Abstract
Background
Plasma miR‐21 is widely investigated as biomarker in many diseases. Recent studies show that miR‐21 participates in the development of systemic lupus erythematosus (SLE). The aim of this study was to evaluate the expression profile of miR‐21 in the plasma of SLE patients.
Methods
Relative quantities of plasma miR‐21 both in SLE patients and healthy controls were determined by relative qRT‐PCR under endogenous and exogenous controls. The diagnostic value of plasma miR‐21 was evaluated in SLE patients. Data of some SLE‐associated clinical parameters were collected.
Results
Eighty participants from Central China were recruited. Forty‐four participants were new‐onset SLE patients and the others were healthy controls. Plasma miR‐21 level in SLE patients was higher than that of healthy controls (P = 0.031). Receiver operating characteristic analysis of plasma miR‐21 revealed an Area Under Curve (AUC) of 0.64 ± 0.06 (95% CI: 0.51–0.76, P = 0.03854) when differentiating SLE from healthy controls. The level of plasma miR‐21 was not associated with the level of white blood cells (P = 0.4284), red blood cells (P = 0.4079), and platelets (P = 0.4961), but significantly correlated with the level of plasma complement C3 (r = −0.5297, P = 0.0004), C4 (r = −0.4732, P = 0.0020), and serum uric acid (r = 0.3932, P = 0.0121) in SLE patients.
Conclusions
Plasma miR‐21 in SLE patients from Central China is overexpressed. Since circulating miR‐21 is aberrantly expressed in many diseases, the applying of it as a disease biomarker should be considered carefully.
Keywords: miR‐21, systemic lupus erythematosus, plasma, biomarker, real‐time polymerase chain reaction
INTRODUCTION
Systemic lupus erythematosus (SLE) is a complex autoimmune disease due to immunopathogenic abnormalities. Immune dysregulation leads to excess production of autoantibodies and immune complexes, excess complement activation, and then immune damage to different organs, which together cause a diverse clinical syndrome in patients, including arthritis, nephritis, and skin rashes. The incidence rate of SLE in Chinese Beijing population is 0.03% 1, while the rate in Chinese Hong Kong population increases to 0.1% 2. Thirty‐six percent of SLE patients would become disabled within 5 years of disease onset 2. Although significant progress has been made, it is difficult to make the early diagnosis of SLE. A number of autoantibodies have been used as biomarkers for the diagnosis of SLE. However, there is no perfect one. Reliable biomarkers are urgently needed for the diagnosis, monitoring, and stratification of SLE.
MicroRNAs (miRNAs), as a notable class of genetic regulators, are a class of endogenous small single‐strand RNAs 21–25 nucleotide in length. Recent studies show that miRNAs are important regulators in the development of SLE and are potential biomarkers for SLE 3. For example, serum miR‐146a, miR‐155, and miR‐200b were lower in SLE patients than in healthy controls 4, 5, 6. miR‐21, one of the early discovered small noncoding RNAs, was widely investigated as potential biomarker in cancer and cardiovascular disease 7, 8, 9, 10, 11. For example, miR‐21 is indicated to be an independent prognostic factor of gastric cancer 12, 13. As surgery can reduce the level of circulating miR‐21 and poor postoperative survival rate always relates with higher level of circulating miR‐21 12, 13, so far, the profile of circulating miR‐21 in autoimmune diseases has been barely studied.
Growing evidence has shown that the aberrant expressed cellular miR‐21 is a contributor of SLE 14, 15. Recently, two studies explored the expression of circulating miR‐21 in SLE patients of different populations, but they got different results 6, 16. The aim of this study was to evaluate the expression profile of miR‐21 in the plasma of SLE patients from Central China.
MATERIAL AND METHODS
Sample Collection
The study was carried out followed the Declaration of Helsinki and approved by Ethical Committee of Wuhan Union Hospital, Huazhong University of Science and Technology. Written informed consents were got from all the participants. SLE patients were recruited from June 2011 to January 2012 at Wuhan Union Hospital of Huazhong University of Science and Technology, China. The SLE patients were diagnosed based on the American College of Rheumatology criteria. All the patients were newly diagnosed and blood samples were collected before any drug therapy. Subjects used as healthy controls were recruited from the Center of Health Examination, Wuhan Union Hospital. No evidence of diseases, especially SLE, other autoimmune diseases, infection diseases, tumors, endocrine diseases, and cardiovascular diseases, was found in these healthy controls.
