Abstract
We undertook a comprehensive comparative analysis of a collection of 30 small (<25 kb) non-conjugative Escherichia coli plasmids previously classified by the gene sharing approach into 10 families, as well as plasmids found in the National Center for Biotechnology Information (NCBI) nucleotide database sharing similar genomic sequences. In total, 302 mobilizable (belonging to 2 MOBrep and 5 MOBRNA families) and 106 non-transferable/relaxase-negative (belonging to three ReLRNA families) plasmids were explored. The most striking feature was the specialization of the plasmid family types that was not related to their transmission mode and replication system. We observed a range of host strain specificity, from narrow E. coli host specificity to broad host range specificity, including a wide spectrum of Enterobacteriaceae . We found a wide variety of toxin/antitoxin systems and colicin operons in the plasmids, whose numbers and types varied according to the plasmid family type. The plasmids carried genes conferring resistance spanning almost all of the antibiotic classes, from those to which resistance developed early, such as sulphonamides, to those for which resistance has only developed recently, such as colistin. However, the prevalence of the resistance genes varied greatly according to the family type, ranging from 0 to 100 %. The evolutionary history of the plasmids based on the family type core genes showed variability within family nucleotide divergences in the range of E. coli chromosomal housekeeping genes, indicating long-term co-evolution between plasmids and host strains. In rare cases, a low evolutionary divergence suggested the massive spread of an epidemic plasmid. Overall, the importance of these small non-conjugative plasmids in bacterial adaptation varied greatly according to the type of family they belonged to, with each plasmid family having specific hosts and genetic traits.
Keywords: Escherichia coli, small non-conjugative plasmids, classification, evolutionary history
Data Summary
Individual plasmid accession numbers can be found in Tables S1 to S11 (available in the online version of this article).
Impact Statement.
Escherichia coli strains contain multiple plasmids that contribute to the dissemination of antibiotic resistance genes. Many of these plasmids are large conjugative plasmids that have been well described, but less is known about the epidemiological contribution of small non-conjugative plasmids encompassing mobilizable and non-transferable plasmids. By taking a gene sharing-based approach on plasmids from a recently published collection and plasmid sequences retrieved from the National Center for Biotechnology Information (NCBI) database, we were able to classify small (<25 kb) non-conjugative plasmids into 10 families and to have a full picture of their evolutionary dynamics and importance in bacterial adaptation. The results indicate that the plasmid families showed host and maintenance system specificity and that the contribution of the small non-conjugative plasmids to bacterial adaptation to diverse and changing environmental conditions such as antibiotic resistance or bacteriocin gene acquisition varied and was specific to the plasmid family they belonged to.
Introduction
Escherichia coli is a commensal inhabitant of the gastrointestinal tract in birds and mammals. It can also cause various intestinal and extraintestinal diseases [1, 2]. Isolates of E. coli are known to contain multiple plasmids that allow the movement of genetic material, including antibiotic resistance genes. Many of them are large conjugative plasmids; however, small non-conjugative plasmids have also been described as contributing to antibiotic resistance [3–8]. These small non-conjugative plasmids encompass mobilizable (MOB) plasmids possessing a relaxase gene but no genes encoding mating pair formation (MPF) at the opposite of conjugative plasmids, and non-transferable plasmids devoid of MPF and relaxase genes called relaxase-negative (RelN) plasmids [8].
Recently, we undertook a comprehensive comparative analysis of the sequences of a large number of diverse plasmids from human and animal E. coli strains isolated over a period spanning before and after the use of third-generation cephalosporins using a gene sharing network approach [8]. This study showed that, although extended spectrum beta-lactamase (ESBL) genes were mainly found on large conjugative plasmids present before the use of third-generation cephalosporins, the small non-conjugative plasmids (<25 kb) also played a role, albeit a modest one, in the diffusion of the ESBL genes. Although their backbones have few genes, the gene sharing approach developed in this previous work showed that these small non-conjugative plasmids belonged to 10 different plasmid families. Fine-scale analysis of these plasmids was not presented [8]. In the present study, we (i) describe the non-conjugative plasmids of our collection belonging to each of the families identified thoroughly by investigating their replication and/or mobilization systems, (ii) use the backbone of each of these 10 plasmid families to search for plasmids sharing similar genomic sequences in the National Center for Biotechnology Information (NCBI) nucleotide database and (iii) undertake a comprehensive comparative analysis of the gene contents and sequences of both our plasmid collection and the plasmids found in the database to obtain a full picture of the evolutionary dynamics of these small plasmids and their importance in bacterial adaptation.
Methods
Plasmids and parental bacterial strains
In our previously described collection of sequenced plasmids [8], 30 plasmids were <25 kb non-conjugative plasmids. The sequences of these small plasmids were obtained either by direct plasmidome sequencing of the parental strains or by sequencing the plasmid contents of recipient strains after having transferred the plasmids from the parental strains either by conjugation in E. coli K-12 J53 Rifr (using a conjugative plasmid present in the native cell as helper) or by electroporation in E. coli K-12 DH10B, using appropriate antibiotic-selective plates (Table S1). The plasmid sequencing and annotation methods, comparative genomic analyses of the sequenced plasmids, global classification of the plasmids and parental strain chromosome phylotyping are detailed in [8].
Plasmid searches in the database
blast searches (performed in October 2018) for the backbone sequences of each plasmid family against the NCBI nucleotide database collection nr/nt were performed with the following parameters: hit length ≥90 % and identity ≥90 % for the query sequences. Redundant plasmids from outbreak strains when documented were discarded.
Plasmid evolutionary history analysis
blast sequence analysis tools were used to confirm the identity of proteins encoded on the plasmids. For each family type of plasmids defined by the comparative genomic analysis and classification, the set of shared genes were retrieved from the NBCI database and concatenated. The concatenated sets of genes were then aligned using BioEdit version 7.2.5 [9] and a tree was built using the program PhyML [10] under the general time-reversible evolution model [11]. Molecular evolutionary analyses including mutational spectrum were conducted using mega version 6 [12].
Results
Characteristics of the 10 plasmid families of non-conjugative plasmids
In our previous collection of 30 small non-conjugative plasmids, 22 were MOB plasmids and 8 were RelN plasmids [8]. These two types of plasmids were further classified according to their type of replication/control system. Among the MOB plasmids, 5 had a protein replication/control system (MOBrep) and 17 had a RNAII/RNAI replication/control system (MOBRNA) like the colE1 plasmid [13, 14]. All eight RelN plasmids had a RNAII/RNAI replication/control system (RelNRNA). The gene sharing approach developed in our previous work followed by a comparative analysis of their complete circularized sequences and in silico plasmid relaxase gene typing (PRaseT) [15] allowed us to define 10 families of plasmids among the 3 groups of plasmids mentioned above. Each plasmid family had a specific backbone that is not shared by any other plasmid family [8].
