Abstract
Malaria was present in most of Europe until the second half of the 20th century, when it was eradicated through a combination of increased surveillance and mosquito control strategies, together with cross-border and political collaboration. Despite the severe burden of malaria on human populations, it remains contentious how the disease arrived and spread in Europe. Here, we report a partial Plasmodium falciparum nuclear genome derived from a set of antique medical slides stained with the blood of malaria-infected patients from Spain’s Ebro Delta, dating to the 1940s. Our analyses of the genome of this now eradicated European P. falciparum strain confirms stronger phylogeographical affinity to present-day strains in circulation in central south Asia, rather than to those in Africa. This points to a longitudinal, rather than a latitudinal, spread of malaria into Europe. In addition, this genome displays two derived alleles in the pfmrp1 gene that have been associated with drug resistance. Whilst this could represent standing variation in the ancestral P. falciparum population, these mutations may also have arisen due to the selective pressure of quinine treatment, which was an anti-malarial drug already in use by the time the sample we sequenced was mounted on a slide.
Keywords: malaria, Plasmodium falciparum, ancient genomics, drug resistance
Data Summary
Plasmodium falciparum reads from Ebro-1944 have been deposited in the European Nucleotide Archive under accession number ERP114811. All modern P. falciparum samples used for the population genomics analyses were reported by the MalariaGEN Plasmodium falciparum Community Project in 2016 [1] and are provided in Table S1 (available in the online version of this article). All software used in the bioinformatic analyses are publicly available. Positions screened for anti-malarial drug resistance are available in Table S2.
Impact Statement.
Malaria is a serious infectious disease affecting over 200 million people annually. The disease is caused by species of parasitic protozoans from the genus Plasmodium, which are transmitted by several species of mosquitoes from the genus Anopheles. Today, Plasmodium is restricted to tropical and subtropical latitudes. However, malaria was historically present in most of Europe, spanning from southern Britain and the Mediterranean to as far north as Finland. Spain represented one of its last footholds, where it persisted until the 1960s. Here, we report a substantial fraction of the genome of a 20th century European Plasmodium falciparum strain isolated from slides stained with the blood of malaria-infected patients in the 1940s. We analyse this genome in the context of worldwide modern strains to trace the historical dispersal of P. falciparum in Europe. We find evidence supporting a longitudinal spread from Asia into Europe, over a latitudinal spread from Africa, as well as variants in anti-malarial resistance genes predating the use of most common anti-malarial drugs. Our work highlights the potential of collections of antique medical slides to open new possibilities in the study of ancient microbial genomics, including malaria.
Introduction
Classical Greek accounts in the fourth and fifth centuries BCE describe people with intermittent fevers and infectious symptoms characteristic of malaria [2]. The Roman author Celsius was able to accurately differentiate the clinical symptoms of Plasmodium vivax versus Plasmodium malariae infections [3]. In contrast, it is unclear whether Plasmodium falciparum, the deadliest form of the pathogen, was already present in classical times. While some authors argue that P. falciparum only spread to southern Europe with the dawn of the Roman Empire, historical accounts suggest it may have affected western and central Italy as early as 400–100 BCE [4], before reaching the Po Delta region of northern Italy around 1000 years later [3]. The spread of this pathogen through Italy in historical times is supported by the discovery of a large infant cemetery at Lugnano (Teverina, Umbria, Italy), dating to approximately 450 CE, and the report of P. falciparum ribosomal DNA sequences obtained from one of these skeletons by means of traditional PCR [5]. The recent retrieval of larger amounts of P. falciparum genetic data, by means of capture baits and second-generation sequencing technologies, from teeth sampled in Velia and Vagnari cemeteries (Italy), directly places this parasite in southern Italy by the beginning of the Roman Imperial period [6].
Genetic analyses of DNA recovered from a unique set of antique microscopy slides (1942–1944), stained with the blood of malaria-infected patients in the Ebro Delta of Spain for immediate diagnostic purposes, allowed us to report the complete mitochondrial DNA (mtDNA) genomes of historical P. falciparum and P. vivax [7]. In this study, we analyse data generated through merging the sequence information derived from four different slides, allowing us to reconstruct the partial nuclear genome of this eradicated European P. falciparum strain.
