Abstract
Curcumin is a natural phytochemical with potent anti-neoplastic properties including modulation of p53. Targeting p53 activity has been suggested as an important strategy in cancer therapy. The purpose of this study was to describe a mechanism by which curcumin restores p53 levels in human cancer cell lines.
HeLa, SiHa, CaSki and MDA-MB-231 cells were exposed to curcumin and a pulse and chase and immunoprecipitation assays were performed. Here we showed that curcumin increases the half-life of p53 by a physical interaction between p53-NQO1 (p53 - NAD(P)H:quinone oxidoreductase 1) proteins after treatment with curcumin. Interestingly, the cell viability assay after treatment with curcumin showed that the cytotoxic activity was selectively higher in cervical cancer cells contained wild type p53 but not in breast cancer cells contained mutated p53. The cytotoxic effect of curcumin in cervical cancer cells was related to the complex p53-NQO1 that avoids the interaction between p53 and its negative regulator ubiquitin ligase E6-associated protein (E6AP). Finally, we demonstrated that in pancreatic epithelioid carcinoma cells (PANC1) that are knockout for NQO1, the reestablishment of NQO1 expression can stabilize p53 in presence of curcumin. Collectively, our findings showed that curcumin is necessary to promote the protein interaction of NQO1 with p53, therefore, it increases the half-life of p53, and permits the cytotoxic effect of curcumin in cancer cells containing wild type p53. Our findings suggest that the use of curcumin may reactivate the p53 pathway in cancer cells with p53 wild-type.
Keywords: P53, Curcumin, NQO1, E6AP, Tumour cell lines
Graphical abstract
Highlights
-
•
Curcumin increases the levels of NQO1, p53 and p21 in tumor cell lines.
-
•
Treatment with curcumin augments the levels of P53 in tumor cell lines through incrementing its half-life in a NQO1 dependent manner.
-
•
Curcumin treatment promotes the interaction between NQO1-p53.
-
•
Curcumin promotes loss of interaction between p53 and E6AP.
1. Introduction
Cancer is a diverse group of diseases characterized by abnormal cells growth and represents an important worldwide problem. Therefore, the search of therapeutic alternatives against tumoral cells has been a major challenge for scientific and commercial interest in the discovery of potent, safe and selective anti-cancer drugs [1]. Among them, antioxidants are molecules with therapeutic potential since they have the ability to neutralize reactive oxygen species but also have been attributed antitumor properties [2,3]. An example of these natural compounds with therapeutic potential is curcumin, a natural phenolic compound obtained from the roots of Curcuma longa. Structurally curcumin is a molecule with polar central and flanking regions separated by a lipophilic methionine segment. Also, it has distinct chemical properties, among these the presence of α,β–unsaturated compounds (Michael acceptor) that facilitate intermolecular interactions with another molecules [4,5]. The anti-tumor activities of curcumin have been demonstrated by some research groups [6,7]. Particularly, it has been shown that it can induce cyclin-dependent kinase inhibitors, therefore this promotes p53 restoration in wild-type and mutant p53 cancer cell lines and represses the growth of numerous cancer cell lines by the phosphatidylinositol 3-kinase pathway (PI3K, AKT), Ras, and β-catenin pathways [8]. Curcumin is also a powerful antioxidant because it activates the Kelch-like ECH-associated protein 1-nuclear factor (erythroid-derived 2)-like 2 pathway (Keap1/Nrf2) under oxidative microenvironment [9]. The Nrf2 pathway is regulated by Keap1 by mediating its degradation, but when cells are exposed to electrophiles or oxidants, Nrf2 is stabilized and translocate into the nucleus, binding to the antioxidant response element (ARE) located in the promoter region of antioxidant genes and upregulates their transcription. Nrf2 functions as a transcriptional factor with multiple targets as NAD(P)H: quinone oxidoreductase 1 (NQO1) [10,11]. This protein has multiple functions including neutralization of reactive oxygen species, detoxification of quinones and stabilization proteins for example; there is a report showing that NQO1 can interact with p53 causing its stabilization [12,13]. p53 is a tumour suppressor gene product that can block cell progression through the cell cycle when deoxyribonucleic acid (DNA) is damaged [14]. It is localized into nucleus, where it functions as transcription factor of DNA sequence-specific in genes as p21. The activation of p53 can activate different gene pathways, resulting in apoptosis, cell-cycle arrest, DNA repair or senescence, among others [15]. Because of its biological importance, mutations in the TP53 tumour suppressor gene are observed in greater than 50% of all human cancers. The vast majority of p53 mutations that are associated with human cancer occur at the region of DNA binding recognition [16]. Moreover, mutant p53 in human cancer is commonly expressed at high levels and is more stable than wild-type p53 [17].
Here, we investigated the mechanism of the activation of p53 mediated by curcumin. We showed that curcumin promotes the complex formation of NQO1-p53 leading to p53 stabilization [18]. High levels of NQO1 are not enough for the p53 stabilization; we demonstrated that the presence of curcumin is necessary to stabilize the p53-NQO1 interaction. Also, this interaction can promote the loss interactions between p53 and its negative regulators. The effect of curcumin on p53 levels is differential between the cancer cell lines because it only has effect on cell viability of HeLa, SiHa and CaSki but not in MDA-MB-231. So curcumin is a molecule with an important therapeutic potential in cancer cells with p53 wild type.