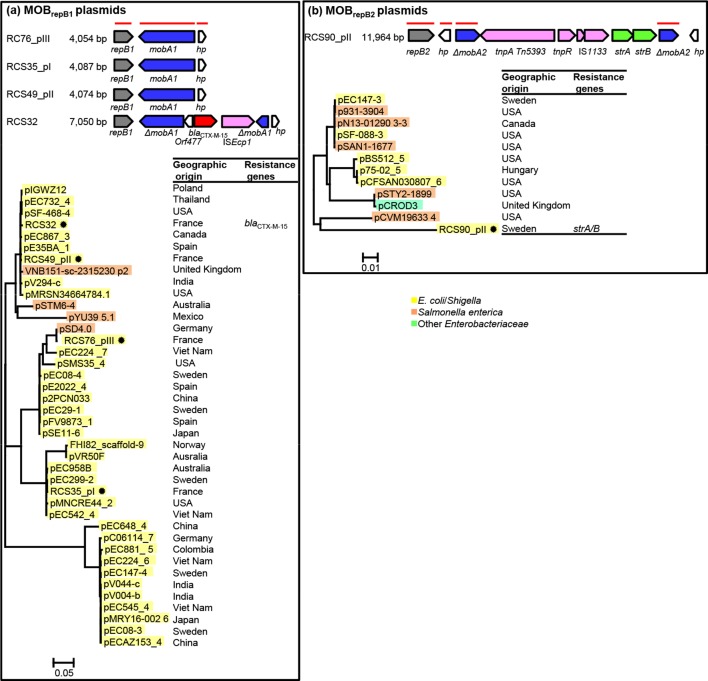
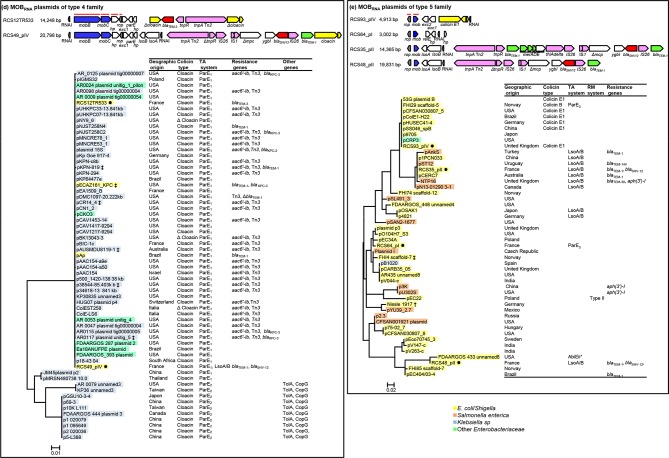
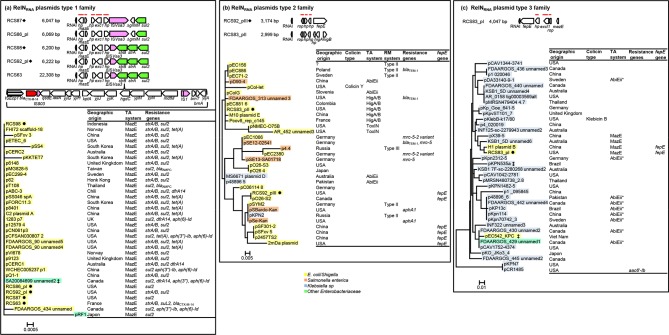
The MOBrep plasmids were found to belong to two families (Fig. 1, Table S1): the MOBrepB1 family of relaxase gene type (RGT) Qu [16] and the MOBrepB2 family, which was not typed by PRaseT. The MOBRNA plasmids were found to belong to five families (Fig. 2, Table S1): the MOBRNA type 1 family of RGT P5-1 (similar to the colE1 plasmid) [14, 17], the MOBRNA type 2 family not typed by PRaseT, the MOBRNA type 3 family of RGT P5-2, the MOBRNA type 4 family of RGT C11 and the MOBRNA type 5 family of RGT P5-3, named by Moran and Hall the ‘NTP16 group’ [18, 19]. The RelNRNA plasmids were found to belong to three families: the RelNRNA type 1 family, named the ‘p9123’ family named by Anantham and Hall [20, 21], the RelNRNA type 2 family and the RelNRNA type 3 family (Fig. 3, Table S1).
Fig. 1.
Linear representations of the MOBrep plasmids of our collection [8] and maximum-likelihood phylogenetic trees of all the MOBrep plasmids, including those from the NCBI database (see main text for details regarding their selection) reconstructed as described in the Methods section. (a) MOBrepB1 plasmids and (b) MOBrepB2 plasmids. Linear representation: replication genes are grey, mobilization genes are blue, IS and transposable elements are pink, ESBL genes are red, other antibiotic resistance genes are green and other genes are white; a horizontal red line indicates genes of the backbones used for phylogenetic analysis. Phylogenetic tree: plasmid IDs are shaded in yellow for E. coli/Shigella sp., orange for S. enterica ssp. enterica and turquoise for other Enterobacteriaceae , not including K. pneumoniae . ✹, plasmids from our collection. Geographical origin and resistance genes are indicated at the right of the trees.
Fig. 2.
Linear representations of the MOBRNA plasmids of our collection [8] and maximum-likelihood phylogenetic trees of all of the MOBRNA plasmids, including those from the NCBI database (see main text for their selection) reconstructed as described in the Methods section. (a) MOBRNAI plasmids of the type 1 family, (b) MOBRNAI plasmids of the type 2 family, (c) MOBRNAI plasmids of the type 3 family, (d) MOBRNAI plasmids of he type 4 family and (e) MOBRNAI plasmids of he type 5 family. Linear representation: replication systems (RNAI and rop gene) are grey, mobilization genes are blue, IS and transposable elements are pink, ESBL genes are red, other antibiotic resistance genes are green, colicin operons are yellow and other genes are white; a horizontal red line indicates the genes of the backbones used for phylogenetic analysis. Phylogenetic tree: plasmid IDs are shaded in yellow for E. coli/Shigella sp., orange for S. enterica ssp. enterica, blue for K. pneumoniae , green for plant pathogen Enterobacteriaceae and turquoise for other Enterobacteriaceae . *, plasmids from our collection. †, plasmids co-integrated in the chromosome. ‡, co-integrated in plasmids. Geographical origin, colicin operons, TA systems, RM systems, resistance genes and others genes are indicated at the right of the trees.
Fig. 3.
Linear representations of the RelNRNAI plasmids from our collection [8] and maximum-likelihood phylogenic trees of all the RelNRNAI plasmids, including those from the NCBI database (see the main text for details regarding their selection) reconstructed as described in the Methods section. (a) RelNRNAI plasmids of the type 1 family, (b) RelNRNAI plasmids of he type 2 family and (c) RelNRNAI plasmids of the type 3 family. Linear representation: replication systems (RNAI and rop gene) are grey, IS are pink, ESBL genes are red, other antibiotic resistance genes are green and other genes are white; a horizontal red line indicates genes of the backbones used for phylogenetic analysis. Phylogenetic tree: plasmid IDs are shaded in yellow for E. coli/Shigella sp., orange for S. enterica ssp. enterica, blue for K. pneumoniae and turquoise for other Enterobacteriaceae . ✹, plasmids from our collection. ‡, co-integrated in plasmids. Geographical origin, colicin operons, TA systems, RM systems, resistance genes and others genes are indicated at the right of the trees.
Plasmid backbone definitions
The backbones of all the MOB plasmids were considered to be the sequences that covered the replication/control system [replication gene or RNAI/RNAII and when present a repressor of primer (rop) gene involved in the regulation of copy number [13]] and the mobilization gene systems. This also included the genes located in between the two systems on the MOBrepB plasmids and the MOBRNA type 4 family plasmids (Figs 1 and 2, Table S2). The backbones of the RelNRNA plasmids were considered to be the sequences that covered the replication/control system (RNAI/RNAII and rop gene when present) and for the RelNRNA type 1 family and RelNRNA type 2 family plasmids, the shared genes (five genes, with the sul2 gene being excluded, and two genes, respectively) and for the single RelNRNA plasmid of type 3 family the two genes in between the RNAI/RNAII and the rop gene, excluding the accessory gene fepE (Fig. 3, Table S2).
