Abstract
Based on the currently proposed algorithms, antibodies specificities (sp‐ANAs) are identified mainly in samples positive for fluorescent antinuclear antibodies (FANA) screening tests. The purpose of the present study was to compare diagnostic performances of FANA and line immune assay (LIA) detecting 15 sp‐ANAs in patients with systemic rheumatic diseases (SRD). In 948 sera from the patients with SRD (n = 590) and non‐SRD (n = 358), we evaluated the fluorescent patterns and intensities in the FANA test, and compared the FANA results with sp‐ANAs against nRNP, Sm, SS‐A, Ro52, SS‐B, Scl‐70, PM/Scl, Jo‐1, CENP B, PCNA, dsDNA, nucleosome, histone, ribosomal‐P, and M2. The sensitivity and specificity was 75.9% and 52.5% of FANA test and 62.0% and 84.4% of sp‐ANAs test for SRD detection. The overall agreement between FANA and sp‐ANAs results was 69.2% (Kappa coefficient; 0.404). According to the clinical diagnosis, the levels of agreement varied from 33.3% to 83.1%. The positive predictive values of each FANA pattern for the detection of sp‐ANAs were less than 50% except for the discrete speckled pattern (91.7%). The 1:100 intensity of FANA as well as the monoreactivity of LIA, anti‐SSA(−)/anti‐Ro52(+), or FANA(−)/sp‐ANAs(+) was associated with non‐SRD. Antibodies against ribosomal‐P or PCNA were specific for systemic lupus eryhthematosus. This study highlights the need for careful interpretation of FANA test results to assess sp‐ANAs and the application of sp‐ANAs tests including less‐common autoantibodies. In patients with clinical suspicion of SRD, screening with both FANA and sp‐ANAs tests could improve diagnostic efficiency. J. Clin. Lab. Anal. 26:307‐314, 2012. © 2012 Wiley Periodicals, Inc.
Keywords: antinuclear antibodies, immunofluorescence, line immunoassay, systemic rheumatic disease, test comparisons
INTRODUCTION
Antinuclear antibodies (ANA) are a hallmark of systemic rheumatic diseases (SRD). The measurement of ANA is used for the screening, diagnosis, and monitoring of rheumatic diseases, and specific categories of antibodies are associated with specific diseases 1, 2. Fluorescent ANA (FANA) test by indirect immunofluorescence on HEp‐2 cells remains the method of choice for ANA screening, and the nuclear or cytoplasmic immunofluorescent patterns are interpreted in order to analyze antibody specificity 3, 4. Although several studies have shown a correlation between FANA patterns and antibody specificities, specificity of ANAs (sp‐ANAs) should be further verified.
Because FANA tests provide increased sensitivity and better standardization, various suggested algorithms comprised the sp‐ANAs tests only in the initial FANA‐positive samples 5, 6, 7. Various commercial tests are available for detecting sp‐ANAs specific for extractable nuclear antigens (ENA) and centromere B (CENP B) However, a previous study reported 1 that about 1–5% of systemic lupus eryhthematosus (SLE) patients does not present ANA. Additionally, there are a number of other important disease‐specific autoantibodies that are not commonly tested in immunology laboratories 8.
The present study was designed to compare the diagnostic performance of FANA and 15 sp‐ANAs detecting line immunoassay (LIA) in clinically well‐defined serum samples. We analyzed the fluorescent patterns and titers from the FANA test and compared these results with those from sp‐ANAs test against nRNP, Sm, SS‐A, Ro52, SS‐B, Scl‐70, PM/Scl, Jo‐1, CENP B, proliferating cell nuclear antigen (PCNA), dsDNA, nucleosome, histone, ribosomal‐P (Rib‐P), and M2. The reliable detection of an increased number of disease‐specific autoantibodies would be beneficial for the accurate and early diagnosis of SRD.