Methods
Sample collection
Four microscope slides, dated between 1942 and 1944, were selected for this study. All were obtained from Dr Ildefonso Canicio’s family collection. Dr Canicio was in charge of the anti-malarial hospital at Sant Jaume d’Enveja (Ebro Delta, Spain), inaugurated in 1925, until his death in 1961. Patients were predominantly local people who worked in the Ebro rice fields and had no history of international travel. The samples consist of four labelled microscopy slides stained with Giemsa – probably not previously fixed – in which parasites were still visible under the microscope. DNA extraction was performed in dedicated ancient DNA (aDNA) laboratories at the Institute of Evolutionary Biology in Barcelona (Spain) and the Centre for GeoGenetics in Copenhagen (Denmark) in 2015 and 2017, respectively, as described in the Supplementary Material.
Ancient sample mapping and assembly
We first analysed the sequenced reads obtained from our eradicated European P. falciparum samples using FastQC (v0.11.7) [8] in order to determine their quality before and after trimming of the adapter sequences. We removed sequencing adapters and reads shorter than 30 bp using cutadapt (v1.3) [9]. We then mapped our reads against the P. falciparum 3D7 and P. vivax Sal1 reference genomes using BWA (v0.7.17) [10] aln, specifying no seeding, a gap open penalty of 2, an edit distance of 0.01 and no trimming; parameters shown to optimize mapping of ancient microbial samples [11]. We removed duplicated reads using Picard (v2.18.6) ‘MarkDuplicates’ and retained all mapped reads with a map quality of at least 30 in SAMtools (v1.6) [12]. As the blood samples had known co-infection with P. vivax, we extracted the sequencing reads mapping more confidently to P. falciparum by selecting those reads that had a lesser edit distance between P. falciparum 3D7 than with P. vivax Sal1 (Fig. S1). The obtained G+C content of the reads matched the expected and extremely low G+C content characteristic of the P. falciparum genome (Fig. S2).
Post-mortem aDNA damage patterns were determined using MapDamage (v2.0.8) [13] (Fig. S3). Most of the P. falciparum reads (0.6294× mean coverage, representing 88 % of the total genome) come from a single slide (labelled CA) sequenced in 2015. The remaining reads come from three other slides with mean genome-wide coverage of 0.0213×, 0.0285× and 0.0314×. The reduced coverage obtained from these slides relates to the overall endogenous sequence content and quality, which may also vary depending on the stage of the parasite’s life cycle when the slides were prepared. To increase the overall coverage, we therefore merged data from all four slides, always calling the dominant allele. We call our resultant composite genome Ebro-1944.
We also generated a reference panel of modern P. falciparum samples from publicly available sequence data [1]. In all cases, we mapped reads using BWA (v0.7.17) [10] mem, before removing duplicated sequence reads and filtering by mapping quality using thresholds as described above. The same procedure was followed for mapping the genome of Plasmodium praefalciparum, which was used as an outgroup. Qualimap (v2.2) [14] was used to generate the mapping metrics for all samples.
Variant calling and dataset creation
To compare Ebro-1944 to current strains in global circulation, we selected modern strains with a mean depth of coverage equal or above 50× and with at least 90 % of the reference genome covered. This filtering strategy resulted in a dataset comprising 206 global samples. We used GATK (v3.7) [15], algorithm UnifiedGenotyper, to call variants, specifying EMIT_ALL_CONFIDENT_SITES and a standard confidence threshold over 50. We subsequently used VCFtools (0.1.14) [16] to filter out positions with less than 40× coverage, heterozygous positions, multi-allelic positions, indels and recombinant sub-telomeric regions (see the Supplementary Material). These samples were later merged using bcftools (v1.3.1) [12] and all variants that were not present in at least three samples were removed leaving 681 486 single nucleotide positions (SNPs). We used these positions to call additional modern samples, which mapped with lower coverage (<50×), applying the same filters. The resultant calls were merged using bcftools. SNPs were called for P. praefalciparum using the same parameters.
For Ebro-1944, we used a different approach to overcome the post-mortem damage associated with aDNA samples. We generated pseudo-haploid calls at the positions identified in the population genetics dataset using SAMtools to call a random base drawn from all possible reads at each site [17]. We then merged all the samples (including Ebro-1944) using bcftools. The complete dataset was filtered for a minimal minor allele frequency of 0.01 and multi-allelic SNPs were removed using VCFtools. All SNPs that were not present in at least 75 % of the samples considered were also removed. This procedure resulted in a final dataset comprising 435 samples and 14 346 bi-allelic SNPs, with 50.85 % of these covered in Ebro-1944.