Plasmids of the 10 families found in the NCBI database
To obtain a more complete picture of the small plasmid families’ distribution and abundance, we searched in the NCBI nucleotide database for plasmids sharing similar backbone sequences to those defined above. We found 378 plasmids with a close hit with the backbones of our 30 plasmids, but their number varied according to the type of family (Tables 1 and S3–12). For the MOBrep plasmid families we found 36 MOBrepB1 plasmids and only 11 MOBrepB2 plasmids. For the MOBRNA plasmids, we found 54 plasmids of type 1, 23 plasmids of type 2, 51 plasmids of type 3, 61 plasmids of type 4 and 44 plasmids of type 5. For the RelNRNA plasmids, we found 30 plasmids of type 1, 31 plasmids of type 2 and 37 plasmids of type 3.
Table 1.
Characteristics of the 408 plasmids from our collection and the NCBI database classified into 10 MOB and RelN families
|
Plasmid characteristics |
MOBrep family type |
MOBRNA family type |
RelNRNA family type |
|||||||
|---|---|---|---|---|---|---|---|---|---|---|
|
MOBrepB1 |
MOBrepB2 |
Type 1 |
Type 2 |
Type 3 |
Type 4 |
Type 5 |
Type 1 |
Type 2 |
Type 3 |
|
|
No. of plasmids |
40 |
12 |
60 |
25 |
54 |
63 |
48 |
35 |
33 |
38 |
|
Plasmid mean size (range), kb |
4 (3.9–5) |
4.2 (3.9–11.9) |
6.7 (4–23.5) |
6.8 (4.2–8.7) |
5.4 (2.9–15.8) |
13.2 (9.1–25.2) |
4.9 (1.9–19.8) |
7.5 (6–38.9) |
6.5 (2.9–19.8) |
5.2 (2.7–10.3) |
|
Co-integrated plasmids (no.)* |
C (1) |
P (1) |
P (6) |
P (1), C (1) |
P (1) |
P (2) |
||||
|
No. of concatenated genes used for molecular analysis (length of the sequence, bp) |
3 (3,123) |
3 (3,026) |
3 (2,107) |
3 (2,409) |
2 (1,823) |
4 (3,407) |
2 (518) |
5 (1,662) |
3 (579) |
3 (920) |
|
Estimate of average evolutionary divergence over all sequence pairs (range, %) |
5.2 (0–10.2) |
3.9 (0–9.5) |
4.5 (0–7.7) |
0.8 (0–1.7) |
7 (0–14.8) |
2.6 (0–10.1) |
2.6 (0–6) |
0.1 (0–0.7) |
1.4 (0–3.7) |
2.2 (0–6.5) |
*C, plasmid integrated in a chromosome; P, plasmid integrated in a plasmid.
Interestingly, we found plasmids integrated on both chromosomes and on other plasmids (Table 1). The plasmids integrated on chromosomes comprised one MOBRNA type 1 family and one MOBRNA type 5 family (integrated in Klebsiella pneumoniae strain TGH10 and E. coli strain Nissle, respectively). The plasmids co-integrated on plasmids belonged to five families and their number varied according to the type of family: one plasmid of the MOBRNA type 3 family, six plasmids of the MOBRNA type 4 family, including the published plasmids ColE-LS6, and pECAZ161_KPC [22, 23], one plasmid of the MOBRNA type 5 family, one plasmid of the RelNRNA type 1 family and two plasmids of the RelNRNA type 3 family. The co-integrated plasmids belonged to all types: conjugative, mobilizable or non-transferable (Tables S7–10 and S12).
Overall characteristics and evolutionary history of the plasmids of each family
We explored the main characteristics of the plasmids of each of the family types, combining the data obtained from the plasmids from our collection and those from the plasmids found in the NCBI nucleotide database, and carried out a phylogenetic analysis. In total, 408 plasmids, 302 mobilizable and 106 non-transferrable, were explored. All of these plasmids were small, with the smallest being the MOBrepB1 plasmids (mean of 4 kb) and the largest being the MOBRNA plasmids of the type 4 family (mean of 13.2 kb) (Table 1). The plasmids were isolated worldwide and we did not observe any group of plasmids isolated from a particular country (Figs 1–3).
To reconstruct the evolutionary history of the plasmids, we first built a tree based on the concatenated coding sequences of the backbones of the plasmids (Figs 1–3, Table 1). This approach allowed the identification of several clades by family, except in two families, the MOBRNA type 4 and RelNRNA type 1 families, where very few variations or no variation was observed.
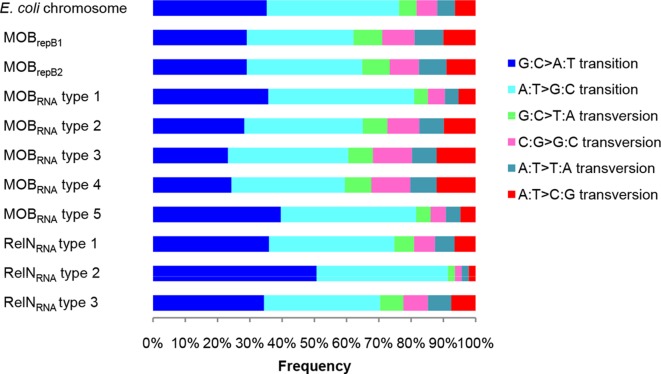
Estimates of the average evolutionary divergence of all sequence pairs within each family of plasmids were then calculated (Table 1). The mean intra-family diversity of the plasmids (MOBrep=4.55 %, MOBRNA=3.5 % and RelNRNA=1.23 %) showed that the MOBrep plasmids have the highest diversity and the RelNRNA the lowest. However, in the MOBRNA plasmid families the plasmids of the type 3 family showed high diversity (7 %), while the plasmids of the type 2 family showed low diversity (0.8 %). Among the plasmids of the RelNRNA families, the plasmids of the type 1 family had the lowest diversity (0.1 %). We compared these data to the evolutionary divergence of E. coli strains by calculating the mean diversity of eight chromosomal housekeeping genes (dinB, icdA, pabB, polB, putP, trpA, trpB and uidA) used for multilocus sequence typing (MLST) of 89 E. coli host strains bearing plasmids, including the 30 plasmids from our collection [8]. The mean diversity obtained was of 2.6 % (0–4.3). To gain insight into the mechanism of mutagenesis, we characterized the mutational spectrum of the single-nucleotide polymorphisms (SNPs) of each plasmid family and compared the results to the mutational spectrum of the E. coli chromosomal genes cited above (Fig. 4). In each plasmid family, we found more than a quarter for each type of transition (25.36–50.66) and ≤12 % (2.2–12) for each of the transversion types, as in the E. coli chromosome genes. In sum, we observed a phylogenetic signal in the backbone of each family type of plasmids that is globally similar to the E. coli housekeeping chromosomal genes.
Fig. 4.
