain was a PVL-positive human variant of MRSA CC398 of south-eastern Asian origin. The collaboration between the local infection control team and NRLAR, as well as the combined use of local and national surveillance data, extensive local screening of patients and hospital staff members, MALDI-TOF MS as a first-line tool for rapid subgrouping of MRSA isolates and whole genome sequencing made it possible to identify the source of the outbreak. These methods also made it possible to detect transmission events between patients and their household members, as well as subsequent spillover into the community.
In addition to the HCW, one of the community cases had links to south-eastern Asia and might have been in contact with five of the outbreak cases (a mother and her four children). However, we deemed it unlikely that the community case was the source of the outbreak, as the mother in question was admitted to the hospital on 2 May 2016, almost 3 months after the first outbreak case (Figure 1 and Table 1). In addition, the outbreak isolates were, on average, more closely related to the isolate from the HCW than to the isolate from the community case (4.4 SNPs vs 6.9 SNPs). Together, these findings support the notion that the HCW acquired the outbreak strain while in Thailand and introduced it into the hospital after the holiday. From there, it spread within the patients’ households and into the local community.
The PVL-positive human variant of MRSA CC398 has been recognised as a relatively frequent cause of infections among individuals in China, both in healthcare settings and in the community. In Europe, however, it has mainly been associated with carriage and infection in people with links to south-eastern Asia, including China, the Philippines, South Korea and Vietnam [7,12-14]. In addition, the type Vb (5C2 and 5) SCCmec element carried by L1 isolates is widely distributed among other community-associated MRSA clones in south-eastern Asia [15,16]. These findings support our assumption that the PVL-positive human variant of MRSA CC398 originates from south-eastern Asia.
Short hospital stays make it difficult to identify HAIs—let alone outbreaks of HAIs—because many cases experience disease onset while in the community. During the outbreak described here, most infections developed after discharge from hospital. In Denmark, the average number of bed-days per hospital admission was 2.91 in 2017 [17], leaving little time for a hospital-acquired MRSA strain to cause clinical symptoms of infection while a patient is hospitalised. This suggests that the annual numbers of hospital-acquired MRSA infections reported in DANMAP might be underestimated.
Previous studies have shown that skin and soft tissue infections (SSTIs) in travellers returning from high-prevalence countries are an important source of antibiotic-resistant S. aureus, including PVL-positive MRSA clones, and that nasal carriage may also play a role [14,18,19]. However, the Danish guidelines for the management of MRSA in healthcare settings only recommend screening HCWs and patients if they have had recent contact with a known MRSA carrier, stayed in a healthcare facility during an MRSA outbreak, been in a high-risk situation outside Denmark (e.g. hospitals, war zones, refugee camps or orphanages) or been in direct or indirect contact with pigs or mink [5]. The HCW, who is suspected to have acquired the outbreak strain while on holiday in Thailand, did not present with any of these risk factors. This suggests that the outbreak strain originated from a community rather than a healthcare reservoir in Thailand. Furthermore, the behaviour of the outbreak strain—with transmission between family members, onward spread into the surrounding community and a clinical picture dominated by SSTIs—resembles the epidemiology of other PVL-positive community-associated MRSA clones, such as CC30 and CC59 in Asia [16], CC80 in Europe [20] and US300 (CC8) in the United States [21] (see [22] also for a recent review on PVL-positive S. aureus). The emergence and rapid spread of community-associated MRSA clones in various parts of the world and the findings of this and other studies [23-25] suggest that international travellers could lead to import of community-associated MRSA and subsequent spread into hospitals. This notion is supported by the finding that imported MRSA clones accounted for an estimated 24% (325/1,356) of all MRSA infections in Denmark in 2016 [3].
Lessons learnt
When the outbreak investigation started, it was not common practice for NRLAR to inform the local clinical microbiology departments about the presence or absence of PVL in submitted MRSA isolates. It is possible that the outbreak and its source could have been detected earlier if the local infection control team had been aware of the unusually high number of mothers and infants testing positive for the PVL-positive human variant of MRSA CC398 in February, March and April 2016. NRLAR has subsequently put this into practice and now informs general practitioners and local clinical microbiology departments of the presence or absence of lukF-PV, together with the spa type and/or the clonal complex.