Population genetics analyses
This dataset was compiled by filtering all modern samples for sites called in Ebro-1944 and retaining all samples that had at least 50 % of these sites covered. This resulted in a dataset of 306 samples and 6 755 SNPs. A principal component analysis (PCA) was performed on this dataset using SmartPCA within the EIG (v.6.0.1) suite of tools [18].
To formally test the relative affinity of Ebro-1944 to South Asian, Oceanian and African strains, we used the full 14 346 biallelic SNP dataset to generate f4 statistics of the form f4(P. praefalciparum, Ebro-1944; X, Y), where we use P. praefalciparum as an outgroup and iterate through all combinations of geographical samples (X and Y) included in our global dataset. This statistic is designed to quantify the covariance in allele frequency differences between P. praefalciparum and Ebro-1944, and X and Y. If P. praefalciparum and Ebro-1944 form a clade with respect to X and Y, then their allele frequency differences should be uncorrelated and f4 will have a value of 0. Deviations from 0, thus, provide the relative affinity of Ebro-1944 to X and Y; positive values indicating a closer relationship to Y relative to X and negative values indicating a closer relationship to X relative to Y. f4 statistics were generated in qpDstat of AdmixTools (v.5.0) [19] and statistical significance was assessed through Z-scores following block jack-knife resampling (Fig. S4).
We additionally sub-sampled the global population genetics dataset more stringently, retaining only the strains with no missingness across the entire alignment, which led to a dataset of 30 strains over 8195 sites (Table S1). We generated patterns of pairwise haplotype sharing across this dataset using Chromopainter v2 [20], specifying a uniform recombination rate of 9.6 kb cM−1 [21] (Fig. S5) and using default estimates of mutation and switch parameters.
We also considered the mitochondrial relationships by placing our Ebro-1944 mtDNA genome (16.1×) in a minimum spanning network together with 435 global strains (Fig. S6). The mtDNA genome was also screened for specific variants restricted to South Asian strains.
Resistance variants screening
A set of variants previously reported in the literature as associated with resistance to different anti-malarial drugs (Table S2) was screened in Ebro-1944 [22–46]. The position of the derived allele was determined using Jvarkit (v.2018.04.05) [47].
Results
Our sequencing of the four antique microscopy slide samples yielded 218 952 P. falciparum reads producing a nuclear genome of 0.67× mean depth and covering 40.99 % of the P. falciparum reference genome 3D7 (Fig. S1). In addition, 1398 reads mapped to the mtDNA genome, generating a mean coverage of 16.1× over 99.43 % of the reference genome. Sequence reads showed damage patterns typical of aDNA (C to T and G to A substitutions at the 5′ and 3′ ends) in ratios of 3.8 and 2.2 %, respectively (Fig. S3). The characteristic aDNA damage of miscoding lesions indicates that our P. falciparum reads are authentically old, deriving from our historic specimen rather than from modern contamination where we would expect no such damage profile.
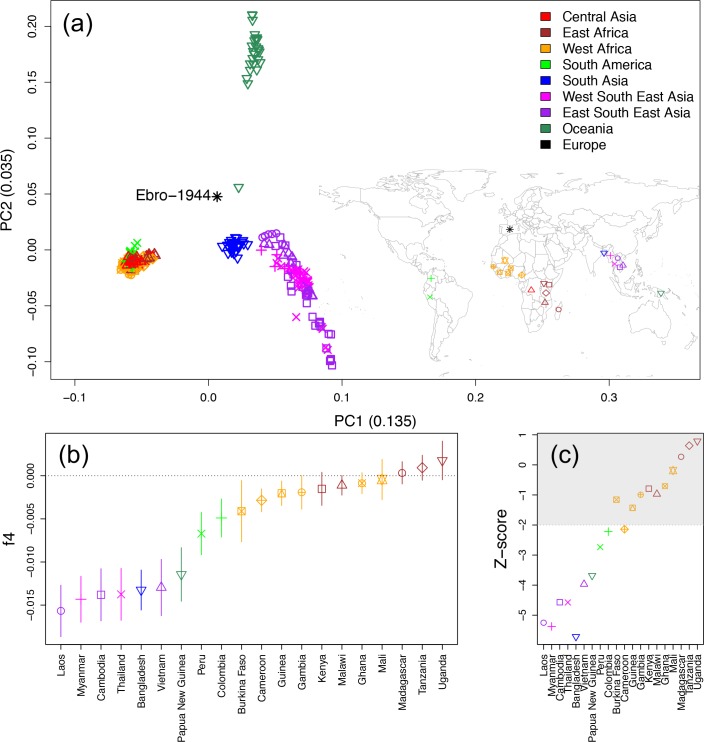
PCA (Fig. 1a), f4 allele sharing statistics (Figs 1b, c, S4) and haplotype sharing analyses (Fig. S5) indicate that the closest genetic affinity of our European strain is to contemporary samples from central south Asia, including those currently in circulation in Laos, Myanmar and Vietnam. We also detect a shared genetic component with samples from Papua New Guinea, in particular PN0008-C (Fig. 1a). Whilst Ebro-1944 is more closely related to current strains from central south Asia than those from Africa, we found a higher proportion of African haplotypes in Ebro-1944 than in any central south Asian samples (two sample t-test P<2.2×10−16). Such a pattern would be consistent with a common origin of the European and central south Asian P. falciparum populations, followed by secondary introgression of African strains into the European population after the split between the European and central south Asian lineages (Fig. S5).