Mutational spectrum of the SNPs of the shared genes of the plasmids in each of the family types and of the chromosomal housekeeping genes from a collection of 89 E. coli host strains bearing plamids [8]. Each colour represents a possible substitution.
Plasmid families have host species specificity
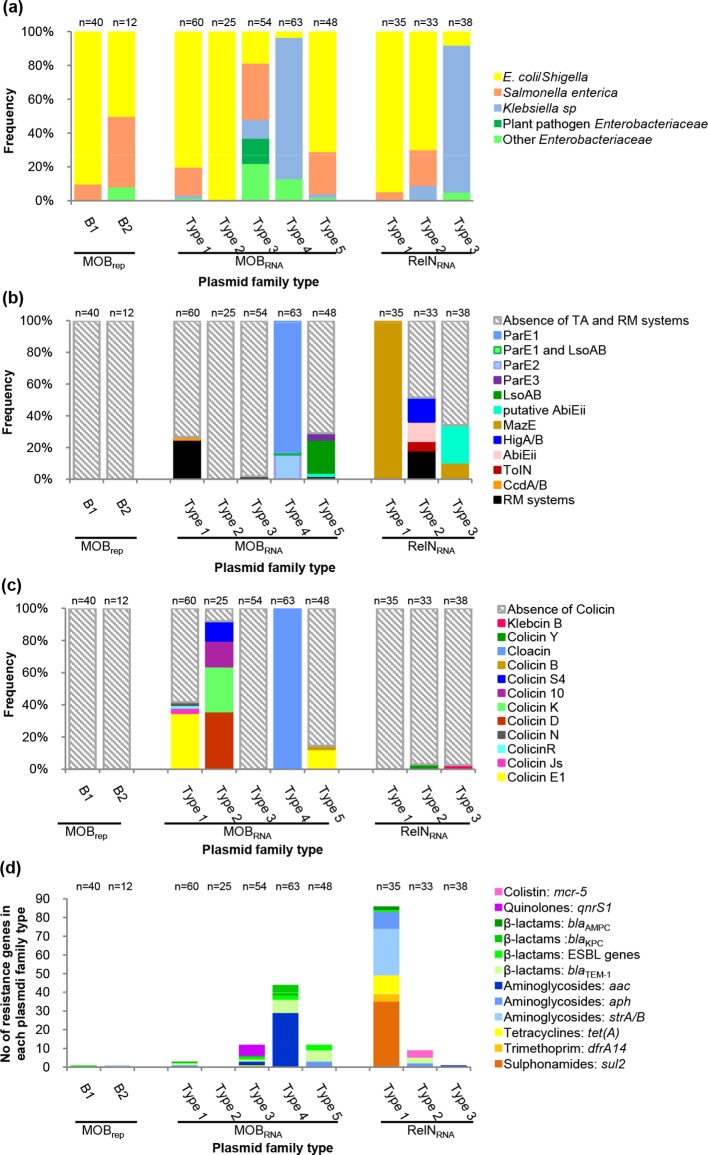
Among the MOBrep plasmids, 90 % of the MOBrepB1 plasmids were isolated from E. coli/Shigella strains and only 10 % were isolated from S. enterica ssp. enterica strains, while the MOBrepB2 plasmids were isolated almost equally from E. coli strains (50 %) and S. enterica ssp. enterica strains (41.6 %) (Figs 1 and 5a Table S3 and S4).
Fig. 5.
Characteristics of the plasmids according to their family type. The numbers of plasmids in each of the families are indicated above the columns of each graph. (a) Frequencies of the plasmids according to the host strain species. (b) Frequencies of the plasmids carrying one of the colicin operon types. (c) Frequencies of the plasmids carrying one of the TA system types, or an RM. (d) Number of resistance genes carried by the plasmids.
Among the MOBRNA plasmids, E. coli/Shigella were the major host species of the plasmids of the type 1 (80 %), type 2 (100 %) and type 5 (70.8 %) families, while S. enterica ssp. enterica was the second most common host species of the plasmids of the type 1 (16.6 %) and type 5 (25 %) families (Figs 2a, b, e and 5a, Tables S5, S6 and S9). K. pneumoniae was the major host species of the plasmids of the type 4 family (80.9 %), while only 6.3 % of the plasmids were isolated from E. coli/Shigella strains and 12.6 % were isolated from various other Enterobacteriaceae ( Citrobacter koseri , Klebsiella aerogenes and Enterobacter cloacae ) (Fig. 2d and Table S8). The plasmids of the type 3 family were ubiquitous. They were isolated from various Enterobateriaceae, including S. enterica ssp. enterica, the predominant species (33.3 %), followed by E. coli/Shigella (18.5 %), K. pneumonia (11.1 %), various other Enterobateriaceae (22.2 %) ( E. cloacae , Enterobacter hormaechei , Citrobacter freundii and Serratia marcescens ) and even plant pathogen Enterobacteriaceae (14.8%) ( Pantoea stewartii , Pectobacterium carotovorum and Tatumella morbirosei ) (Fig. 2c, Table S7).
For the MOBrep and MOBRNA plasmids of our collection for which the background of the E. coli parental strains inferred by the phylogenic group and the Pasteur Institute MLST scheme was available, we observed very diverse phylogenetic backgrounds not linked to plasmid types (Fig. S1 and Table S1).
Among the RelNRNA plasmids, E. coli/Shigella was the most common host species of the plasmids of the type 1 (94.2 %) and type 2 (69.6 %) families, while S. enterica ssp. enterica was the host species of only 5.7 % and 21.2 % of the plasmids of these two families, respectively (Fig. 3a, b and 5a, Table S10 and S11). K. pneumonia was the major host species of the plasmids of the type 3 family (86.8 %), while E. coli/Shigella were the host species for only 7.8 % of them (Figs 3c and 5a, Table S12). In these three families, the plasmids from our collection were only isolated from E. coli phylogroup A strains, but of different sequence types (Fig. S1 and Table S1). In contrast, the plasmids found in the database were isolated from E. coli strains of various phylogroups (data not shown) [5, 24, 25], showing that there was no correlation between the background of the E. coli host strain and dissemination of these types of plasmid.
In sum, the plasmids were only found in Enterobacteriaceae , but, depending on the family type, the plasmids showed host species specificity. A continuum of host spectrum was observed from E. coli -specific plasmids (MOBRNA type 2) to broad-range plasmids (MOBRNA type 3), without any link with the main groups of plasmids (MOBrep, MOBRNA and RelNRNA) (Fig. 5a).
Toxin/antitoxin (TA) systems have plasmid family specificity
We looked for genes implicated in plasmid maintenance, i.e. TA and restriction modification (RM) systems (Fig. 5b) [26, 27]. RM systems were rare and found on plasmids belonging to the MOBRNA type 1 (15/60, 25%), type 3 (1/54, 1.8 %) and type 5 (1/48, 2.1 %) families and on plasmids belonging to the RelNRNA type 2 family (6/33, 18.1 %) (Fig. 5b). The plasmids carried a type II RM system, except for one plasmid of the RelNRNA type 2 family that carried a type III RM system (Fig. 3b, Table S10). Each of these plasmids was devoid of a TA system.