MATERIALS AND METHODS
Patients
The samples used in this study consisted of 948 sera that were sent to the clinical immunology laboratory at Seoul St. Mary's hospital for both FANA and sp‐ANAs tests. The specimens were from 590 new or follow‐up patients with various SRD: SLE; n = 154, Sjögren's syndrome (Sjogren; n = 87), systemic sclerosis (SS; n = 25), mixed connective tissue disease (MCTD; n = 101), overlap syndrome (n = 136), polymyositis/dermatomyositis (PM/DM; n = 12), rheumatoid arthritis (RA; n = 51), vasculitis (n = 24), and 358 patients with non‐SRD according to the medical record review. Of the 24 patients with vasculitis, two patients had idiopathic small‐vessel vasculitis and 22 patients had Behcet's disease. The 358 sera from patients with non‐SRD were requested by the physicians in Rheumatology, Dermatology, Internal Medicine, Neurology, and Oncology in the course of routine clinical practice. In these 358 patients, the diagnosis of SRD was excluded or not confirmed by the medical record assessment using a demographic data, related clinical features, and final diagnosis. This study was approved by the Institutional Review Board of Seoul St. Mary's Hospital.
FANA Test
Serum samples were initially diluted 1:100 and tested using mosaic HEp‐20–10/Liver (Monkey) kit (EUROIMMUN AG. Lübeck, Germany). The positive samples, the observed fluorescence patterns, and end‐point serum dilutions that produced positive results were described. Fluorescence patterns were divided into six categories: homogeneous, speckled, nucleolar, discrete speckled (DS), mixed pattern, and others including cytoplasmic, nuclear dots, cell division, and nuclear membrane patterns. The two authors (SA Lee and EJ Oh) evaluated the FANA slides in a blinded manner without any clinical information and achieved an agreement in pattern and titer of the results.
Sp‐ANAs Test
Sp‐ANAs were identified with LIA (ANA Profile 3, EUROIMMUN AG. Lübeck, Germany). This assay simultaneously identifies 15 different autoantibodies against nRNP, Sm, SS‐A, Ro52, SS‐B, Scl‐70, PM/Scl, Jo‐1, CENP B, PCNA, dsDNA, nucleosome, histone, Rib‐P, and M2. All assays were performed and interpreted according to the manufacturers’ instructions.
Statistical Analysis
Statistical analyses were performed with SPSS version 12.0 (SPSS, Chicago, IL). Agreement between the FANA and LIA results was assessed using Kappa coefficient (0.001–0.2 indicated slight concurrence, 0.201–0.4 indicated fair agreement, 0.401–0.6 showed moderate agreement, 0.601–0.8 indicated substantial concurrence, and 0.801–0.999 showed excellent agreement). All P‐values were two‐tailed and P‐values <0.05 were considered to be statistically significant.
RESULTS
FANA Test
Among the 948 sera samples, 618 (65.2%) were positive for FANA. The most common patterns were speckled (28.2%), homogeneous (24.8%), and mixed (17.6%) pattern. The frequency of a cytoplasmic pattern was 6.5% among the FANA‐positive sera. Figure 1 shows the frequency of FANA patterns according to the clinical diagnosis. Among SLE patients, homogeneous, speckled, and mixed patterns were most frequently found. DS or speckled fluorescent patterns were common in patients with SS. In sera from the patients with Sjogren, overlap syndrome, and PM/DM, a speckled pattern was most common.
Figure 1.
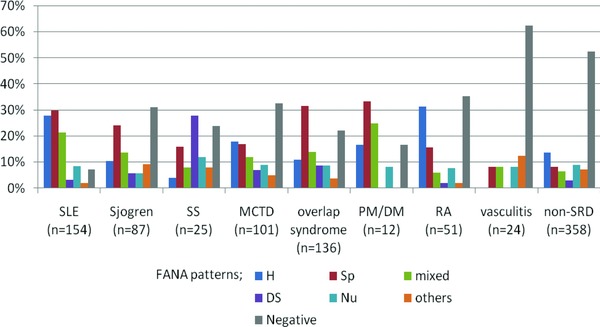
Frequency of FANA patterns according to clinical diagnoses.