Fig. 1.
(a) PCA with worldwide sample locations (inset map). (b) f4-statistics under the test relationship f4(P. praefalciparum, Ebro-1944, X, Democratic Republic of Congo), where X iterates through the geographical sampling locations of our included modern P. falciparum strains. A more negative f4 value indicates a closer relationship of Ebro-1944 to X relative to strains sampled from the Democratic Republic of Congo. (c) Z-scores under the test relationship f4(P. praefalciparum, Ebro-1944, X, Colombia) assessed through block jack-knife resampling. All possible f4 topologies were tested (Fig. S4), with Ebro-1944 showing a consistent closer affinity to strains sampled from central south Asia.
The close genetic affinity of the nuclear genome to strains in circulation in central south Asia is further confirmed at the mtDNA level (Fig. S6). Ebro-1944 carries two Indian-specific mtDNA substitutions at positions 276 and 2763 [48]. One additional mutation (position 725) is also present in our sample and is shared with nine contemporary Indian samples and two African samples included in our modern reference dataset (Table S1). We observe mutations at these three positions across all four of our sampled slides, suggesting the strains combined in Ebro-1944 are phylogenetically similar (Table S3). Unfortunately, these three mutations are not covered in a previously published partial strain retrieved from Roman cemetery sites [6]. Additionally, none of our reads overlapped with the ribosomal DNA fragment of European P. falciparum retrieved in a previous work [5].
We screened the Ebro-1944 nuclear genome for the presence of 117 variants that have been previously associated with anti-malarial drug resistance (Table S2). We achieved sequence coverage of at least one read at 62 positions. Of these, only two mutations (H191Y and I876V) displayed the resistance-associated allele (Table S2). Both alleles are located in the multidrug resistance protein 1 encoding gene (pfmrp1), which encodes an ABC family transporter, and has been associated to alterations in quinine, chloroquine, artemisinin, piperaquine and primaquine sensitivity [49]. Although one of these mutations, H191Y, is a C to T substitution that is characteristic of post-mortem DNA deamination, and might, thus, be expected in ancient samples, we believe this to be authentic as the transition is present in two overlapping reads, and is not located at the end of the reads where damage tends to accumulate [50]. While the second variant, I876V, is only covered by a single read, it is an A to G substitution and is, thus, not a common DNA damage motif. To provide additional support for the presence of these derived alleles, we imputed these positions in our partial genome by using a reference panel of modern strains carrying the H191Y and I876V variants (see the Supplementary Material). Interestingly, while the latter variant is distributed worldwide, the former is currently restricted to Asia and Oceania, consistent with an Asian dispersal of P. falciparum into Europe.
Discussion
We showed that an eradicated European P. falciparum strain from the 20th century is most closely related to extant strains from central south Asia, such as those currently in circulation in Laos, Myanmar and Vietnam. Although we detect some evidence for secondary introgression of African P. falciparum into the extinct European population, the significantly stronger affinity to Asian strains argues against a direct African origin [4] of European P. falciparum, and points instead to a migration event between Europe and Asia.
We cannot infer the directionality of the migration between Europe and Asia from the genetic evidence alone. A recent expansion of P. falciparum from Europe to Asia and Oceania, coinciding with European colonial expansion, might be conceivable given the well-characterized role of colonialism in the widespread dissemination of other major infectious diseases, such as Mycobacterium tuberculosis lineage 4, the globally distributed agent of tuberculosis [51].