No MOBrep family plasmid carried a TA system. In the MOBRNA type 1 family only one plasmid, RCS82_pII, carried a TA system, a CcdA/B system. This TA system, which is frequently found in IncF plasmids [28], was part of a 14 900 bp sequence of IncF plasmid origin acquired by means of an IS629 (Fig. 2a). All the plasmids (63/63) of the MOBRNA type 4 family carried TA systems belonging to the ParE/D family. These systems encoded two homologous types of toxin containing the COG3668 domain that we arbitrarily named ParE1 and ParE2. Thereby, among the plasmids of the MOBRNA type 4 family, 53 (84.1 %) encoded ParE1 and the last 10 (15.8 %) ParE2. In addition to the ParE1 system, RCS49_pIV, a plasmid belonging to our collection, carried an LsoAB system. All but two of the ParE1 plasmids clustered closely in a single branch of the phylogenetic tree, revealing a group of highly conserved plasmids that had been isolated worldwide (Fig. 2d). Interestingly, a copG gene coding for a ribbon–helix–helix protein (COG3904), and a tolA gene coding for a cell envelope integrity protein (TIGR02794), were found exclusively on all the 10 ParE2 plasmids. Among plasmids of the MOBRNA type 5 family, 13/48 (27.1 %) carried a TA system: 10 an LsoA/B system, 1 a putative AbiEii system (pfam13304) and 2 a ParE/D system encoding a ParE toxin containing a COG4679 domain that we named ParE3. Although they were isolated from distant areas, most of the LsoA/B plasmids clustered in a main branch of the phylogenic tree but were not closely related (Fig. 2e).
All the plasmids of the RelNRNA type 1 family carried a MazE TA system. Among the plasmids of the RelNRNA type 2 family, 12/33 (36.3 %) carried a TA system: 6 a HigA/B system, 4 an AbiEii system (pfam08843) and 2 a ToxIN system. Among the plasmids of the RelNRNA type 3 family, 13/38 (34.2 %) carried a TA system: 4 a MazE system and 9 a putative AbiEii system (pfam13304) that differed from the one carried by a plasmid of the MOBRNA type 5 family.
In sum, only the plasmids belonging to three types of family were devoid of TA and RM systems: the plasmids of the two MOBrep families and the MOBRNA type 2 family. The number of plasmids that carry a TA system and the type of TA system varied according to the family type (Fig. 5b). The plasmids carried only one TA system, with the exception of one plasmid of the MOBRNA type 4 family that carried two. With the exception of the MazE and the LsoAB systems that were each found on plasmids belonging to two different families, all the other types of TA systems were unique to one family type (Fig. 5b).
Plasmids carried accessory genes that give selective advantage to the host cells
We next looked for genes that give the host strain-selective advantage and found various colicin operons and antibiotic determinants (Fig. 5c and d).
Colicin operons have plasmid family specificity
Among the plasmids of the MOBRNA type 1 family, 25/48 (41.6 %) carried a colicin operon: 21 a colicin E1, 2 a colicin Js, 1 a colicin R and 1 a colicin N. If the majority of the plasmids that carried a colicin E1 operon were clustered in one of the five main branches of the phylogenetic tree, this operon was also found on plasmids clustered in three other branches (Fig. 2a). All but 2 plasmids (23/25, 92 %) of the MOBRNA type 2 family carried a colicin operon: 9 a colicin D, 7 a colicin K, 4 a colicin 10 and 3 a colicin S4. No strong association between the type of colicin and the distribution of the plasmids in the phylogenetic tree was evidenced (Fig. 2b). All of the plasmids of the MOBRNA type 4 family (63/63) carried a colacin operon (Figs 2d and 5b). Among the plasmids of the MOBRNA type 5 family, 7/48 (14.5 %) carried a colicin operon: 6 a colicin E1 operon and 1 a colicin B. All of these colicin plasmids were found in a cluster of highly conserved plasmids (Fig. 2e). In each of the RelNRNA families of type 2 and type 3 only one plasmid carried a colicin operon, a colicin Y operon and a klebicin B operon, respectively (Fig. 5c).
In sum, colicin operons were found almost exclusively in MOBRNA plasmids (except MOBRNA plasmids of the type 3 family) and their number varied according to the type of family. Only the colicin E1 operon was found on plasmids of two different families. Each of the other types of colicin operon was unique to one family and the MOBRNA type 4 family had specificity for the cloacin operon (Fig. 5c).
MOBRNA plasmids of the type 4 family and RelNRNA plasmids of the type 1 family are highly implicated in the dissemination of antibiotic resistance determinants
In 4 families the plasmids carried few antibiotic resistance determinants: 1/40 (2.5 %) of the MOBrepB1 plasmids, 1/12 (8.3 %) of the MOBrepB2 plasmids, 2/60 (3.2 %) of the plasmids of the MOBRNA type 1 family and 1/33 (2.6 %) of the plasmids of the RelNRNA type 3 family. In 3 families the number of plasmids carrying antibiotic resistance determinants was higher: 9/48 (18.7 %) of the plasmids of the MOBRNA type 5 family, 11/54 (20.3 %) of the plasmids of the MOBRNA type 3 family and 8/33 (24.2 %) of the plasmids of the RelNRNA type 2 family. In the last two other families, the number of plasmids carrying antibiotic resistance determinants was the highest: 36/63 (57.1 %) of the plasmids of the MOBRNA type 4 family and 35/35 (100 %) of the plasmids of the RelNRNA type 1 family (Fig. 5d and S2).
Overall, the plasmids carried various genes encoding resistance to almost all the classes of antibiotics used to treat human infections: aminoglycosides, β-lactams (including 3GC and carbapenems), quinolones, colistin, tetracycline, sulphonamides and trimethoprim. (Figs 1–3 and 5d). The aminoglycoside resistance genes were the most frequently found, followed by the β-lactam resistance genes (Fig. 5d and S3). However, the frequency of each of the resistance determinants varied according to the type of plasmids (Figs 1–3 and 5d). We next focused on the two plasmid families carrying the highest number of resistance determinants, the MOBRNA type 4 family and the RelNRNA type 1 family, and on the plasmid families carrying resistance genes implicated in therapeutic challenges (ESBL genes, carbapenemase genes and resistance genes conferring resistance to fluoroquinolones and to colistin).
In the MOBRNA type 4 family, of the 36 plasmids carrying antibiotic determinants, 29 carried an aac6′-Ib-Tn1331 transposon [29, 30], associated 4 times with a bla KPC gene. These aac6′-Ib-Tn1331 plasmids and the seven other plasmids of this family that carried other resistance determinants (bla TEM-1, bla TEM-3, bla SHV12 and bla KPC genes) belonged a cluster of highly conserved ParE1 plasmids isolated worldwide as described above (Fig. 2d), suggesting clonal dissemination of these plasmids.
All the plasmids of the RelNRNA type 1 family carried a sul2 gene (sul2-ISVsa3 configuration) [31] and most of these plasmids (28/35, 80%) also carried streptomycin resistance genes, strA/B (sul2-strA-strB-ΔISVsa3 configuration) [3, 21] (Fig. 3a). The sul2-ISVsa3 configuration has been predicted to be the precursor of the sul2-strA-strB-ΔISVsa3 configuration by Anantham and Hall [20]. In addition, these plasmids had acquired various other resistance genes: tet(A) (n=9) [32], dfrA14 cassette (n=2) [20], aph(3'')-Ib (n=4), aph(6)-Id (n=5), bla AMPC (n=2) and bla CTX-M-14 (n=1) [8] (Fig. 3a).