2. Materials and methods
2.1. Chemicals and reagents
3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT), dicumarol, cycloheximide (CHX), dimethyl sulfoxide (DMSO), curcumin (C1386), protease inhibitor cocktail tablets EDTA-free (S8830), protein G sepharose (GE28), Trizma base (T1503), sodium chloride (NaCl S9888) were purchased from Sigma-Aldrich (St. Louis, MO, USA). Pierce BCA Protein Assay Kit (23225) and lipofectamine plus transfection reagent (15338100) were from ThermoFisher (Waltham, MA, USA) Nonidet P-40 (CAS 68412-54-4), anti-p53 mouse monoclonal antibody (DO-1), anti-NQO1 mouse monoclonal antibody (H9), anti-E6AP (E4) mouse monoclonal antibody, anti-lamin A/C (2A1) mouse monoclonal antibody, anti-glyceraldehyde-3-phosphate dehydrogenase (GAPDH, L8) goat polyclonal antibody, donkey anti-goat IgG-HRP (sc-2020), and goat anti-mouse IgG-horse radish peroxidase (HRP, sc-2005) were purchased from Santa Cruz Biotechnology (Dallas, TX, USA). Dulbecco's Modified, Eagle Medium high glucose (DMEM 11965–084) and fetal bovine serum (10500056) were from GIBCO.
2.2. Cell lines and culture
Cell lines HeLa, SiHa and CaSki were cultured in DMEM supplemented with 10% fetal bovine serum. MDAMB-231 cells were cultured in Dulbecco's Modified Eagle Medium Nutrient Mixture (DMEM, GIBCO, 11320–033) supplemented with 10% fetal bovine serum. All cell lines were cultured at 37°C in a 5% CO2 incubator.
2.3. Western blot
The cells samples lysates were extracted with lysis buffer composed of 50 mM Tris, pH 7.6, 150 mM NaCl, 1% Nonidet P-40, 10 mM sodium phosphate, and a complete tablet protease Inhibitor Cocktail per 100 ml of buffer, and the protein concentration in the lysates was quantified using an enhanced bicinchoninic acid protein assay kit with bovine serum albumin as a standard. The total protein extract will be used for western blot analysis. Equal amounts of total protein were subjected to 10% sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) and transferred into a nitrocellulose membrane, followed by incubation overnight to 4°C using the following dilution of primary antibodies: anti-p53 (1:100), anti NQO1 (1:1000), anti-MDM2 (1:500), anti-E6AP (1:1000), anti-lamin A/C (1:500), anti-GAPDH (1:1000) and following by incubation with secondary antibody in blocking solution 1 h room temperature; anti-mouse (1:10000), anti-goat (1:20000) finally protein expression levels were visualized with Li-COR C-DiGit chemiluminescence western blot scanner and UVP Imaging system.
2.4. Pulse and chase assays
The cells were seeds in p35 plates at density of 1.5 × 105 cells/plate and treated with curcumin at concentration of 20 μM for 24 h, the treatment with curcumin was removed and the cells were washed with PBS, continuing with the treatment with CHX with a concentration of 50 μg/ml as previously reported [19,20], the CHX treatment is a standard protein synthesis inhibitor and aliquots of cells were collected every then minutes starting on 0 min, 10 min, 20 min so on until 60 min immediately following addition of the compound cells were lysed with lysis buffer composed of 50 mM Tris, pH 7.6, 150 mM NaCl, 1% Nonidet P-40, 10 mM sodium phosphate, and a complete tablet protease Inhibitor Cocktail per 100 ml of buffer, p53 protein abundance at each time point was analyzed, by western blot as described.
2.5. Immunoprecipitation assays
The cells were seeded in p60 plates at density of 2.5 × 105 cells/plate and treated with 20 μM curcumin for 24 h and then lysed, after centrifugation, the clear cell lysate was separated from the pellet of cell and then incubated 2 h at 4°C with 50 μl Protein G sepharose, 1 μg of antibodies and 0.5 mg/ml RNAse A, after incubation the sepharose beads coupled to protein G were washed 20 times with lysis buffer. For western blot, the resulting immunoprecipitates were resolved by SDS-PAGE, then the gel contents were transferred to a nitrocellulose membrane and probed with specific antibodies, finally protein expression levels were visualized with Li-COR C-DiGit chemiluminescence western blot scanner (LI-COR Biosciences, Lincoln, Nebraska USA).
2.6. Cell viability assays
Cell viability was determined by the MTT (M2128) assay [21]. HeLa, SiHa, CaSki and MDA-MB-231 cells (5 × 104 cells/well) were seeded in a 96-well plate and treated with 10 μM and 20 μM curcumin (C1386) for 24 h. After addition 10 μl per well MTT (5 mg/ml) solution the cells were incubated at 37°C for 4 h, the formazan crystals were dissolved using 50 μl DMSO [41]. The cell viability was determined by measuring the absorbance at 570 nm on a Synergy H1 Hybrid Multi-Mode Microplate Reader (BioTek, Winooski, VT, USA).
2.7. Transfection of PANC1 cells with NQO1 plasmids
PANC1 cells (Null to NQO1 were obtained from American Type Culture Collection (ATCC CRL-1469) were transient transfected using Lipofectamine plus transfection reagent with 1 μg pCDNA3 NQO1 or 1 μg pCDNA3 (Empty Vector) the plasmids were obtained from addgene nonprofit plasmid repository. The results were obtained from three separate biological replicates. The cells transfected and PANC1 wild type (non transfected) were treated with curcumin, pulse and chase assay was performed. The expression levels of p53 protein were analyzed by western blot as described above.