However, a migration of P. falciparum from Europe to Asia does not sit well with historical evidence of the arrival of the parasite in Europe during antiquity [2–6]. Given that our extinct European strain shares significantly more alleles with extant Asian rather than African strains, the arrival of P. falciparum malaria parasites into Europe likely took place from the Asian sub-continent rather than spreading from Africa via the Mediterranean during the Roman Empire.
Plausible historical migrations responsible for a spread into Europe from the East include the extensive commercial exchanges and movement between people of various ethnicities in the Achaemenid Empire (550-330 BCE). Alternatively, P. falciparum might have reached Europe during the subsequent Hellenistic period, connecting India with the Mediterranean, following the conquest of the Achaemenid Empire by Alexander the Great.
The availability of a strain pre-dating most of the currently used anti-malarial drugs allowed us to look for the presence of resistance variants that may inform on the spread of such resistances in the future. The two resistance variants we observed in the pfmrp1 gene could be explained by standing variation for drug-resistance mutations in P. falciparum or may have arisen following the use of quinine for over three centuries; chloroquines were not introduced in Spain until 1948 and initially only in the African colonies.
Our results provide novel insights into the evolution and past demography of one of the world’s deadliest pathogens, which could not have been reached by studying the genomes of extant strains alone. Additional genomic evidence from both medical collections and ancient remains will be needed to reconstruct more precise timings and routes for the spread of malaria into Europe, and could also help in determining the emergence and drivers of resistance to anti-malarial drugs.
Data bibliography
1. The Plasmodium falciparum reference genome used was P. falciparum 3D7, assembly ASM276v2.
2. The Plasmodium vivax reference genome used was P. vivax Sal-1, assembly ASM241v2.
3. Details of the Plasmodium falciparum samples used in the population genetics analyses can be found under the study accession number PRJEB2136. Accession numbers for each sample are described in Table S1.
4. The sequence of Plasmodium praefalciparum can be found under accession number SAMEA2464702.
Supplementary Data
Funding information
C. L. -F. is supported by Obra Social ‘La Caixa’, Secretaria d’Universitats i Recerca Programme del Departament d’Economia i Coneixement de la Generalitat de Catalunya (GRC 2017 SGR 880) and by FEDER-MINECO (PGC2018-095931-B-100). L. v. D. and F. B. acknowledge financial support from the MRC Newton Fund UK–China NSFC initiative (grants MR/P007597/1 and 81661138006).
Acknowledgements
We are grateful to the descendants of Dr Canicio, Miquel and Ildefons Oliveras, for sharing their slides with us.
Author contributions
F. B., M. T. P. G. and C. L.-F. conceived the research; C. A. and R. E. collected the samples and associated information; C. C., M. S.-V. and R. F. performed experimental procedures; T. d.-D., L. v. D. and P. G. performed computational analysis; S. H. provided information on resistance variants; T. d.-D., L. v. D., F. B., M. T. P. G. and C. L.-F wrote the paper.
Conflicts of interest
The authors declare that there are no conflicts of interest.
Ethical statement
Experimental protocols have been approved by the Clinical Research Ethics Committee of Institut Hospital del Mar d'Investigacions Mèdiques (CEIC-PSMAR).
Footnotes
Abbreviations: aDNA, ancient DNA; mtDNA, mitochondrial DNA; PCA, principal component analysis; SNP, single nucleotide position.
All supporting data, code and protocols have been provided within the article or through supplementary data files. Supplementary material is available with the online version of this article.