Among FANA‐positive samples, the most prevalent intensity was 1:100‐positivity (37.2%). This was found in about half of the FANA‐positive samples from the patients with RA, vasculitis, and non‐SRD. However, 57.1% and 52.9% of the sera samples from the patients with SLE or overlap syndrome, respectively, showed high titers (≥1:400) of FANA‐positivity (Fig. 2). When titers of 1:100 and 1:200 were set as cut‐off values, the sensitivity and specificity for detecting SRD were 75.9% and 52.5% for 1:100 titer, and 55.9% and 83.8% for 1:200 titer. The sensitivities of FANA varied (37.5–92.9%) according to the clinical diagnosis of SRD (Table 1); the highest sensitivity was observed for SLE patients (92.9%) and the lowest sensitivity was seen for the diagnosis of vasculitis (37.5%). Of 170 FANA‐positive (≥1:100) sera from the patients with non‐SRD, 45(26.5%) sera showed speckled or mixed speckled pattern.
Figure 2.
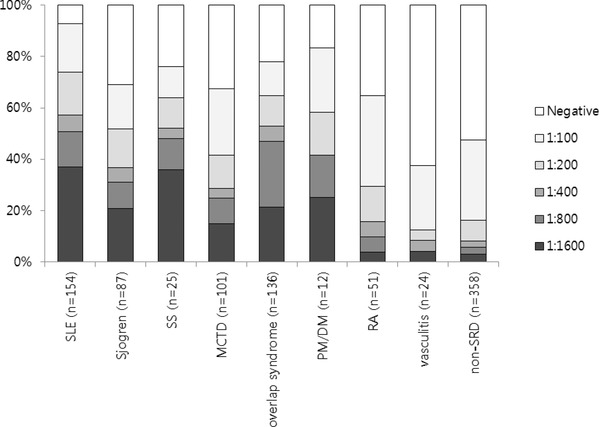
Distribution of FANA results for different dilutions according to clinical diagnoses.
Table 1.
Diagnostic Performance of FANA and sp‐ANAs Tests for Detecting Systemic Rheumatic Diseases
| Clinical diagnosis | No. of cases | FANA(+) | sp‐ANAs(+) | FANA(+) or sp‐ ANAs(+) | FANA(+) and sp‐ ANAs(+) | |
|---|---|---|---|---|---|---|
| Sensitivity, (CI) | SRD | 590 | 75.9 (72.2–79.3) | 62.0 (58.0–65.9) | 81.0 (77.6–84.1) | 56.9 (52.8–61.0) |
| Specificity, (CI) | 52.5 (47.2–57.8) | 84.4 (80.1–87.9) | 47.5 (42.2–52.8) | 89.4 (85.6–92.3) | ||
| PPV, (CI) | 72.5 (68.8–75.9) | 86.7 (83.0–89.7) | 71.8 (68.2–75.1) | 89.8 (86.2–92.6) | ||
| NPV, (CI) | 57.0 (51.4–62.3) | 57.4 (53.1–61.7) | 60.3 (54.3–66.0) | 55.7 (51.6–59.8) | ||
| Sensitivity, (CI) | SLE | 154 | 92.9 (87.3–96.2) | 76.0 (68.3–82.3) | 94.2 (88.9–97.1) | 74.7 (66.9–81.2) |
| Sjogren | 87 | 69.0 (58.0–78.2) | 66.7 (55.7–76.2) | 77.0 (66.5–85.1) | 58.6 (47.6–68.9) | |
| SS | 25 | 76.0 (54.5–89.8) | 56.0 (35.3–75.0) | 76.0 (54.5–89.4) | 56.0 (35.3–75.0) | |
| MCTD | 101 | 67.3 (57.2–76.1) | 49.5 (39.5–59.6) | 74.3 (64.4–82.2) | 42.6 (32.9–52.8) | |
| Overlap | 136 | 77.9 (69.9–84.4) | 72.8 (64.4–79.9) | 83.8 (76.3–89.4) | 66.9 (58.3‐74.6) | |
| PM/DM | 12 | 83.3 (50.9–97.1) | 33.3 (11.3–64.6) | 91.7 (59.8–99.6) | 25.0 (6.7–57.2) | |
| RA | 51 | 64.7 (50.0–77.2) | 41.2 (27.9–55.8) | 74.5 (60.1–85.2) | 31.4 (19.5–46.0) | |