We found ESBL genes (2 bla CTX-M, 4 bla SHV and 2 bla TEM) on eight plasmids distributed in five families belonging to the three groups of families (Figs 1a–3a) and eight carbapenemase genes of the bla KPC type on plasmids distributed in three of the MOBRNA families, most of them belonging to the MOBRNA type 4 family, as seen above (Fig. 2a, c and d).
The gene encoding fluoroquinolone resistance, qnrS1 (Fig. 2c) was only found in six plasmids of the MOBRNA type 3 family (Fig. 2c). These plasmids, all originating from S. enterica serovar Typhimurium isolated in distant geographical areas, were clustered closely in the phylogenetic tree, showing the worldwide clonal dissemination of these plasmids, as previously observed [33–36].
The genes encoding colistin resistance, mcr-5 and the mcr-5.2 variant [37, 38], were only found on four plasmids of the RelNRNA type 2 family. These plasmids clustered in the same branch of the tree independently of the host strain species ( E. coli or S. enterica serovar paratyphi B), showing the dissemination of this plasmid among different species (Fig. 3b).
In sum, antibiotic resistance determinants were found in plasmids of all the families, except plasmids of the MOBRNA type 2 family. These plasmids are able to acquire genes of resistance to various antibiotic classes as they are marketed. Two groups of plasmids that seemed to easily acquire resistance genes were identified: a highly conserved cluster of plasmids of the MOBRNA type 4 family and the plasmids of the RelNRNA type 1 family.
Discussion
In this study, we provided a framework for the classification of small non-conjugative plasmids in E. coli . Thanks to a previous work using the gene sharing approach on a limited number of plasmids [8] coupled to the increasing number of plasmid nucleotide sequences in public databases, we gained insights into the main characteristics and evolutionary history of these plasmids.
The most striking feature of our work was the specificity of each of the plasmid family types. All of the plasmids were only found in Enterobacteriaceae . However, among this family of bacteria, they showed host range specificity according to the plasmid family type they belonged to. We found plasmid family types with a narrow host range that seemed to be well adapted to E. coli or K. pneumoniae and family plasmid types with a broad host range including a wide spectrum of species of Enterobacteriaceae, even plant pathogen species, as described for the MOBRNA plasmids of the type 3 family.
Among the accessory genes carried by plasmids, we found maintenance systems, RM and TA systems, and genes that conferred a selective advantage to the bacterial host, such as colicin operons and determinants of resistance to antibiotics. The various types of TA systems and colicin operons that we identified were not present in all plasmid families, but when present, in most cases they were specific to a plasmid family type. The plasmids carried various genes conferring resistance to almost all the antibiotic classes, from those to which resistance developed early, such as sulphonamides and tetracycline, to those to which resistance has only developed recently, such as colistin (mcr genes) [39]. As for the previous accessory genes, we observed plasmid family specificity. Indeed, the prevalence of the resistant plasmids varied according to the family type from 0 to 100 %. Two types of plasmids highly implicated in the dissemination of antibiotic resistance determinants were highlighted: a cluster of highly conserved aac6′-Ib-Tn1331 plasmids in the MOBRNA plasmids of the type 4 family and the RelNRNA plasmids of the type 1 family all carried at least a sul2 gene, with both the resistance markers, aac6′-Ib-Tn1331 and the sul2 gene, being specific to the two types of plasmids, respectively. For both family types, all the plasmids carried a specific TA system, and these systems are known to play a crucial role in the dissemination and evolution of antibiotic resistance by maintaining multi-resistant plasmids [40].
The plasmids were able to disseminate other adaptive genes specific to a family type, as illustrated by the presence of a gene fepE coding for a ferric enterobactin transport gene, on plasmids of the RelNRNA type 2 and 3 families (Fig. 3b and c), or the presence of a gene copG coding for a ribbon–helix–helix protein, and a gene tolA coding for a cell envelope integrity protein on plasmids of the MOBRNA type 4 family (Fig. 2d). Many other genes were carried by the plasmids, possibly with a role in the adaptation of the host strains to changing environmental conditions, but most of them were of unknown function or had functions that are not yet well described in these plasmids.
The evolutionary history of the plasmid families showed variable nucleotide divergence that was however globally in line with the divergence observed in E. coli housekeeping chromosomal genes. Similarly, the mutational spectrum did not reveal a high mutation rate footprint in any of the families and it was similar to the chromosomal one. Taken together, these data indicated long-term co-evolution of the small plasmids with bacteria, not at the strain level, but at the species, genus or family level, according to the host range. Rarely, we observed low evolutionary divergence, suggesting the massive spread of an epidemic plasmid, as illustrated by the aac6′-Ib-Tn1331 plasmid cluster of the MOBRNA type 4 family (Fig. 2d).
Our work showed some limitations. First, the diversity of the type of plasmids that we found is far from being exhaustive, as the classification was based on the characterization of a set of only 30 E. coli non-conjugative plasmids of <25 kb identified during a comparative analysis of the sequences of a plasmid collection [8]. Other small non-conjugative plasmids, mobilizable or non-transferrable, have been described in E. coli strains [41–44] that were not present in our collection. Second, as the backbones of these small plasmids contain few genes, the evolutionary history was based on a limited number of genes – as few as two for some types of plasmids – which could hinder the signal. Nevertheless, our data will be helpful to expand knowledge on the diversity of these small non-conjugative plasmids and our approach can be applied to large epidemiological studies.
Overall, the importance of the role of these small non-conjugative plasmids in bacterial adaptation varied greatly according to the type of family they belong to. Furthermore, when they play a role, each of the plasmid families has its own host and genetic traits.
Data bibliography
1. Sequence data for the 30 E. coli plasmids of our collection are available in in the European Nucleotide Archive (www.ebi.ac.uk/ena/) Genoscope CEA, Bioproject PRJEB24625 (2018) and accession numbers are listed in Table S1.
2. References and accession numbers for global plasmids used in the analyses are listed in Tables S2 to S11 and are available at the NCBI database (https://www.ncbi.nlm.nih.gov/).
3. Fineran PC, Blower TR, Foulds IJ, Humphreys DP, Lilley KS et al. The phage abortive infection system, ToxIN, functions as a protein-RNA toxin-antitoxin pair. Proc Natl Acad Sci USA 2009;106:894-899. DOI 10.1073/pnas.0808832106.
4. Dy RL, Przybilski R, Semeijn K, Salmond GP, Fineran PC. A widespread bacteriophage abortive infection system functions through a Type IV toxin-antitoxin mechanism. Nucleic Acids Res 2014;42:4590-4605. DOI 10.1093/nar/gkt1419.
5. Rijavec M, Budic M, Mrak P, Muller-Premru M, Podlesek Z et al. Prevalence of ColE1-like plasmids and colicin K production among uropathogenic Escherichia coli strains and quantification of inhibitory activity of colicin K. Appl Environ Microbiol 2007;73:1029-1032. DOI 10.1128/AEM.01780-06.
6. Cascales E, Buchanan SK, Duche D, Kleanthous C, Lloubes R et al. Colicin biology. Microbiol Mol Biol Rev 2007;71:158-229. DOI 10.1128/MMBR.00036-06.
7. Riley MA, Cadavid L, Collett MS, Neely MN, Adams MD et al. The newly characterized colicin Y provides evidence of positive selection in pore-former colicin diversification. Microbiology 2000;146 (Pt 7):1671-1677. DOI 10.1099/00221287-146-7-1671.