References
- 1.MalariaGEN Plasmodium falciparum Community Project Genomic epidemiology of artemisinin resistant malaria. 2016 doi: 10.7554/eLife.08714. https://elifesciences.org/articles/08714 [DOI] [PMC free article] [PubMed]
- 2.Jones WHS. Malaria, a Neglected Factor in the History of Greece and Rome. Cambridge: Cambridge University Press; 1907. pp. 97–102. [Google Scholar]
- 3.Sallares R, Bouwman A, Anderung C. The spread of malaria to southern Europe in antiquity: new approaches to old problems. Med Hist. 2004;48:311–328. doi: 10.1017/S0025727300007651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.De Zulueta J. Malaria and Mediterranean history. Parassitologia. 1973;15:1–15. [PubMed] [Google Scholar]
- 5.Sallares R, Gomzi S. Biomolecular archaeology of malaria. Ancient Biomolecules. 2001;3:195–213. [Google Scholar]
- 6.Marciniak S, Prowse TL, Herring DA, Klunk J, Kuch M, et al. Plasmodium falciparum malaria in 1st–2nd century CE southern Italy. Current Biology. 2016;26:R1220–R1222. doi: 10.1016/j.cub.2016.10.016. [DOI] [PubMed] [Google Scholar]
- 7.Gelabert P, Sandoval-Velasco M, Olalde I, Fregel R, Rieux A, et al. Mitochondrial DNA from the eradicated European Plasmodium vivax and P. falciparum from 70-year-old slides from the Ebro Delta in Spain. Proc Natl Acad Sci USA. 2016;113:11495–11500. doi: 10.1073/pnas.1611017113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Andrews S. FastQC, a quality control tool for high throughput sequence data. 2010 http://www.Bioinformatics.Babraham.Ac.Uk/Projects/Fastqc/
- 9.Martin M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet.Journal. 2011;17:10–12. [Google Scholar]
- 10.Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Key FM, Posth C, Krause J, Herbig A, Bos KI. Mining metagenomic data sets for ancient DNA: recommended protocols for authentication. Trends Genet. 2017;33:508–520. doi: 10.1016/j.tig.2017.05.005. [DOI] [PubMed] [Google Scholar]
- 12.Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jónsson H, Ginolhac A, Schubert M, Johnson PLF, Orlando L, et al. mapDamage2.0: fast approximate Bayesian estimates of ancient DNA damage parameters. Bioinformatics. 2013;29:1682–1684. doi: 10.1093/bioinformatics/btt193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.García-Alcalde F, Okonechnikov K, Carbonell J, Cruz LM, Götz S, et al. Qualimap: evaluating next-generation sequencing alignment data. Bioinformatics. 2012;28:2678–2679. doi: 10.1093/bioinformatics/bts503. [DOI] [PubMed] [Google Scholar]
- 15.Van der Auwera GA, Carneiro MO, Hartl C, Poplin R, del Angel G, et al. From FastQ data to high-confidence variant calls: the Genome Analysis Toolkit best practices pipeline. Curr Protoc Bioinformatics. 2013;43:11.10.1–11.10.33. doi: 10.1002/0471250953.bi1110s43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Danecek P, Auton A, Abecasis G, Albers CA, Banks E, et al. The variant call format and VCFtools. Bioinformatics. 2011;27:2156–2158. doi: 10.1093/bioinformatics/btr330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Green RE, Krause J, Briggs AW, Maricic T, Stenzel U, et al. A draft sequence of the Neandertal genome. Science. 2010;328:710–722. doi: 10.1126/science.1188021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Price AL, Patterson NJ, Plenge RM, Weinblatt ME, Shadick NA, et al. Principal components analysis corrects for stratification in genome-wide association studies. Nat Genet. 2006;38:904–909. doi: 10.1038/ng1847. [DOI] [PubMed] [Google Scholar]
- 19.Patterson N, Moorjani P, Luo Y, Mallick S, Rohland N, et al. Ancient admixture in human history. Genetics. 2012;192:1065–1093. doi: 10.1534/genetics.112.145037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lawson DJ, Hellenthal G, Myers S, Falush D. Inference of population structure using dense haplotype data. PLoS Genet. 2012;8:e1002453. doi: 10.1371/journal.pgen.1002453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jiang H, Li N, Gopalan V, Zilversmit MM, Varma S, et al. High recombination rates and hotspots in a Plasmodium falciparum genetic cross. Genome Biol. 2011;12:R33. doi: 10.1186/gb-2011-12-4-r33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Foote SJ, Kyle DE, Martin RK, Oduola AM, Forsyth K, et al. Several alleles of the multidrug-resistance gene are closely linked to chloroquine resistance in Plasmodium falciparum . Nature. 1990;345:255–258. doi: 10.1038/345255a0. [DOI] [PubMed] [Google Scholar]