Two milliliters of blood samples were collected at early morning. The samples were centrifuged at 3,000 g for 5 min and the supernatants were transferred into Eppendorf tubes for another centrifugation at 12,000 g for 10 min. The final supernatants were stored at −80°C until use. All the samples were processed within 4 h after collection.
miRNA Extraction
The extraction of plasma miRNA was carried out by miRcute miRNA isolation kit (Tiangen Biotech Co., Beijing, China) according to the manufacturer's protocol. Briefly, 400 μl plasma was mixed with same volume of denaturing solution. Five microliters of synthetic cel‐miR‐39 (Caenorhabditis elegans miRNA, 5 fmol/μl stock solution) was added. Cel‐miR‐39 was used as an exogenous reference 17. miRNAs were eluted by 30 μl RNase‐free water.
Reverse Transcription
miRcute miRNA first‐strand cDNA synthesis kit (Tiangen Biotech Co., Beijing, China) was used to synthesize the cDNA of miRNA. The adding poly(A) reaction was in a 20 μl volume: 11.6 μl RNase‐free ddH2O, 2 μl 10× reverse‐transcription buffer, 0.4 μl poly(A) polymerase (5 U/μl), 4 μl 5× rATP solution, and 2 μl miRNAs. The mixture reacted at 37°C for 60 min. Two microliters of products were used for reverse transcription and the other products were stored at −80°C. The reverse transcription was also in a 20 μl volume: 11.5 μl RNase‐free ddH2O, 1 μl RNasin (40 U/μl), 2 μl 10× RT buffer, 2 μl 10× RT primer, 1 μl dNTP mixture (2.5 mM each), 0.5 μl Quant RTase, and 2 μl poly(A) reaction product. The reaction condition was at 37°C for 60 min. The synthesized cDNAs were stored at −80°C until use.
Quantitative RT‐PCR Analyses
miRcute miRNA qPCR detection kit (Tiangen Biotech Co., Beijing, China) was used for quantitative RT‐PCR analysis. The reaction mixture for qRT‐PCR was in a 20 μl volume including 7.2 μl ddH2O, 10 μl 2× miRcute miRNA premix (including SYBR and ROX), 0.4 μl reverse primer (10 μM), 0.4 μl forward primer (10 μM), and 2 μl cDNA. This mixture was aliquoted into duplicate. The reactions conditions were as follows: 94°C for 2 min, followed by 40 cycles of 94°C for 2 sec and 60°C for 34 sec. The reactions were performed on ABI step‐one (Applied Biosystems, FosterCity, California). Cycle thresholds (CT) were automatically set. The specific primer sequences were the following: for miR‐21—GGGTAGCTTATCAGACTGATGTTGAA, for U6b—ACGCAAATTCGTGAAGCGTT, and for cel‐miR‐39—CACCGGGTGTAAATCAGCTTG. Both U6b and cel‐miR‐39 were used as reference controls 17, 18, 19. According to Tomasetti's description 19, the expression of miR‐21 was normalized to U6b and cel‐miR‐39 (ΔCTmiR‐21, CTmiR‐21‐CTU6b‐CTcel‐miR39). Higher ΔCTmiR‐21 means lower level of miR‐21. The relative level of miR‐21 in SLE compared to healthy control was calculated according to the 2−ΔΔCT method 20. Quantitative RT‐PCR reactions were performed in duplicate and the mean values were recorded.
Statistical Analysis
The normalized concentrations of miR‐21 were expressed as mean ± SD. t‐Test was used to evaluate the difference of miR‐21 concentrations between SLE patients and healthy controls. The associations between miR‐21 concentration and the clinical parameters of SLE were evaluated by Spearman's rank correlation test. Receiver operating characteristics (ROC) curves were performed to evaluate the diagnostic efficiency. P‐value of less than 0.05 was considered statistically significant. All probabilities were two‐tailed. SPSS 13.0 (SPSS Inc.) was used to make statistical analysis.