8. Zankari E, Hasman H, Cosentino S, Vestergaard M, Rasmussen S et al. Identification of acquired antimicrobial resistance genes. J Antimicrob Chemother 2012;67:2640-2644. DOI 10.1093/jac/dks26.
9. Siguier P, Perochon J, Lestrade L, Mahillon J, Chandler M. ISfinder: the reference centre for bacterial insertion sequences. Nucleic Acids Res 2006;34:D32-36. DOI 10.1093/nar/gkj014.
Supplementary Data
Funding information
This work was supported by a grant from the Agence Nationale de la Recherche (grant ANR-10-GENM-0012) to C. B. This work was also partially supported by a grant from the Fondation pour la Recherche Médicale to E. D. (Equipe FRM 2016, grant number DEQ20161136698).
Conflicts of interest
The authors declare that there are no conflicts of interest.
Ethical statement
There are no ethical considerations applicable to the work presented.
Footnotes
Abbreviations: ESBL, extended-spectrum beta-lactamase; IP, Institut Pasteur; MLST, multilocus sequence typing; MOB, mobilizable; MPF, mating pair formation; PRaseT, plasmid relaxase gene typing; RelN, relaxase negative; RGT, relaxase gene type; RM, restriction modification; SNP, single nucleotide polymorphism; ST, sequence type; TA, toxin-antitoxin.
All supporting data, code and protocols have been provided within the article or through supplementary data files. Twelve supplementary tables and three supplementary figures are available with the online version of this article.
References
- 1.Croxen MA, Finlay BB. Molecular mechanisms of Escherichia coli pathogenicity. Nat Rev Microbiol. 2010;8:26–38. doi: 10.1038/nrmicro2265. [DOI] [PubMed] [Google Scholar]
- 2.Tenaillon O, Skurnik D, Picard B, Denamur E. The population genetics of commensal Escherichia coli . Nat Rev Microbiol. 2010;8:207–217. doi: 10.1038/nrmicro2298. [DOI] [PubMed] [Google Scholar]
- 3.Handford CL, Stang CT, Raivio TL, Dennis JJ. The contribution of small cryptic plasmids to the antibiotic resistance of enteropathogenic Escherichia coli E2348/69. Can J Microbiol. 2009;55:1229–1239. doi: 10.1139/W09-079. [DOI] [PubMed] [Google Scholar]
- 4.Smillie C, Garcillán-Barcia MP, Francia MV, Rocha EPC, de la Cruz F. Mobility of plasmids. Microbiol Mol Biol Rev. 2010;74:434–452. doi: 10.1128/MMBR.00020-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brolund A, Franzén O, Melefors O, Tegmark-Wisell K, Sandegren L. Plasmidome-analysis of ESBL-producing Escherichia coli using conventional typing and high-throughput sequencing. PLoS One. 2013;8:e65793. doi: 10.1371/journal.pone.0065793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.de Toro M, Garcilláon-Barcia MP, De La Cruz F. Plasmid diversity and adaptation analyzed by massive sequencing of Escherichia coli Plasmids. Microbiol Spectr. 2014;2 doi: 10.1128/microbiolspec.PLAS-0031-2014. [DOI] [PubMed] [Google Scholar]
- 7.Stephens CM, Adams-Sapper S, Sekhon M, Johnson JR, Riley LW. Genomic analysis of factors associated with low prevalence of antibiotic resistance in extraintestinal pathogenic Escherichia coli sequence type 95 strains. mSphere. 2017;2 doi: 10.1128/mSphere.00390-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Branger C, Ledda A, Billard-Pomares T, Doublet B, Fouteau S, et al. Extended-spectrum β-lactamase-encoding genes are spreading on a wide range of Escherichia coli plasmids existing prior to the use of third-generation cephalosporins. Microb Genom. 2018;4 doi: 10.1099/mgen.0.000203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hall TA. BioEdit: a user-friendly biological sequence alignment editor and analysis program for window 95/98/NT. Nucleic Acids Symposium Series. 1999;41:95–98. [Google Scholar]
- 10.Guindon S, Gascuel O. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst Biol. 2003;52:696–704. doi: 10.1080/10635150390235520. [DOI] [PubMed] [Google Scholar]
- 11.Tavaré S. Some probabilistic and statistical problems in the analysis of DNA sequences. In American Mathematical Society: Lectures on Mathematics in the Life Sciences. 1986;17:57–86. [Google Scholar]
- 12.Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol. 2013;30:2725–2729. doi: 10.1093/molbev/mst197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cesareni G, Muesing MA, Polisky B. Control of ColE1 DNA replication: the rop gene product negatively affects transcription from the replication primer promoter. Proc Natl Acad Sci U S A. 1982;79:6313–6317. doi: 10.1073/pnas.79.20.6313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chan PT, Ohmori H, Tomizawa J, Lebowitz J. Nucleotide sequence and gene organization of ColE1 DNA. J Biol Chem. 1985;260:8925–8935. [PubMed] [Google Scholar]
- 15.Compain F, Poisson A, Le Hello S, Branger C, Weill F-X, et al. Targeting relaxase genes for classification of the predominant plasmids in Enterobacteriaceae . Int J Med Microbiol. 2014;304:236–242. doi: 10.1016/j.ijmm.2013.09.009. [DOI] [PubMed] [Google Scholar]
- 16.Zaleski P, Wolinowska R, Strzezek K, Lakomy A, Plucienniczak A. The complete sequence and segregational stability analysis of a new cryptic plasmid pIGWZ12 from a clinical strain of Escherichia coli . Plasmid. 2006;56:228–232. doi: 10.1016/j.plasmid.2006.05.004. [DOI] [PubMed] [Google Scholar]
- 17.Varsaki A, Moncalián G, Garcillán-Barcia MdelP, Drainas C, de la Cruz F. Analysis of ColE1 MbeC unveils an extended ribbon-helix-helix family of nicking accessory proteins. J Bacteriol. 2009;191:1446–1455. doi: 10.1128/JB.01342-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cannon PM, Strike P. Complete nucleotide sequence and gene organization of plasmid NTP16. Plasmid. 1992;27:220–230. doi: 10.1016/0147-619X(92)90024-5. [DOI] [PubMed] [Google Scholar]
- 19.Moran RA, Hall RM. Analysis of pCERC7, a small antibiotic resistance plasmid from a commensal ST131 Escherichia coli, defines a diverse group of plasmids that include various segments adjacent to a multimer resolution site and encode the same NikA relaxase accessory protein enabling mobilisation. Plasmid. 2017;89:42–48. doi: 10.1016/j.plasmid.2016.11.001. [DOI] [PubMed] [Google Scholar]
- 20.Anantham S, Hall RM. pCERC1, a small, globally disseminated plasmid carrying the dfrA14 cassette in the strA gene of the sul2-strA-strB gene cluster. Microb Drug Resist. 2012;18:364–371. doi: 10.1089/mdr.2012.0008. [DOI] [PubMed] [Google Scholar]
- 21.Enne VI, Bennett PM, Livermore DM, Hall LMC. Enhancement of host fitness by the sul2-coding plasmid p9123 in the absence of selective pressure. J Antimicrob Chemother. 2004;53:958–963. doi: 10.1093/jac/dkh217. [DOI] [PubMed] [Google Scholar]