- 23.Price RN, Cassar C, Brockman A, Duraisingh M, van Vugt M, et al. The pfmdr1 gene is associated with a multidrug-resistant phenotype in Plasmodium falciparum from the Western border of Thailand. Antimicrob Agents Chemother. 1999;43:2943–2949. doi: 10.1128/AAC.43.12.2943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dahlström S, Ferreira PE, Veiga MI, Sedighi N, Wiklund L, et al. Plasmodium falciparum multidrug resistance protein 1 and artemisinin-based combination therapy in Africa. J Infect Dis. 2009;200:1456–1464. doi: 10.1086/606009. [DOI] [PubMed] [Google Scholar]
- 25.Rottmann M, McNamara C, Yeung BKS, Lee MCS, Zou B, et al. Spiroindolones, a potent compound class for the treatment of malaria. Science. 2010;329:1175–1180. doi: 10.1126/science.1193225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Setthaudom C, Tan-ariya P, Sitthichot N, Khositnithikul R, Suwandittakul N, et al. Role of Plasmodium falciparum chloroquine resistance transporter and multidrug resistance 1 genes on in vitro chloroquine resistance in isolates of Plasmodium falciparum from Thailand. Am J Trop Med Hyg. 2011;85:606–611. doi: 10.4269/ajtmh.2011.11-0108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Veiga MI, Ferreira PE, Jörnhagen L, Malmberg M, Kone A, et al. Novel polymorphisms in Plasmodium falciparum ABC transporter genes are associated with major ACT antimalarial drug resistance. PLoS One. 2011;6:e20212. doi: 10.1371/journal.pone.0020212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Takala-Harrison S, Clark TG, Jacob CG, Cummings MP, Miotto O, et al. Genetic loci associated with delayed clearance of Plasmodium falciparum following artemisinin treatment in Southeast Asia. Proc Natl Acad Sci USA. 2013;110:240–245. doi: 10.1073/pnas.1211205110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gupta B, Xu S, Wang Z, Sun L, Miao J, et al. Plasmodium falciparum multidrug resistance protein 1 (pfmrp1) gene and its association with in vitro drug susceptibility of parasite isolates from north-east Myanmar. J Antimicrob Chemother. 2014;69:2110–2117. doi: 10.1093/jac/dku125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Vaidya AB, Morrisey JM, Zhang Z, Das S, Daly TM, et al. Pyrazoleamide compounds are potent antimalarials that target Na+ homeostasis in intraerythrocytic Plasmodium falciparum . Nat Commun. 2014;5:5521. doi: 10.1038/ncomms6521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Miotto O, Amato R, Ashley EA, MacInnis B, Almagro-Garcia J, et al. Genetic architecture of artemisinin-resistant Plasmodium falciparum . Nat Genet. 2015;47:226–234. doi: 10.1038/ng.3189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pelleau S, Moss EL, Dhingra SK, Volney B, Casteras J, et al. Adaptive evolution of malaria parasites in French Guiana: reversal of chloroquine resistance by acquisition of a mutation in pfcrt . Proc Natl Acad Sci USA. 2015;112:11672–11677. doi: 10.1073/pnas.1507142112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Callaghan PS, Siriwardana A, Hassett MR, Roepe PD. Plasmodium falciparum chloroquine resistance transporter (PfCRT) isoforms PH1 and PH2 perturb vacuolar physiology. Malar J. 2016;15:186. doi: 10.1186/s12936-016-1238-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Reed MB, Saliba KJ, Caruana SR, Kirk K, Cowman AF. Pgh1 modulates sensitivity and resistance to multiple antimalarials in Plasmodium falciparum . Nature. 2000;403:906–909. doi: 10.1038/35002615. [DOI] [PubMed] [Google Scholar]