RESULTS
The Clinical Characteristics of Participants
Forty‐four newly diagnosed SLE patients and thirty‐six healthy controls were recruited in this study (Table 1). All of them were from Central China. Healthy controls were age‐ and sex‐matched with SLE patients. Blood samples were collected before any drug intervention. All the patients' Systemic Lupus Erythematosus Disease Activity Index (SLEDAI) values are equal or above 10.
Table 1.
Clinical Characteristics of the Participants
| SLE | Healthy controls | |
|---|---|---|
| No. of cases | 44 | 36 |
| No. men/no. women | 4/40 | 4/32 |
| Age, median (range) years | 39(18–67) | 38(20–65) |
| Disease manifestations, number (%) | ||
| Renal disease | 28(64) | 0 |
| Vasculitis | 13(30) | 0 |
| Arthritis | 11(25) | 0 |
| Rash | 5(11) | 0 |
| Alopecia | 8(18) | 0 |
| Mucosal ulcers | 10(23) | 0 |
| Serositis | 7(16) | 0 |
| Leukopenia | 12(27) | 0 |
| Thrombocytopenia | 6(14) | 0 |
| Fever | 8(18) | 0 |
| Visual disturbance | 0 | 0 |
| Anti‐dsDNA positive, number (%) | 25(57) | 0 |
| SLEDAI | 13.91 ± 3.70 | 0 |
| Proteinuria (mg/24 h) | 1,899.58 ± 2,389.02 | N/A |
| Serum creatinine (μmol/l) | 72.84 ± 37.22 | 64.57 ± 20.79 |
| Urea nitrogen (mmol/l) | 6.93 ± 3.75 | 5.47 ± 1.19 |
| Uric acid (μmol/l) | 360.70 ± 138.60 | 316.02 ± 56.32 |
| Cystatin C (mg/l) | 2.00 ± 0.98 | 0.83 ± 0.31 |
| C3 (g/l) | 0.55 ± 0.34 | 1.15 ± 0.34 |
| C4 (g/l) | 0.12 ± 0.11 | 0.28 ± 0.10 |
The quantity data are expressed as mean ± SD, except where indicated otherwise.
N/A, not available.
The Plasma Levels of miR‐21 in SLE Patients and Normal Controls
The plasma level of miR‐21 was higher in SLE patients than in normal controls (33.31 ± 3.39 (95% CI: 32.28–34.34, P = 0.031) vs. 31.41 ± 3.47 (95% CI: 30.23–32.58, P = 0.031); Fig. 1). Compared with the normal controls, on average, SLE patients had a 3.73‐fold increase in miR‐21 level. The ROC curves analysis showed that the AUC value was 0.64 ± 0.06 (95% CI: 0.51–0.76, P = 0.03676; Fig. 2). When the relative value of plasma miR‐21 reaches 33.07, the corresponding sensitivity was 52.27% and the corresponding specificity was 72.22%.
Figure 1.
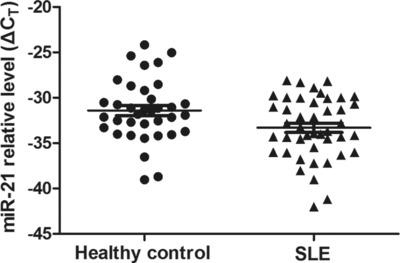
The level of plasma miR‐21 in healthy controls and SLE patients. ΔCT = CTmiR21‐CTU6B‐CTcel‐miR39, higher ΔCT means lower level of miR‐21.
Figure 2.
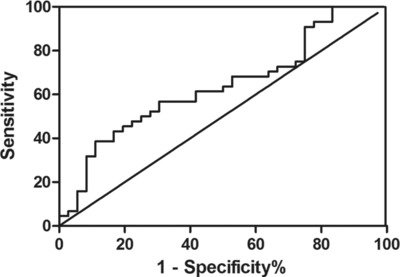
ROC curve analysis using plasma miR‐21 for discriminating SLE patients from healty controls. Plasma miR‐21 yielded an AUC of 0.64 (95% CI: 0.51–0.76, P = 0.039).
The Correlation Between miR‐21 and Blood Cells
In order to determine whether the blood cells (white blood cell, red blood cell, and platelet) could influence the concentration of plasma miR‐21, data of differentiating and counting of blood cells were collected. Our results demonstrated that miR‐21 was not associated with the counts of white blood cells (P = 0.2288), red blood cells (P = 0.8098), or platelets (P = 0.9230) in all the samples. The concentration of miR‐21 was not also associated with the blood cell count in healthy controls (data not shown).