2.8. Statistical analysis
Results are expressed as mean ± SD. Statistical tests were performed using GraphPad PRISM version 6.0c. The ANOVA test with Tukey was applied to compare the means of groups, a p < 0.05 was significant.
3. Results
3.1. Curcumin treatment increases the levels of NQO1, p53 and p21
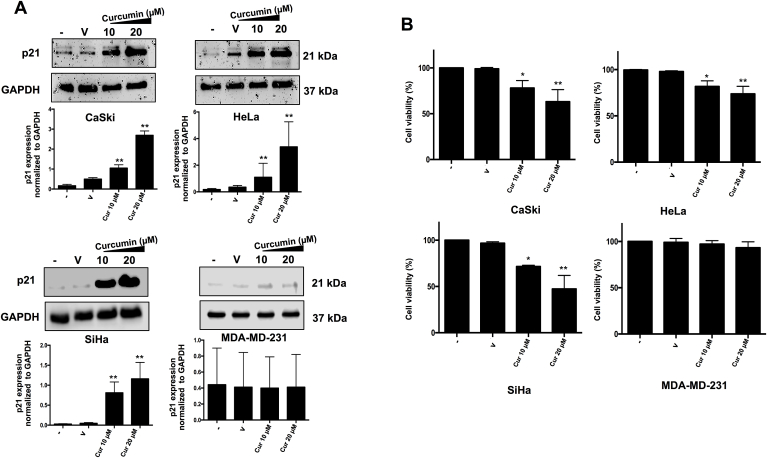
Previous studies have shown that curcumin is capable to activate the Keap1/Nrf2 pathway in order to increase the expression of phase II enzymes including NQO1 [22]. To determine the participation of curcumin on the Keap1/Nrf2 in our model, we evaluate the activation of the Keap1/Nrf2 pathway through the translocation of Nrf2 to the nucleus in Hela, CasKi and SiHa cervical cancer cells lines and MDA-MB-231 breast cancer cells in response to curcumin treatment. We confirmed the immunodetection of Nrf2 in the nuclear extract after 20 μM of curcumin treatment in all cell lines (data not shown) and observed the overexpression of NQO1 (Fig. 1A). Therefore, we determine that curcumin induces the nuclear translocation of Nrf2, an event that indicates the activation of the Keap1/Nrf2 pathway and the increased levels of NQO1 that is consistent with the Nrf2 nuclear translocation. Then, we observed that curcumin treatment also increases the p53 levels (Fig. 1B). All together, these results indicate that curcumin may have an important role in the activating the p53 pathway. To evaluate the functionality of p53 pathway in response to curcumin treatment the expression of its target p21 was evaluated. We showed that curcumin increases the expression of p21 in cervical cancer cell lines with p53 wild-type genotype (CaSki, HeLa and SiHa) meanwhile, this effect was not observed in MDA-MB-231 cell line that has mutated p53 (Fig. 2A) which has decreased p53 activity. These results indicate that curcumin activates the Keap1/Nrf2 pathway in our cellular model with wild-type p53 and leads to the activation of p53 pathway.
Fig. 1.
Effect of curcumin (Cur) on NQO1, p53 levels in CaSki, HeLa, SiHa and MDA-MB-231 cells. CaSki, HeLa, SiHa and MDA-MB-231 cells were treated with 10 μM and 20 μM of curcumin and (A) NAD(P)H: quinone oxidoreductase 1 (NQO1, 31 kDa) and (B) p53 were detected by immunoblotting. The cells were treated for 24 h. The values are means ± SD of three independent experiments. The negative sign (−) represents without treatment, the V represents Vehicle. Statistical differences in A and B were determined using one-way ANOVA and Tukey's multiple comparison test; (*) p < 0.005, (**) p < 0.001 (−) vs V, (−) vs 10 μM of curcumin and (−) vs 20 μM of curcumin.
Fig. 2.
Effect of curcumin (Cur) on p21 levels and cell viability in CaSki, HeLa, SiHa and MDA-MB-231 cells. CaSki, HeLa, SiHa and MDA-MB-231 cells were treated for 24 h with 10 μM and 20 μM of curcumin and (A) p21 (21 kDa) levels were detected by immunoblotting and (B) Cell viability was evaluated using MTT. The values are means ± SD of three independent experiments. The negative sign (−) represents without treatment, the V represents Vehicle. Statistical differences in A and B were determined using one-way ANOVA and Tukey's multiple comparison test; (*) p < 0.005, (**) p < 0.001 (−) vs V, (−) vs 10 μM of curcumin and (−) vs 20 μM of curcumin.
In order to evaluate the effect of curcumin on the cell viability a MTT assay was performed. Cells were treated with 10 μM and 20 μM of curcumin; non-treated cells (−) and cells treated with vehicle (V) were used as control. Curcumin treatment induces a significant decrease on the cell viability was observed at the p53 wild type cervical cancer cell lines. Interestingly, at the same concentrations, MDA-MB-231 cells with p53 mutated were resistant to the cytotoxic effect of curcumin. These results suggest that the cytotoxic effect is related with the activation of the p53 wild type (Fig. 2B).