- 22.Villa L, Capone A, Fortini D, Dolejska M, Rodríguez I, et al. Reversion to susceptibility of a carbapenem-resistant clinical isolate of Klebsiella pneumoniae producing KPC-3. J Antimicrob Chemother. 2013;68:2482–2486. doi: 10.1093/jac/dkt235. [DOI] [PubMed] [Google Scholar]
- 23.Stoesser N, Sheppard AE, Peirano G, Anson LW, Pankhurst L, et al. Genomic epidemiology of global Klebsiella pneumoniae carbapenemase (KPC)-producing Escherichia coli . Sci Rep. 2017;7:5917. doi: 10.1038/s41598-017-06256-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Iguchi A, Thomson NR, Ogura Y, Saunders D, Ooka T, et al. Complete genome sequence and comparative genome analysis of enteropathogenic Escherichia coli O127:H6 strain E2348/69. J Bacteriol. 2009;191:347–354. doi: 10.1128/JB.01238-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Haugum K, Johansen J, Gabrielsen C, Brandal LT, Bergh K, et al. Comparative genomics to delineate pathogenic potential in non-O157 Shiga toxin-producing Escherichia coli (STEC) from patients with and without haemolytic uremic syndrome (HUS) in Norway. PLoS One. 2014;9:e111788. doi: 10.1371/journal.pone.0111788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Van Melderen L, Saavedra De Bast M. Bacterial toxin–antitoxin systems: more than selfish entities? PLoS Genet. 2009;5:e1000437. doi: 10.1371/journal.pgen.1000437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kulakauskas S, Lubys A, Ehrlich SD. Dna restriction-modification systems mediate plasmid maintenance. J Bacteriol. 1995;177:3451–3454. doi: 10.1128/jb.177.12.3451-3454.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mnif B, Vimont S, Boyd A, Bourit E, Picard B, et al. Molecular characterization of addiction systems of plasmids encoding extended-spectrum beta-lactamases in Escherichia coli . J Antimicrob Chemother. 2010;65:1599–1603. doi: 10.1093/jac/dkq181. [DOI] [PubMed] [Google Scholar]
- 29.García-Fernández A, Villa L, Carta C, Venditti C, Giordano A, et al. Klebsiella pneumoniae ST258 producing KPC-3 identified in italy carries novel plasmids and OmpK36/OmpK35 porin variants. Antimicrob Agents Chemother. 2012;56:2143–2145. doi: 10.1128/AAC.05308-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Warburg G, Hidalgo-Grass C, Partridge SR, Tolmasky ME, Temper V, et al. A carbapenem-resistant Klebsiella pneumoniae epidemic clone in Jerusalem: sequence type 512 carrying a plasmid encoding aac(6')-Ib . J Antimicrob Chemother. 2012;67:898–901. doi: 10.1093/jac/dkr552. [DOI] [PubMed] [Google Scholar]
- 31.Haneda T, Okada N, Miki T, Danbara H. Sequence analysis and characterization of sulfonamide resistance plasmid pRF-1 from Salmonella enterica serovar Choleraesuis. Plasmid. 2004;52:218–224. doi: 10.1016/j.plasmid.2004.07.002. [DOI] [PubMed] [Google Scholar]
- 32.Hartman AB, Essiet II, Isenbarger DW, Lindler LE. Epidemiology of Tetracycline Resistance Determinants in Shigella spp. and Enteroinvasive Escherichia coli: Characterization and Dissemination of tet(A)-1 . J Clin Microbiol. 2003;41:1023–1032. doi: 10.1128/JCM.41.3.1023-1032.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kehrenberg C, Hopkins KL, Threlfall EJ, Schwarz S. Complete nucleotide sequence of a small qnrS1-carrying plasmid from Salmonella enterica subsp. enterica Typhimurium DT193. J Antimicrob Chemother. 2007;60:903–905. doi: 10.1093/jac/dkm283. [DOI] [PubMed] [Google Scholar]
- 34.Wu J-J, Ko W-C, Chiou C-S, Chen H-M, Wang L-R, et al. Emergence of Qnr determinants in human Salmonella isolates in Taiwan. J Antimicrob Chemother. 2008;62:1269–1272. doi: 10.1093/jac/dkn426. [DOI] [PubMed] [Google Scholar]
- 35.Akiyama T, Khan AA. Isolation and characterization of small qnrS1-carrying plasmids from imported seafood isolates of Salmonella enterica that are highly similar to plasmids of clinical isolates. FEMS Immunol Med Microbiol. 2012;64:429–432. doi: 10.1111/j.1574-695X.2011.00921.x. [DOI] [PubMed] [Google Scholar]
- 36.Campos MJ, Palomo G, Hormeño L, Herrera-León S, Domínguez L, et al. Prevalence of quinolone resistance determinants in non-typhoidal Salmonella isolates from human origin in Extremadura, Spain. Diagn Microbiol Infect Dis. 2014;79:64–69. doi: 10.1016/j.diagmicrobio.2014.01.010. [DOI] [PubMed] [Google Scholar]
- 37.Hammerl JA, Borowiak M, Schmoger S, Shamoun D, Grobbel M, et al. mcr-5 and a novel mcr-5.2 variant in Escherichia coli isolates from food and food-producing animals, Germany, 2010 to 2017. J Antimicrob Chemother. 2018;73:1433–1435. doi: 10.1093/jac/dky020. [DOI] [PubMed] [Google Scholar]
- 38.Borowiak M, Fischer J, Hammerl JA, Hendriksen RS, Szabo I, et al. Identification of a novel transposon-associated phosphoethanolamine transferase gene, mcr-5, conferring colistin resistance in d-tartrate fermenting Salmonella enterica subsp. enterica serovar Paratyphi B. J Antimicrob Chemother. 2017;72:3317–3324. doi: 10.1093/jac/dkx327. [DOI] [PubMed] [Google Scholar]
- 39.Wise MG, Estabrook MA, Sahm DF, Stone GG, Kazmierczak KM. Prevalence of mcr-type genes among colistin-resistant Enterobacteriaceae collected in 2014-2016 as part of the INFORM global surveillance program. PLoS One. 2018;13:e0195281. doi: 10.1371/journal.pone.0195281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yang QE, Walsh TR. Toxin–antitoxin systems and their role in disseminating and maintaining antimicrobial resistance. FEMS Microbiol Rev. 2017;41:343–353. doi: 10.1093/femsre/fux006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Burian J, Stuchlík S, Kay WW. Replication control of a small cryptic plasmid of Escherichia coli . J Mol Biol. 1999;294:49–65. doi: 10.1006/jmbi.1999.3266. [DOI] [PubMed] [Google Scholar]
- 42.Hiraga S, Sugiyama T, Itoh T. Comparative analysis of the replicon regions of eleven ColE2-related plasmids. J Bacteriol. 1994;176:7233–7243. doi: 10.1128/jb.176.23.7233-7243.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Labar AS, Millman JS, Ruebush E, Opintan JA, Bishar RA, et al. Regional dissemination of a trimethoprim-resistance gene cassette via a successful transposable element. PLoS One. 2012;7:e38142. doi: 10.1371/journal.pone.0038142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Moran RA, Hall RM. pBuzz: A cryptic rolling-circle plasmid from a commensal Escherichia coli has two inversely oriented oriTs and is mobilised by a B/O plasmid. Plasmid. 2019;101:10–19. doi: 10.1016/j.plasmid.2018.11.001. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.