The Relationship Between Plasma miR‐21 in SLE Patients and the Clinical Parameters Associated With SLE
The levels of several clinical parameters associated with SLE were analyzed. The concentration of miR‐21 was negatively correlated with the levels of C3 (r = −0.5297, P = 0.0004) and C4 (r = −0.4732, P = 0.0020; Fig. 3A and B). In addition, significant correlation was observed between C3 and C4 (r = 0.8179, P < 0.0001). The plasma level of miR‐21 was also correlated with uric acid (r = 0.3932, P = 0.0121; Fig. 3 C). However, plasma level of miR‐21 was not associated with creatinine (P = 0.4461), 24‐h urine protein (P = 0.3361), cystatin C (P = 0.0770), Glomerular Filtration Rate (GFR) (P = 0.1314), urinary cast (P = 0.7590), hematuria (P = 0.4609), pyuria (P = 0.7030), autoantibody (P = 0.0923), anti‐dsDNA (P = 0.2856), red blood cells (P = 0.4079), white blood cells (P = 0.4284), platelet (P = 0.4961), or SLEDAI (P = 0.7242).
Figure 3.
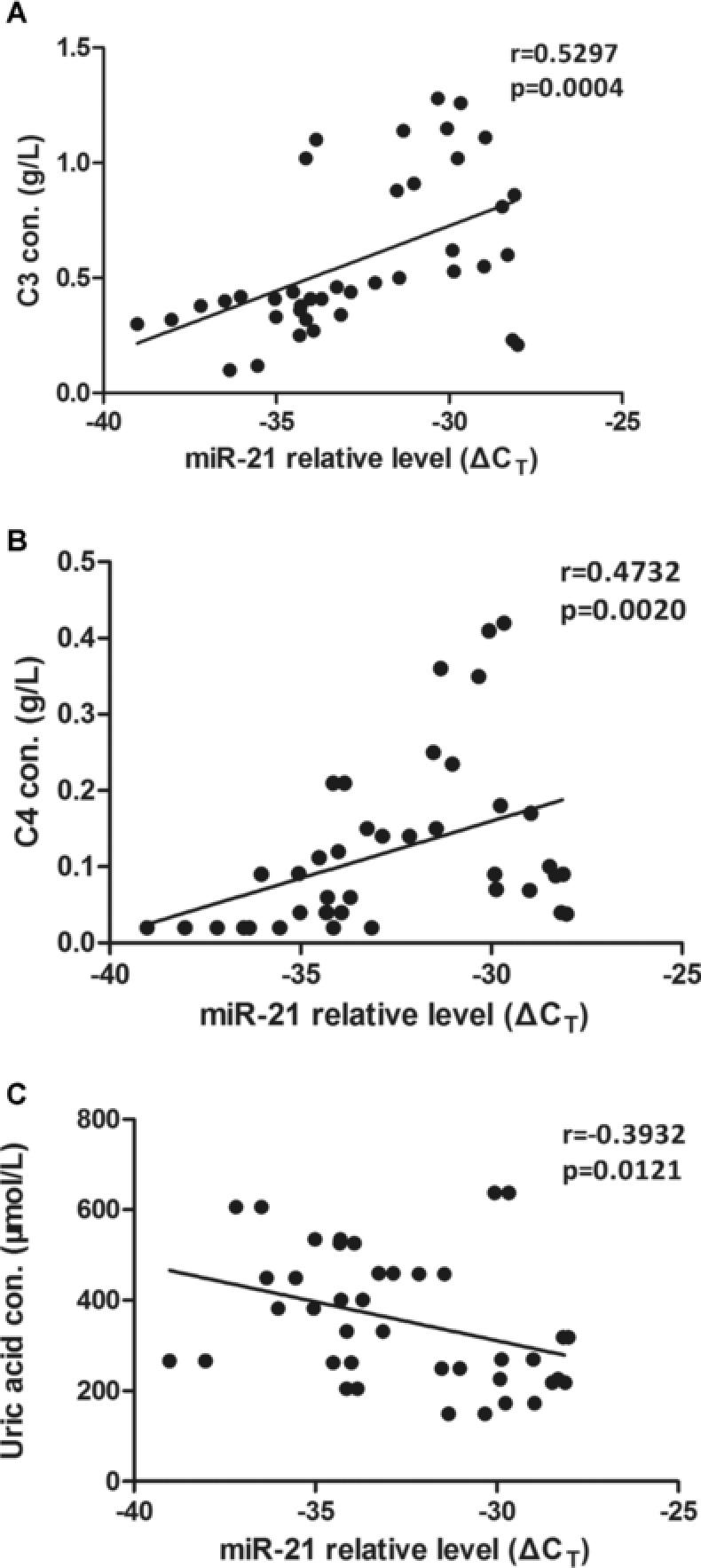
Correlations between plasma miR‐21 and clinical characteristics of SLE. The results were from the analysis by Spearman's rank correlation coefficient. (A) Decreased miR‐21 (increased ΔCT) correlated with C3. (B) Association between C4 and plasma miR‐21. (C) Correlation between plasma miR‐21 and uric acid. Con., concentration
DISCUSSION
Plasma miR‐21 is widely investigated as a biomarker of tumors 8, 13, 21 and cardiovascular diseases 11. Moreover, the level of plasma miR‐21 is also associated with age and inflammation 22. In our study, the concentration of plasma miR‐21 was significantly higher in SLE patients than in healthy controls, and the level of plasma miR‐21 correlated with serum uric acid, complement component C3, and C4. In addition, plasma miR‐21 did not associate with the amount of white blood cell, red blood cell, or platelet.
Previous study has indicated that the expression of miRNA varies among different populations 23. In the present study, we collected the Central Chinese as participants and found that the level of plasma miR‐21 in SLE patients was significantly higher than that in healthy controls. Interestingly, not only the expression trend, but also the change folds are similar with another report based on South China subjects (3.73‐fold in this study compared tofourfold increase reported by Wang et al.) 6. It may further confirm the expression characteristic of plasma