3.2. Curcumin increases the half life time of p53
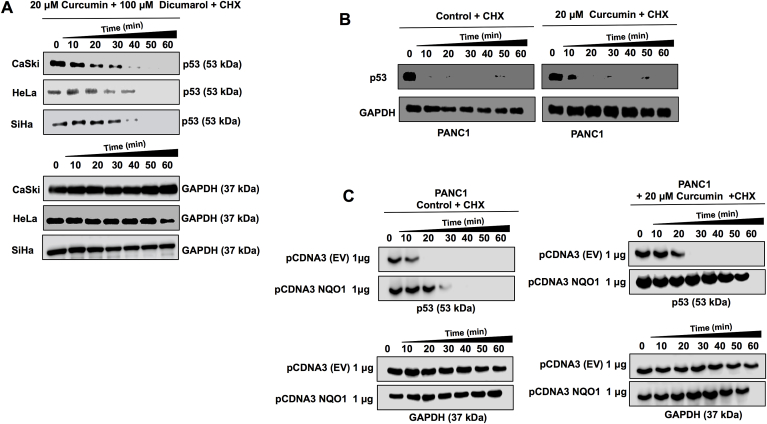
It is widely reported that the half-life time of p53 is 20 min in physiological conditions [23]. In order to evaluate the effect of curcumin on the half-life of p53 we analyzed its stability after treatments with curcumin or cycloheximide, a well-known inhibitor of protein synthesis [24]. It was found that p53 stability of HeLa, SiHa, CaSki and MDA-MB-231 cells without treatment with curcumin was 20 min (Fig. 3A), however when cells were treated with 20 μM curcumin, p53 protein was detected until 60 min (Fig. 3B). These results suggest that curcumin is able to enhance the p53 half-life of tumor cells.
Fig. 3.
Curcumin stabilizes p53. CaSki, HeLa, SiHa and MDA-MB-231 cells were treated with 20 μM of curcumin for 24 h and the half-life of p53 (53 kDa) was measured with a pulse and chase experiment with cycloheximide (50 μg/ml), the protein extraction was performed every 10 min (0–60) and revealed by immunoblot, the stability of p53 was evaluated in (A) control cells (without curcumin) and in (B) cells treated with curcumin.
3.3. Curcumin promotes the interaction between p53 and NQO1
The NQO1 interaction with p53 under stress oxidative conditions has also been observed [25,26]. In order to evaluate whether curcumin promotes the physical interaction of p53 with NQO1, immunoprecipitation assays were performed. In these assays, p53 was inmunoprecipitated and then NQO1 was revealed by western blot assay. Non-treated cells were used as control. The input material was evaluated against p53 and NQO1 in increasing amounts of protein (2.5 and 5%). It was found that NQO1 physically interact with p53 when HeLa, SiHa and CaSki cells were treated with curcumin (Fig. 4A). These results suggested that NQO1-p53 complex could be stabilizing p53 in response to curcumin treatment for altered cell viability.
Fig. 4.
Curcumin promotes interaction between p53 and NQO1. CaSki, HeLa and SiHa cells were treated with 20 μM curcumin for 24 h and NQO1 (31 kDa) was detected by immunoblot analysis following immunoprecipitation of p53 (53 kDa) from cell lysates. The input material was evaluated against p53 and NQO1 in increasing amounts of protein (2.5 and 5%). (A) The p53-NQO1 interaction after treatment with curcumin was evaluated. Cell lines were treated for 24 h with 20 μM curcumin and p53 (53 kDa) was detected by immunoblot analysis following immunoprecipitation of E6AP (100 kDa). The effect of curcumin in the interaction between E6AP-p53 is shown. (B) The input material was evaluated against p53 and E6AP in increasing amounts of protein (2.5 and 5%).
In order to show that the effect of curcumin could avoid the interaction between p53 and its negative regulators ubiquitin ligase E6-associated protein (E6AP) a immunoprecipitation assay was performed (Fig. 4B). In order to address this question cells with wild type p53 were used for this purpose because we did not observe any effect on cell viability in MDA-MB-231. To evaluate the change in the direct interaction between E6AP and p53 in response to curcumin treatment, E6AP protein was inmunoprecipitated and then p53 was revealed by western blot assay. Non-treated cells were used as control. The input material was evaluated against p53 and E6AP in increasing amounts of protein (2.5 and 5%). Results showed that curcumin treatment induces loss of interaction between p53 and E6AP (Fig. 4B). All together, our results showed that curcumin induces loss in the E6AP-p53 interaction and favors interaction of NQO1-p53 protein complex to promote cytotoxicity on the cervical cancer cells with p53 wild type.
3.4. Curcumin is necessary for complex NQO1-p53 and p53 stabilization
To determine whether the enzymatic activity of NQO1 is required for p53 stability when curcumin is present, the cells were treated with curcumin and after they were treated 4 h with 100 μM of dicumarol (a NQO1 activity inhibitor). To evaluate p53 stability a pulse and chase assay was performed. Results showed that p53 half-life time was less than 60 min when the cells were treated with dicumarol (Fig. 5A). This suggests that stabilization of p53 by NQO1 depends on an intact enzymatic activity of NQO1. In order to demonstrate if NQO1 is responsible for p53 stability, we used pancreatic epithelioid carcinoma cells (PANC1), a cell line null to NQO1 gene [27,28]. We first treated wild type cells PANC1 (non-transfected) with curcumin and interestingly, observed no stabilization of p53 (Fig. 5B), suggesting that the presence of curcumin is not sufficient to increase the half-life of p53. NQO1 expression was reestablished by exogenous expression of NQO1 messenger cloned in a plasmid (PCDNA-NQO1). After 48 h of transfection, pulse and chase assay was performed in PANC1 transfected cells, but without curcumin treatment, and again the stabilization of p53 was not observed (Fig. 5C) even when NQO1 protein was expressed, suggesting that the presence of NQO1 is not enough to increase the half-life of p53. Surprisingly, when transfected cells were then treated with 20 μM curcumin, stabilization of p53 was indeed observed (Fig. 5C). This result clearly showed that curcumin is necessary to promote the stabilization of p53, via interaction with the NQO1 protein.