- 35.Mishra N, Bharti RS, Mallick P, Singh OP, Srivastava B, et al. Emerging polymorphisms in falciparum Kelch 13 gene in Northeastern region of India. Malar J. 2016;15:4–9. doi: 10.1186/s12936-016-1636-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wang Z, Cabrera M, Yang J, Yuan L, Gupta B, et al. Genome-wide association analysis identifies genetic loci associated with resistance to multiple antimalarials in Plasmodium falciparum from China-Myanmar border. Sci Rep. 2016;6:33891.. doi: 10.1038/srep33891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ye R, Hu D, Zhang Y, Huang Y, Sun X, et al. Distinctive origin of artemisinin-resistant Plasmodium falciparum on the China-Myanmar border. Sci Rep. 2016;6:20100. doi: 10.1038/srep20100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kobasa T, Talundzic E, Sug-aram R, Boondat P, Goldman IF, et al. Emergence and spread of kelch13 mutations associated with artemisinin resistance in Plasmodium falciparum parasites in 12 Thai provinces from 2007 to 2016. Antimicrob Agents Chemother. 2018;62:e02141-17. doi: 10.1128/AAC.02141-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ross LS, Dhingra SK, Mok S, Yeo T, Wicht KJ, et al. Emerging Southeast Asian PfCRT mutations confer Plasmodium falciparum resistance to the first-line antimalarial piperaquine. Nat Commun. 2018;9:3314.. doi: 10.1038/s41467-018-05652-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mu J, Ferdig MT, Feng X, Joy DA, Duan J, et al. Multiple transporters associated with malaria parasite responses to chloroquine and quinine. Mol Microbiol. 2003;49:977–989. doi: 10.1046/j.1365-2958.2003.03627.x. [DOI] [PubMed] [Google Scholar]
- 41.Pickard AL, Wongsrichanalai C, Purfield A, Kamwendo D, Emery K, et al. Resistance to antimalarials in Southeast Asia and genetic polymorphisms in PfMDR1. Antimicrob Agents Chemother. 2003;47:2418–2423. doi: 10.1128/AAC.47.8.2418-2423.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Durrand V, Berry A, Sem R, Glaziou P, Beaudou J, et al. Variations in the sequence and expression of the Plasmodium falciparum chloroquine resistance transporter (Pfcrt) and their relationship to chloroquine resistance in vitro. Mol Biochem Parasitol. 2004;136:273–285. doi: 10.1016/j.molbiopara.2004.03.016. [DOI] [PubMed] [Google Scholar]
- 43.Happi CT, Gbotosho GO, Folarin OA, Akinboye DO, Yusuf BO, et al. Polymorphisms in Plasmodium falciparum dhfr and dhps genes and age related in vivo sulfadoxine-pyrimethamine resistance in malaria-infected patients from Nigeria. Acta Trop. 2005;95:183–193. doi: 10.1016/j.actatropica.2005.06.015. [DOI] [PubMed] [Google Scholar]
- 44.Sidhu ABS, Valderramos SG, Fidock DA. Pfmdr1 mutations contribute to quinine resistance and enhance mefloquine and artemisinin sensitivity in Plasmodium falciparum . Mol Microbiol. 2005;57:913–926. doi: 10.1111/j.1365-2958.2005.04729.x. [DOI] [PubMed] [Google Scholar]
- 45.Echeverry DF, Holmgren G, Murillo C, Higuita JC, Björkman A, et al. Polymorphisms in the pfcrt and pfmdr1 genes of Plasmodium falciparum and in vitro susceptibility to amodiaquine and desethylamodiaquine. Am J Trop Med Hyg. 2007;77:1034–1038. doi: 10.4269/ajtmh.2007.77.1034. [DOI] [PubMed] [Google Scholar]
- 46.Dahlström S, Veiga MI, Mårtensson A, Björkman A, Gil JP. Polymorphism in PfMRP1 (Plasmodium falciparum multidrug resistance protein 1) amino acid 1466 associated with resistance to sulfadoxine-pyrimethamine treatment. Antimicrob Agents Chemother. 2009;53:2553–2556. doi: 10.1128/AAC.00091-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pierre L. JVarkit: java-based utilities for bioinformatics. 2015.
- 48.Tyagi S, Pande V, Das A. New insights into the evolutionary history of Plasmodium falciparum from mitochondrial genome sequence analyses of Indian isolates. Mol Ecol. 2014;23:2975–2987. doi: 10.1111/mec.12800. [DOI] [PubMed] [Google Scholar]
- 49.Raj DK, Mu J, Jiang H, Kabat J, Singh S, et al. Disruption of a Plasmodium falciparum multidrug resistance-associated protein (PfMRP) alters its fitness and transport of antimalarial drugs and glutathione. J Biol Chem. 2009;284:7687–7696. doi: 10.1074/jbc.M806944200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Briggs AW, Stenzel U, Johnson PLF, Green RE, Kelso J, et al. Patterns of damage in genomic DNA sequences from a Neandertal. Proc Natl Acad Sci USA. 2007;104:14616–14621. doi: 10.1073/pnas.0704665104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Brynildsrud OB, Pepperell CS, Suffys P, Grandjean L, Monteserin J, et al. Global expansion of Mycobacterium tuberculosis lineage 4 shaped by colonial migration and local adaptation. Sci Adv. 2018;4:eaat5869. doi: 10.1126/sciadv.aat5869. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.