The outbreak investigation might also have been more timely if there was an automatic outbreak detection system that could have linked the 21 mothers and infants who had been discharged before the start of the outbreak investigation. As no such system is in place, these cases were first identified during a retrospective review of the LIVA database in early May 2016. SSI has laid out plans for a national, real-time, electronic surveillance system based on data from MiBa, a nationwide, automatically updated database containing all clinical microbiology and strain typing results from hospitals, general practitioners and national reference laboratories [26], and MiBAlert, a warning system that generates an alert in the electronic medical records of individuals carrying multidrug-resistant bacteria [27]. This planned surveillance system will be a valuable tool for rapid outbreak detection across healthcare sectors and will overcome the obstacles involved in detecting cases outside hospital settings.
Conclusions
This study describes the investigation of an unusual hospital outbreak caused by the PVL-positive human variant of MRSA CC398 of south-eastern Asian origin. The use of MALDI-TOF MS as a first-line tool for subgrouping of MRSA isolates allowed accurate case detection and identification of the MRSA CC398 outbreak. Whole genome sequencing facilitated detection of transmission pathways within and outside the hospital, and is now used routinely during outbreak investigations in Denmark. The data presented here and previous findings suggest that international travellers returning from high-prevalence countries could be an important source of PVL-positive community-associated MRSA. Identification and screening of individuals with a history of travel-related SSTI, particularly HCWs, may therefore be taken into consideration during the next evaluation of the Danish MRSA guidelines.
Acknowledgements
We would like to thank the staff in the maternity and neonatal units at Kolding Hospital, the infection control team at Vejle Hospital and the staff at the National Reference Laboratory for Antimicrobial Resistance for their invaluable contribution to the outbreak investigation.
Conflict of interest: None declared.
Authors’ contributions: All authors fulfil ICMJE authorship criteria. JKM, CØ and MAK were members of the local infection control team, initiated the outbreak investigation and performed epidemiological investigations as well as initial genotypic and phenotypic characterisation of the MRSA isolates. ARL, CHM and JL were members of the National Reference Laboratory for Antimicrobial Resistance, analysed national surveillance data, and performed extended genotypic characterisation of the MRSA isolates. JL conducted whole genome sequencing and phylogenetic analysis of the isolates. JKM and JL prepared the initial manuscript. CØ, MAK, ARL and CHM contributed to the subsequent editorial revisions. All authors proofread the article.
References
- 1. Cassini A, Högberg LD, Plachouras D, Quattrocchi A, Hoxha A, Simonsen GS, et al. Attributable deaths and disability-adjusted life-years caused by infections with antibiotic-resistant bacteria in the EU and the European Economic Area in 2015: a population-level modelling analysis. Lancet Infect Dis. 2019;19(1):56-66. 10.1016/S1473-3099(18)30605-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.European Centre for Disease Prevention and Control (ECDC). Surveillance of antibiotic resistance in Europe 2016. Annual Report of the European Antimicrobial Resistance Surveillance Network (EARS-Net). Stockholm: ECDC; 2017. Available from: https://ecdc.europa.eu/sites/portal/files/documents/AMR-surveillance-Europe-2016.pdf
- 3.Danish Integrated Antimicrobial Resistance Monitoring and Research Programme. DANMAP 2016 - Use of antimicrobial agents and occurrence of antimicrobial resistance in bacteria from food animals, food and humans in Denmark. Copenhagen: Statens Serum Institut, National Veterinary Institute and National Food Institute, Technical University of Denmark; October 2017. [Accessed 15 Nov 2018]. Available from: https://www.danmap.org/-/media/arkiv/projekt-sites/danmap/danmap-reports/danmap-2016/danmap_2016_web.pdf?la=en