Fig. 5.
Curcumin increases p53 stability in a NQO1 dependent manner. Pulse and chase analysis were performed in cell lines treated for 4 h with 20 μM curcumin 24 h and 100 μM dicumarol (NQO1 Inhibitor). (A) the effect of the inhibition of NQO1 activity by dicumarol on p53 stability is shown. Pancreatic carcinoma cells null to NQO1 (PANC1 cells) wild type and transfected with 1 μg pCDNA3 EV (empty vector) or with 1 μg of pCDNA3-NQO1 (to express the complete NQO1 protein) were treated with 20 μM for 24 h and after pulse and chase of p53 (53 kDa) was performed. The chase of proteins was performed every 10 min and revealed by immunoblot, the effect of treatment with curcumin on p53 stability in (B) absence of NQO1 (PANC1 WT or PANC1+ pCDNA3) and when (C) NQO1 levels are restored is shown. CHX = cycloheximide.
4. Discussion
Tumor suppressor p53 plays a central role in protecting the genome and preventing cell transformation [29]. However, normal cells must maintain p53 levels under tight control to prevent death. Despite mutations in the p53 gene occurring in 50% of all cancers, cervical cancer cells as HeLa, SiHa and CaSki cells retain functional wild-type p53 [30,31] while, MDA-MB-231 cells has a loss-of-function mutation in TP53 gene [32]. Therefore, cancer cell lines with wild type p53 can reactivate downstream pathways and consequently promote cell death and decrease cell viability [33]. Hence, p53 restoration may be a promising therapeutic strategy, however, stability of p53 is an important factor for the reactivation pathway. The main negative regulator of p53 is MDM2, however in HPV-positive cancer cell lines like HeLa, SiHa and CaSki, viral oncoprotein E6 forms a complex with E6AP and their physical interaction with wild-type p53 promotes its degradation via the ubiquitin pathway [34]. The half-life of the p53 protein is short (20 min); during periods of cellular stress, the p53 protein is regulated by a negative feedback mechanism. Here we show that curcumin increases the half-life of p53 up to 60 min and present data on the role of NQO1 in protecting p53 from degradation which are consistent with data previously reported [35]. However, in this work we demonstrated that curcumin is necessary to promote NQO1-mediated stabilization of p53. NQO1 plays an important role in this stabilization because it can physically interact with p53. NQO1 is even considered an anticancer enzyme since it protects cells from oxidative stresses [36]. Thus, the use of dietary compounds to induce the expression of NQO1 has emerged as a promising strategy for cancer prevention [37]. Compounds as curcumin had the ability to translocate Nrf2 which in turn bind to ARE in the promoter region of antioxidant enzymes and increase the expression of phase II antioxidant proteins such as NQO1 [38,39]. The increase in NQO1 levels is one of the results of the translocation of Nrf2 to the nucleus induced by curcumin. NQO1 is a protein that can form complexes with other proteins to stabilize them, including p53.
In this report, we propose that curcumin is necessary to induce NQO1 and promote its interaction with p53, which in turn results in an increase of the p53 stability, contrary to previous reports [40,41]. In 2005, Tsvetkov et al. showed that curcumin and dicoumarol destabilized p53 in M1 mouse myeloid leukemic cells transfected with a plasmid containing a temperature-sensitive p53 [135 Ala → Val] (M1-t-p53). M1-t-p53 protein has a wild-type conformation and activity at 32°C, but not at 37°C. In addition, they used mouse A31N-ts20 cells which had a temperature-sensitive E1 ubiquitin activating enzyme, that is inactivated at 39°C and causes accumulation of p53. They observed degradation of M1-t-p53 by low doses of curcumin (20 μM) and dicumarol (100 μM) in mouse cells growth at two different temperature conditions (39 °C or 32 °C). However, the same authors showed degradation of WT p53 in the p53-null cell line HCT116 from human colorectal cancer cells [40], In that report, cells were transfected with human WT p53 and treated with high doses of curcumin (60 μM) and dicumarol (300 μM) [40]. In our report we used cell lines derived from human cervical cancer; HeLa, SiHa, and CaSki. These cell lines exhibit a low basal wild-type p53 expression due to the presence of the Human Papillomavirus (HPV) E6 oncoprotein, therefore, in this case the mechanism for degradation of P53 is dependent on the formation of a complex between E6 and the E6AP protein. Finally, it is important to emphasize that we used low doses of curcumin (20 μM) and dicumarol (100 μM).
On the other hand, Zeekpudsa et al. in 2014 reported an increased expression of p53 when NQO1 activity is inhibited by dicoumarol. The authors used two Opisthorchis viverrini-related cholangiocarcinoma cell lines (KKU-100 and KKU-M214) with high and low NQO1 expression levels, respectively [41]. To decrease NQO1 levels, the authors used a specific NQO1 siRNA. On the contrary, in our work we used human cervical cancer cell lines with wild-type p53 and importantly, we use curcumin, which plays a central role in directing the interaction between NQO1 and p53 to promote its stabilization. It is worth to mention that in the report of Zeekpudsa et al., curcumin was not used. Our results indeed demonstrate that curcumin is necessary to promote the maintenance of an environment in which p53 and NQO1 can interact, and therefore increase the half-life of p53.