miR‐21 in Chinese SLE patients, although current detection methods often give different results of plasma miRNA levels 24, 25. To further explore the potential of plasma miR‐21 as a SLE biomarker, we carried out the ROC analysis. Our data suggest that plasma miR‐21 has a weak sensitivity and a moderate specificity to distinguish SLE patients from healthy controls (sensitivity = 52.27%, specificity = 72.22%). Since plasma miR‐21 is also upregulated in rheumatoid arthritis 6, the application of plasma miR‐21 in SLE diagnosis needs to be interpreted with caution.
Serum complement C3/C4 levels negatively correlate with the activity of SLE. Our data showed that plasma miR‐21 negatively correlated with the levels of complement C3/C4 (P = 0.0004/0.0020). Currently, there is no direct experimental evidence to explain the mechanism on the relationship between plasma miR‐21 and complement C3/C4 levels. Studies have shown that miR‐21 downregulates the expression of Programmed Cell Death Protein 4 (PDCD4) and consequently indirectly upregulates the expression Interleukin 10 (IL‐10) 26. IL‐10 in SLE patient promotes B‐cell differentiation and autoantibody production 27. Autoantibody can activate classic complement activation pathway and consume C3/C4 simultaneously. So miR‐21 may influence both plasma complement C3/C4 levels from PDCD4‐IL10‐B cell–autoantibody–complement activation pathway.
Our data supported the relationship between plasma miR‐21 and complement C3/C4 levels, serum uric acid, but these data did not suggest the correlation between plasma miR‐21 and the activity of SLE (P = 0.7242). Other studies also found miRNAs correlated with SLE activity related parameters but not with SLEDAI score 4, 5. Although miR‐21 was overexpressed in CD4+ T cells from active SLE patients (14; 15), miR‐21 is also upregulated in B cells derived from quiescent lupus patients 28. Plasma miRNA may originate from blood cells and/or organs. Kidney is the most popular organ influenced during active SLE. However, no miR‐21 was detected in kidney biopsies from lupus nephritis patients 28, 29. Based on the above evidence, we speculate that the level of plasma miR‐21 may not correlate with the activity of SLE. Since the exact origin and the function of circulating miRNA are not clear, it should be cautious to discuss the mechanism of relationship between circulating miRNA and other parameters.