- 4. Böcher S, Skov RL, Knudsen MA, Guardabassi L, Mølbak K, Schouenborg P, et al. The search and destroy strategy prevents spread and long-term carriage of methicillin-resistant Staphylococcus aureus: results from the follow-up screening of a large ST22 (E-MRSA 15) outbreak in Denmark. Clin Microbiol Infect. 2010;16(9):1427-34. 10.1111/j.1469-0691.2010.03137.x [DOI] [PubMed] [Google Scholar]
- 5.Danish Health Authority. Guidance on preventing the spread of MRSA. 3rd edition. Copenhagen: Danish Health Authority; December 2016. [Accessed 15 Nov 2018]. Available from: https://www.sst.dk/da/sygdom-og-behandling/smitsomme-sygdomme/mrsa/~/media/F3F52EC1C6A94C6080F50F435DA02E59.ashx
- 6. Østergaard C, Møller JK. Subdivision of MRSA CC398 isolates using MALDI-TOF MS. Int J Med Microbiol. 2018;308(4):476-85. 10.1016/j.ijmm.2018.04.001 [DOI] [PubMed] [Google Scholar]
- 7. Larsen J, Petersen A, Sørum M, Stegger M, van Alphen L, Valentiner-Branth P, et al. Meticillin-resistant Staphylococcus aureus CC398 is an increasing cause of disease in people with no livestock contact in Denmark, 1999 to 2011. Euro Surveill. 2015;20(37):30021. 10.2807/1560-7917.ES.2015.20.37.30021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.European Committee on Antimicrobial Susceptibility Testing (EUCAST). Breakpoint tables for interpretation of MICs and zone diameters. Version 8.1. Växjö: EUCAST; May 2018. [Accessed 15 Nov 2018]. Available from: http://www.eucast.org/fileadmin/src/media/PDFs/EUCAST_files/Breakpoint_tables/v_8.1_Breakpoint_Tables.pdf
- 9. Østergaard C, Hansen SG, Møller JK. Rapid first-line discrimination of methicillin resistant Staphylococcus aureus strains using MALDI-TOF MS. Int J Med Microbiol. 2015;305(8):838-47. 10.1016/j.ijmm.2015.08.002 [DOI] [PubMed] [Google Scholar]
- 10. Larsen AR, Stegger M, Sørum M. spa typing directly from a mecA, spa and pvl multiplex PCR assay-a cost-effective improvement for methicillin-resistant Staphylococcus aureus surveillance. Clin Microbiol Infect. 2008;14(6):611-4. 10.1111/j.1469-0691.2008.01995.x [DOI] [PubMed] [Google Scholar]
- 11. Stegger M, Liu CM, Larsen J, Soldanova K, Aziz M, Contente-Cuomo T, et al. Rapid differentiation between livestock-associated and livestock-independent Staphylococcus aureus CC398 clades. PLoS One. 2013;8(11):e79645. 10.1371/journal.pone.0079645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Yu F, Chen Z, Liu C, Zhang X, Lin X, Chi S, et al. Prevalence of Staphylococcus aureus carrying Panton-Valentine leukocidin genes among isolates from hospitalised patients in China. Clin Microbiol Infect. 2008;14(4):381-4. 10.1111/j.1469-0691.2007.01927.x [DOI] [PubMed] [Google Scholar]
- 13. Welinder-Olsson C, Florén-Johansson K, Larsson L, Oberg S, Karlsson L, Ahrén C. Infection with Panton-Valentine leukocidin-positive methicillin-resistant Staphylococcus aureus t034. Emerg Infect Dis. 2008;14(8):1271-2. 10.3201/eid1408.071427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Nurjadi D, Fleck R, Lindner A, Schäfer J, Gertler M, Mueller A, et al. Import of community-associated, methicillin-resistant Staphylococcus aureus to Europe through skin and soft-tissue infection in intercontinental travellers, 2011-2016. Clin Microbiol Infect. 2019;25(6):739-46. 10.1016/j.cmi.2018.09.023 [DOI] [PubMed] [Google Scholar]
- 15. Boyle-Vavra S, Ereshefsky B, Wang CC, Daum RS. Successful multiresistant community-associated methicillin-resistant Staphylococcus aureus lineage from Taipei, Taiwan, that carries either the novel Staphylococcal chromosome cassette mec (SCCmec) type VT or SCCmec type IV. J Clin Microbiol. 2005;43(9):4719-30. 10.1128/JCM.43.9.4719-4730.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Chuang YY, Huang YC. Molecular epidemiology of community-associated meticillin-resistant Staphylococcus aureus in Asia. Lancet Infect Dis. 2013;13(8):698-708. 10.1016/S1473-3099(13)70136-1 [DOI] [PubMed] [Google Scholar]
- 17.Statistics Denmark. IND05: Admissions, bed-days and hospital patients by region, number of bed-days, age and sex. Copenhagen: Statistics Denmark; 2018. [Accessed 15 Nov 2018]. Available from: https://www.statbank.dk/IND05.