Finally, our data also showed that the physical interaction between NQO1 and p53 promote its loss of interaction with its negative regulator E6AP. Moreover, NQO1 exhibits catalytic enzyme properties, first reported by Ernster and Navazio in 1958 [42], that we here shown necessary for the stabilization of p53, since dicumarol, an inhibitor of NQO1 activity, inhibits the stabilization of p53, consistent with the previously reported [43,44].
We here propose a model in which p53 stability is determined by two effectors acting together: curcumin and NQO1 (Fig. 6). Curcumin activates the Keap1/Nrf2 pathway and promote the increase of NQO1 levels and, in the presence of curcumin, NQO1 can interact physically with p53 and promote its stability. Also, the formation of the NQO1-p53 complex promote the loss of interaction between p53 and its negative regulator E6AP. In cancer cells with wild-type p53, like those positive for HPV, the stabilization of p53 promotes a reactivation of the p53 pathway, and therefore decrease cell viability.
Fig. 6.
Schematic model of effects of curcumin in the p53 stabilization. Curcumin activates the Kelch-like ECH-associated protein 1-nuclear factor (erythroid-derived 2)-like 2 pathway (Keap1/Nrf2) pathway; Nrf2 is translocated into the nucleus and binds to the antioxidant response element (ARE) sequences increasing the expression of NAD(P)H: quinone oxidoreductase 1 (NQO1). NQO1 is translated and binds to p53 promoting the loss of interaction between p53 and its negative regulators. Curcumin also stabilizes the interaction between NQO1-p53. In cells with wild-type p53 (WT p53) the p53 pathway is activated and has an effect on cell viability whereas in cells with mutated p53 (MT p53) there is only accumulation of p53 without effect on cell viability.
5. Conclusions
Our results demonstrate the importance of curcumin in the regulation of p53 stability. In this work we demonstrate that curcumin treatment increases p53 levels and provides an appropriate cellular environment for p53 and NQO1 to interact. At the same time this interaction promotes the loss of interaction of p53 with its negative regulator E6AP, the negative regulator of p53 when HPV E6 oncoproteins are present. In our work, HeLa, SiHa and CaSki tumor-derived cell lines that have wild-type p53 and downstream pathways intact, therefore curcumin may be a potential therapeutic agent for tumors with wild type p53.
Author's contributions
“CCPM, ESR, and AGC conceived the study, designed the experimental strategy, analyzed the results, and drafted the manuscript. CCPM. EAO, and VAV carried out molecular biology studies and performed statistical analysis. EOS and JPC participated in drafting the discussion and helped to revise the manuscript. All authors read and approved the final manuscript.”
Funding
This work was supported by Consejo Nacional de Ciencia y Tecnología (CONACyT No. 253804, to AG-C) and by Fundación Miguel Alemán, A.C. (to AG-C).
Conflicts of interest
The authors declare no conflicts of interests.
Acknowledgments
Carlos César Patiño Morales is a doctoral student from Programa de Doctorado en Ciencias Biomédicas, Universidad Nacional Autónoma de México (UNAM) and received fellowship from CONACYT. CVU 416000. We thank to the Unidad de Investigación Biomédica en Cáncer, Universidad Nacional Autónoma de México-Instituto Nacional de Cancerología (Mexico City) and to the Departamento de Ciencias Naturales, Universidad Autónoma Metropolitana, Unidad Cuajimalpa (Mexico City).
References
- 1.Kanti B.P., Syed I.R. Plant polyphenols as dietary antioxidants in human health and disease. Oxidat. Med. Cell. Longev. 2009;2(5):270–278. doi: 10.4161/oxim.2.5.9498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chikara S., Nagaprashantha L.D., Singhal J., Horne D., Awasthi S., Singhal S.S. Oxidative stress and dietary phytochemicals: role in cancer chemoprevention and treatment. Cancer Lett. 2018;413:122–134. doi: 10.1016/j.canlet.2017.11.002. [DOI] [PubMed] [Google Scholar]
- 3.Houghton C.A., Fassett R.G., Coombes J.S. Sulforaphane and other nutrigenomic Nrf2 activators: can the clinician's expectation Be matched by the reality? Oxidat. Med. Cell. Longev. 2016:785718. doi: 10.1155/2016/7857186. 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Heger M., van Golen R.F., Broekgaarden M., Michel M.C. The molecular basis for the pharmacokinetics and pharmacodynamics of curcumin and its metabolites in relation to cancer. Pharmacol. Rev. 2013;66(1):222–307. doi: 10.1124/pr.110.004044. [DOI] [PubMed] [Google Scholar]
- 5.Nabavi S.F., Nabavi S.M., Daglia M., Tamilselvam R., Bishayee A., Pazoki-toroudi H., Kasi P.D. Molecular targets of curcumin for cancer therapy: an updated review. Tumor Biol. 2016;37(10):13017–13028. doi: 10.1007/s13277-016-5183-y. [DOI] [PubMed] [Google Scholar]