Our data also show that the concentration of plasma miR‐21 is not correlated with the amounts of white blood cells, red cells, and platelets. Previous studies have shown that the expression level of miR‐21 increases in CD4+ T cells from SLE patients 14, 15. The reason for plasma miR‐21 level not associated with white blood cells could be explained by that the plasma miR‐21 is selectively secreted or produced by other cells within the body 30. Mitchell's study also showed that circulating miRNA was not associated with blood cell counts 10. The results that blood cell counts do not influence the level of miR‐21 will benefit the miR‐21 as a plasma biomarker.
CONCLUSIONS
Plasma miR‐21 is overexpressed in SLE patients from Central China and correlated with serum C3, C4, and uric acid, which are parameters indicating the activity of SLE. Studies based on larger samples are needed to further describe the expression profile of plasma miR‐21 in SLE patients. As plasma miR‐21 is aberrantly expressed in many diseases, it's use in a specific disease should be cautious.
REFERENCES
- 1. Li R, Sun J, Ren LM, et al. 2012. Epidemiology of eight common rheumatic diseases in China: A large‐scale cross‐sectional survey in Beijing. Rheumatology (Oxford) 51(4):721–729. [DOI] [PubMed] [Google Scholar]
- 2. Mok CC. 2011. Epidemiology and survival of systemic lupus erythematosus in Hong Kong Chinese. Lupus 20(7):767–771. [DOI] [PubMed] [Google Scholar]
- 3. Shen N, Liang D, Tang Y, de Vries N, Tak PP. 2012. MicroRNAs—Novel regulators of systemic lupus erythematosus pathogenesis. Nat Rev Rheumatol 8(12):701–709. [DOI] [PubMed] [Google Scholar]
- 4. Wang G, Tam L, Li EK, et al. 2011. Serum and urinary free microRNA level in patients with systemic lupus erythematosus. Lupus 20(5):493–500. [DOI] [PubMed] [Google Scholar]
- 5. Wang G, Tam LS, Li EK, et al. 2010. Serum and urinary cell‐free MiR‐146a and MiR‐155 in patients with systemic lupus erythematosus. J Rheumatol 37(12):2516–2522. [DOI] [PubMed] [Google Scholar]
- 6. Wang H, Peng W, Ouyang X, Li W, Dai Y. 2012. Circulating microRNAs as candidate biomarkers in patients with systemic lupus erythematosus. Transl Res 160(3):198–206. [DOI] [PubMed] [Google Scholar]
- 7. Lawrie CH, Gal S, Dunlop HM, et al. 2008. Detection of elevated levels of tumour‐associated microRNAs in serum of patients with diffuse large B‐cell lymphoma. Br J Haematol 141(5):672–675. [DOI] [PubMed] [Google Scholar]
- 8. Qi P, Cheng SQ, Wang H, Li N, Chen YF, Gao CF. 2011. Serum MicroRNAs as biomarkers for hepatocellular carcinoma in chinese patients with chronic hepatitis B virus infection. PLoS One 6(12):e28486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Zhou J, Yu L, Gao X, et al. 2011. Plasma microRNA panel to diagnose hepatitis B virus‐related hepatocellular carcinoma. J Clin Oncol 29(36):4781–4788. [DOI] [PubMed] [Google Scholar]
- 10. Mitchell PS, Parkin RK, Kroh EM, et al. 2008. Circulating microRNAs as stable blood‐based markers for cancer detection. Proc Natl Acad Sci USA 105(30):10513–10518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Zile MR, Mehurg SM, Arroyo JE, Stroud RE, DeSantis SM, Spinale FG. 2011. Relationship between the temporal profile of plasma microRNA and left ventricular remodeling in patients after myocardial infarction. Circ Cardiovas Genet 4(6):614–619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Tsujiura M, Ichikawa D, Komatsu S, et al. 2010. Circulating microRNAs in plasma of patients with gastric cancers. Br J Cancer 102(7):1174–1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Komatsu S, Ichikawa D, Tsujiura M, et al. 2013. Prognostic impact of circulating miR‐21 in the plasma of patients with gastric carcinoma. Anticancer Res 33(1):271–276. [PubMed] [Google Scholar]