- 18. Zanger P, Nurjadi D, Schleucher R, Scherbaum H, Wolz C, Kremsner PG, et al. Import and spread of Panton-Valentine Leukocidin-positive Staphylococcus aureus through nasal carriage and skin infections in travelers returning from the tropics and subtropics. Clin Infect Dis. 2012;54(4):483-92. 10.1093/cid/cir822 [DOI] [PubMed] [Google Scholar]
- 19. Nurjadi D, Friedrich-Jänicke B, Schäfer J, Van Genderen PJ, Goorhuis A, Perignon A, et al. Skin and soft tissue infections in intercontinental travellers and the import of multi-resistant Staphylococcus aureus to Europe. Clin Microbiol Infect. 2015;21(6):567.e1-10. 10.1016/j.cmi.2015.01.016 [DOI] [PubMed] [Google Scholar]
- 20. Köck R, Becker K, Cookson B, van Gemert-Pijnen JE, Harbarth S, Kluytmans J, et al. Methicillin-resistant Staphylococcus aureus (MRSA): burden of disease and control challenges in Europe. Euro Surveill. 2010;15(41):19688. 10.2807/ese.15.41.19688-en [DOI] [PubMed] [Google Scholar]
- 21. David MZ, Daum RS. Community-associated methicillin-resistant Staphylococcus aureus: epidemiology and clinical consequences of an emerging epidemic. Clin Microbiol Rev. 2010;23(3):616-87. 10.1128/CMR.00081-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Saeed K, Gould I, Esposito S, Ahmad-Saeed N, Ahmed SS, Alp E, et al. Panton-Valentine leukocidin-positive Staphylococcus aureus: a position statement from the International Society of Chemotherapy. Int J Antimicrob Agents. 2018;51(1):16-25. 10.1016/j.ijantimicag.2017.11.002 [DOI] [PubMed] [Google Scholar]
- 23. Orendi JM, Coetzee N, Ellington MJ, Boakes E, Cookson BD, Hardy KJ, et al. Community and nosocomial transmission of Panton-Valentine leucocidin-positive community-associated meticillin-resistant Staphylococcus aureus: implications for healthcare. J Hosp Infect. 2010;75(4):258-64. 10.1016/j.jhin.2010.03.023 [DOI] [PubMed] [Google Scholar]
- 24. Ali H, Nash JQ, Kearns AM, Pichon B, Vasu V, Nixon Z, et al. Outbreak of a South West Pacific clone Panton-Valentine leucocidin-positive meticillin-resistant Staphylococcus aureus infection in a UK neonatal intensive care unit. J Hosp Infect. 2012;80(4):293-8. 10.1016/j.jhin.2011.12.019 [DOI] [PubMed] [Google Scholar]
- 25. Shore AC, Tecklenborg SC, Brennan GI, Ehricht R, Monecke S, Coleman DC. Panton-Valentine leukocidin-positive Staphylococcus aureus in Ireland from 2002 to 2011: 21 clones, frequent importation of clones, temporal shifts of predominant methicillin-resistant S. aureus clones, and increasing multiresistance. J Clin Microbiol. 2014;52(3):859-70. 10.1128/JCM.02799-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Voldstedlund M, Haarh M, Mølbak K, the MiBa Board of Representatives C, MiBa Board of Representatives The Danish Microbiology Database (MiBa) 2010 to 2013. Euro Surveill. 2014;19(1):20667. 10.2807/1560-7917.ES2014.19.1.20667 [DOI] [PubMed] [Google Scholar]
- 27. Olesen B, Anhøj J, Rasmussen KP, Mølbak K, Voldstedlund M. MiBAlert-a new information tool to fight multidrug-resistant bacteria in the hospital setting. Int J Med Inform. 2016;95:43-8. 10.1016/j.ijmedinf.2016.09.001 [DOI] [PubMed] [Google Scholar]