- 6.Devassy J.G., Nwachukwu I.D., Jones P.J.H. Curcumin and cancer: barriers to obtaining a health claim. Nutr. Rev. 2015;73(3):155–165. doi: 10.1093/nutrit/nuu064. [DOI] [PubMed] [Google Scholar]
- 7.Goel A., Kunnumakkara A.B., Aggarwal B.B. Curcumin as “Curecumin”: from kitchen to clinic. Biochem. Pharmacol. 2008;75(4):787–809. doi: 10.1016/j.bcp.2007.08.016. [DOI] [PubMed] [Google Scholar]
- 8.Mishra A., Das B.C. Curcumin as an anti-human papillomavirus and anti-cancer compound. Future Oncol. 2015;11(18):2487–2490. doi: 10.2217/fon.15.166. [DOI] [PubMed] [Google Scholar]
- 9.Prasad S., Gupta S.C., Tyagi A.K., Aggarwal B.B. Curcumin, a component of golden spice: from bedside to bench and back. Biotechnol. Adv. 2014;32(6):1053–1064. doi: 10.1016/j.biotechadv.2014.04.004. [DOI] [PubMed] [Google Scholar]
- 10.Theodore M., Kawai Y., Yang J., Kleshchenko Y., Reddy S.P., Villalta F., Arinze I.J. Multiple nuclear localization signals function in the nuclear import of the transcription factor Nrf2. J. Biol. Chem. 2008;283(14):8984–8994. doi: 10.1074/jbc.M709040200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Richardson B.G., Jain A.D., Speltz T.E., Moore T.W. Non-electrophilic modulators of the canonical Keap1/Nrf2 pathway. Bioorg. Med. Chem. Lett. 2015;25(11):2261–2268. doi: 10.1016/j.bmcl.2015.04.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Stefanson A.L., Bakovic M. Dietary regulation of Keap1/Nrf2/ARE pathway: focus on plant-derived compounds and trace minerals. Nutrients. 2014;6(9):3777–3801. doi: 10.3390/nu6093777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Asher G., Lotem J., Sachs L., Kahana C., Shaul Y. Mdm-2 and ubiquitin-independent p53 proteasomal degradation regulated by NQO1. Cell. 2002;99(20):13125–13130. doi: 10.1073/pnas.202480499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sullivan K.D., Gallant-behm C.L., Henry R.E., Fraikin J., Joaquín M. vol. 1825. 2013. pp. 229–244. (The P53 Circuit Board Kelly). (2) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Blattner C. Regulation of p53: the next generation. Cell Cycle. 2008;7(20):3149–3153. doi: 10.4161/cc.7.20.6921. [DOI] [PubMed] [Google Scholar]
- 16.Haller J., Haller J.S. Analysis of TP53 mutation status in human cancer cell lines: a reassessment bernard. Shadow Medicine. 2015;35(6):153–160. doi: 10.1002/humu.22556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Scheffner M., Munger K., Byrne J.C., Howley P.M. The state of the p53 and retinoblastoma genes in human cervical carcinoma cell lines. Proc. Natl. Acad. Sci. 2006;88(13):5523–5527. doi: 10.1073/pnas.88.13.5523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Siegel D., Anwar A., Tang L.J., Pietenpol J.A., Dehn D., Ross D., Kepa J.K. Interaction of human NAD(P)H:quinone oxidoreductase 1 (NQO1) with the tumor suppressor protein p53 in cells and cell-free systems. J. Biol. Chem. 2003;278(12):10368–10373. doi: 10.1074/jbc.M211981200. [DOI] [PubMed] [Google Scholar]
- 19.Yewdell J.W., Lacsina J.R., Rechsteiner M.C., Nicchitta C.V. Out with the old, in with the new? Comparing methods for measuring protein degradation. Cell Biol. Int. 2011;35(5):457–462. doi: 10.1042/CBI20110055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hochstrasser M. Ubiquitin, proteasomes, and the regulation of intracellular protein degradation. Curr. Opin. Cell Biol. 1995;7(2):215–223. doi: 10.1016/0955-0674(95)80031-x. [DOI] [PubMed] [Google Scholar]
- 21.Langdon S.P. Cell sensivity assays: the MTT assay. Methods Mol. Biol. 2003;731:237–245. doi: 10.1007/978-1-61779-080-5_20. [DOI] [PubMed] [Google Scholar]
- 22.Pulido-Moran M., Moreno-Fernandez J., Ramirez-Tortosa C., Ramirez-Tortosa M.C. Curcumin and health. Molecules. 2016;21(3):1–22. doi: 10.3390/molecules21030264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sullivan K.D., Galbraith M.D., Andrysik Z., Espinosa J.M. Mechanisms of transcriptional regulation by p53. Cell Death Differ. 