- 14. Stagakis E, Bertsias G, Verginis P, et al. 2011. Identification of novel microRNA signatures linked to human lupus disease activity and pathogenesis: miR‐21 regulates aberrant T cell responses through regulation of PDCD4 expression. Ann Rheum Dis 70(8):1496–1506. [DOI] [PubMed] [Google Scholar]
- 15. Pan W, Zhu S, Yuan M, et al. 2010. MicroRNA‐21 and microRNA‐148a contribute to DNA hypomethylation in lupus CD4+ T cells by directly and indirectly targeting DNA methyltransferase 1. J Immunol 184(12):6773–6781. [DOI] [PubMed] [Google Scholar]
- 16. Carlsen AL, Schetter AJ, Nielsen CT, et al. 2013. Circulating microRNA expression profiles associated with systemic lupus erythematosus. Arthritis Rheum 65(5):1324–1334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Kroh EM, Parkin RK, Mitchell PS, Tewari M. 2010. Analysis of circulating microRNA biomarkers in plasma and serum using quantitative reverse transcription‐PCR (qRT‐PCR). Methods 50(4):298–301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Huang Z, Huang D, Ni S, Peng Z, Sheng W, Du X. 2010. Plasma microRNAs are promising novel biomarkers for early detection of colorectal cancer. Int J Cancer 127(1):118–126. [DOI] [PubMed] [Google Scholar]
- 19. Tomasetti M, Staffolani S, Nocchi L, et al. 2012. Clinical significance of circulating miR‐126 quantification in malignant mesothelioma patients. Clin Biochem 45(7–8):575–581. [DOI] [PubMed] [Google Scholar]
- 20. Livak KJ, Schmittgen TD. 2001. Analysis of relative gene expression data using real‐time quantitative PCR and the 2(‐Delta Delta C(T)) method. Methods 25(4):402–408. [DOI] [PubMed] [Google Scholar]
- 21. Tomimaru Y, Eguchi H, Nagano H, et al. 2012. Circulating microRNA‐21 as a novel biomarker for hepatocellular carcinoma. J Hepatol 56(1):167–175. [DOI] [PubMed] [Google Scholar]
- 22. Olivieri F, Spazzafumo L, Santini G, et al. 2012. Age‐related differences in the expression of circulating microRNAs: miR‐21 as a new circulating marker of inflammaging. Mech Ageing Dev 133(11–12):675–685. [DOI] [PubMed] [Google Scholar]
- 23. Huang RS, Gamazon ER, Ziliak D, et al. 2011. Population differences in microRNA expression and biological implications. RNA biology 8(4):692–701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Koshiol J, Wang E, Zhao Y, Marincola F, Landi MT. 2010. Strengths and limitations of laboratory procedures for microRNA detection. Cancer Epidemiol Biomarkers Prev 19(4):907–911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. McDonald JS, Milosevic D, Reddi HV, Grebe SK, Algeciras‐Schimnich A. 2011. Analysis of circulating microRNA: Preanalytical and analytical challenges. Clin Chem 57(6):833–840. [DOI] [PubMed] [Google Scholar]
- 26. Sheedy FJ, Palsson‐McDermott E, Hennessy EJ, et al. 2010. Negative regulation of TLR4 via targeting of the proinflammatory tumor suppressor PDCD4 by the microRNA miR‐21. Nat Immunol 11(2):141–147. [DOI] [PubMed] [Google Scholar]
- 27. Beebe AM, Cua DJ, de Waal Malefyt R. 2002. The role of interleukin‐10 in autoimmune disease: Systemic lupus erythematosus (SLE) and multiple sclerosis (MS). Cytokine Growth Factor Rev 13(4–5):403–412. [DOI] [PubMed] [Google Scholar]
- 28. Te JL, Dozmorov IM, Guthridge JM, et al. 2010. Identification of unique microRNA signature associated with lupus nephritis. PLoS One 5(5):e10344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Dai Y, Sui W, Lan H, Yan Q, Huang H, Huang Y. 2009. Comprehensive analysis of microRNA expression patterns in renal biopsies of lupus nephritis patients. Rheumatol Int 29(7):749–754. [DOI] [PubMed] [Google Scholar]
- 30. Jones CI, Zabolotskaya MV, King AJ, et al. 2012. Identification of circulating microRNAs as diagnostic biomarkers for use in multiple myeloma. Br J Cancer 107(12):1987–1996. [DOI] [PMC free article] [PubMed] [Google Scholar]