2018;25(1):133–143. doi: 10.1038/cdd.2017.174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Buchanan B.W., Lloyd M.E., Engle S.M., Rubenstein E.M. Cycloheximide chase analysis of protein degradation in “Saccharomyces cerevisiae”. J. Vis. Exp. 2016;(110):1–9. doi: 10.3791/53975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Das L., Vinayak M. Long term effect of curcumin in restoration of tumour suppressor p53 and phase-II antioxidant enzymes via activation of Nrf2 signalling and modulation of inflammation in prevention of cancer. PLoS One. 2015;10(4):1–22. doi: 10.1371/journal.pone.0124000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kama R., Shaul Y., Lotem J., Sachs L., Asher G. NQO1 stabilizes p53 through a distinct pathway. Proc. Natl. Acad. Sci. 2002;99(5):3099–3104. doi: 10.1073/pnas.052706799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shieh B., Yan C., Ross D., Siegel D., Kepa J.K. Role for NAD(P)H:quinone oxidoreductase 1 and manganese-dependent superoxide dismutase in 17-(Allylamino)-17-demethoxygeldanamycin-Induced heat shock protein 90 inhibition in pancreatic cancer cells. J. Pharmacol. Exp. Ther. 2010;336(3):874–880. doi: 10.1124/jpet.110.176438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Traver R.D., Siegel D., Beall H.D., Phillips R.M., Gibson N.W., Franklin W.A., Ross D. Characterization of a polymorphism in NAD(P)H: quinone oxidoreductase (DT-diaphorase) Br. J. Canc. 1997;75(1):69–75. doi: 10.1038/bjc.1997.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lavin M.F., Gueven N. The complexity of p53 stabilization and activation. Cell Death Differ. 2006;13(6):941–950. doi: 10.1038/sj.cdd.4401925. [DOI] [PubMed] [Google Scholar]
- 30.Hollstein M., Rice K., Greenblatt M.S., Soussi T., Fuchs R., Sørlie T., Harris C.C. Database of p53 gene somatic mutations in human tumors and cell lines. Nucleic Acids Res. 1994;22(17):3551–3555. [PMC free article] [PubMed] [Google Scholar]
- 31.Srivastava S., Tong Y.A., Devadas K., Zou Z., Chen Y., Pirouo K.F., Chang E.H. The status of the p53 gene in human papilloma virus positive or negative cervical carcinoma cell lines. Oncol. Res. 1992;13(7):1273–1275. doi: 10.1093/carcin/13.7.1273. [DOI] [PubMed] [Google Scholar]
- 32.Huovinen M., Loikkanen J., Myllynen P., Vähäkangas K.H. Characterization of human breast cancer cell lines for the studies on p53 in chemical carcinogenesis. Toxicol. In Vitro. 2011;25(5):1007–1017. doi: 10.1016/j.tiv.2011.03.018. [DOI] [PubMed] [Google Scholar]
- 33.Goodwin E.C., DiMaio D. Repression of human papillomavirus oncogenes in HeLa cervical carcinoma cells causes the orderly reactivation of dormant tumor suppressor pathways. Proc. Natl. Acad. Sci. 2002;97(23):12513–12518. doi: 10.1073/pnas.97.23.12513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lane D.P., Cheok C.F., Lain S. P53-Based cancer therapy. Cold Spring Harbor Perspect. Biol. 2010;1–24 doi: 10.1101/cshperspect.a001222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Oh E.T., Park H.J. Implications of NQO1 in cancer therapy. BMB Reports. 2015;48(11):609–617. doi: 10.5483/BMBRep.2015.48.11.190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mizumoto A., Ohashi S., Kamada M., Saito T., Nakai Y., Baba K., Muto M. Combination treatment with highly bioavailable curcumin and NQO1 inhibitor exhibits potent antitumor effects on esophageal squamous cell carcinoma. J. Gastroenterol. 2019;54(8):687–698. doi: 10.1007/s00535-019-01549-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Braicu C., Atanasov A.G., Berindan-Neagoe I., Nabavi S.M., Vladimirov B., Mehterov N., Sarafian V. Nutrigenomics in cancer: revisiting the effects of natural compounds. Semin. Cancer Biol. 2017;46:84–106. doi: 10.1016/j.semcancer.2017.06.011. [DOI] [PubMed] [Google Scholar]
- 38.Kunnumakkara A.B., Bordoloi D., Padmavathi G., Monisha J., Roy N.K., Prasad S., Aggarwal B.B. Curcumin, the golden nutraceutical: multitargeting for multiple chronic diseases. Br. J. Pharmacol. 2017;174(11):1325–1348. doi: 10.1111/bph.13621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Taguchi K., Yamamoto M. The KEAP1–NRF2 system in cancer. Front. Oncol. 2017;7:85. doi: 10.3389/fonc.2017.00085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tsvetkov P., Asher G., Reiss V., Shaul Y., Sachs L., Lotem J. Inhibition of NAD(P)H:quinone oxidoreductase 1 activity and induction of p53 degradation by the natural phenolic compound curcumin. Proc. Natl. Acad. Sci. 2005;102(15):5535–5540. doi: 10.1073/pnas.0501828102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zeekpudsa P., Kukongviriyapan V., Senggunprai L., Sripa B. Suppression of NAD (P) H-quinone oxidoreductase 1 enhanced the susceptibility of cholangiocarcinoma cells to chemotherapeutic agents. J. Exp. Clin. Cancer Res. 2014;33(1):1–13. doi: 10.1186/1756-9966-33-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.L, Navazio F. Soluble diaphorase in animal tissues. Acta Chem. Scand. 1958;12:595–602. [Google Scholar]
- 43.Asher G., Dym O., Tsvetkov P., Adler J., Shaul Y. The crystal structure of NAD(P)H quinone oxidoreductase 1 in complex with its potent inhibitor dicoumarol. Biochemistry. 2006;45(20):6372–6378. doi: 10.1021/bi0600087. [DOI] [PubMed] [Google Scholar]
- 44.Levine A.J., Oren M. The first 30 years of p53: growing ever more complex. Nat. Rev. Cancer. 2010;9(10):749–758. doi: 10.1038/nrc2723. [DOI] [PMC free article] [PubMed] [Google Scholar]