| Vasculitis | 24 | 37.5 (19.6–59.2) | 12.5 (3.3–33.5) | 37.5 (19.6–59.2) | 12.5 (2.1‐22.5) |
Sp‐ANAs Test
Of 948 sera samples, 422 (44.5%) had one and more sp‐ANAs out of the 15 sp‐ANAs. Among the 422 sp‐ANAs‐positive samples, 36.5% showed monoreactivity on the LIA. Samples from non‐SRD group had significantly higher rates of monoreactivity on the LIA than the samples from SRD patients (73.2% vs. 30.9%, respectively; P < 0.001). The sensitivity and specificity of the sp‐ANAs test for detecting SRD was 62.0% and 84.4%, respectively. Among the sp‐ANAs‐positive sera sample (Table 2), the most frequently observed sp‐ANAs specificities were against SS‐A (24.6%) and Ro52 (23.4%). Out of the 285 anti‐SS‐A or anti‐Ro52‐positive sera, 170 (59.6%) were both anti‐SS‐A and anti‐Ro52‐positive, 63 (22.1%) sera reacted only against SS‐A, and 52 (18.2%) sera reacted only against Ro52. In sera with anti‐SS‐A or anti‐Ro52 antibodies, the frequency of anti‐SS‐A(−)/anti‐Ro52(+) was higher among patients with non‐SRD than those with SLE or Sjogren (32.0% vs. 6.6% (P = 0.001) or 11.5% (P = 0.025), respectively). Sp‐ANAs against nucleosomes (29.8%), histones (29.0%), and dsDNA (24.2%) were relatively common among patients with SLE. Antibodies against PCNA were detected only in sera from SLE patients and 12 (85.7%) out of 14 anti‐Rib‐P‐positive sera were from SLE patients (Table 2).
Table 2.
Number (%) of Samples With Positive sp‐ANAs Results According to Different Clinical Diagnoses
| Sp‐ANA | SLE | Sjogren | SS | MCTD | Overlap syndrome | PM/DM | RA | Vasculitis | SRD | Non‐SRD |
|---|---|---|---|---|---|---|---|---|---|---|
| n = 154 | n = 87 | n = 25 | n = 101 | n = 136 | n = 12 | n = 51 | n = 24 | n = 590 | n = 358 | |
| Negative | 37 (24.0) | 29 (33.3) | 11 (44.0) | 51 (50.5) | 37 (27.2) | 8 (66.7) | 30 (58.8) | 21 (87.5) | 224 (38.0) | 302 (84.8) |
| Positive | 117(76.0) | 58 (66.7) | 14 (56.0) | 50 (49.5) | 99 (72.8) | 4 (33.3) | 21 (87.5) | 3 (12.5) | 366 (62.0) | 56 (15.6) |
| M2 | 4 (2.6) | 3 (3.4) | 0 (0.0) | 3 (3.0) | 2 (1.5) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 12 (2.0) | 2 (0.6) |
| Rib‐P | 12 (7.9) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 2 (1.5) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 14 (2.4) | 0 (0.0) |
| Histone | 45 (29.8) | 3 (2.3) | 0 (0.0) | 9 (8.9) | 13 (9.6) | 1 (8.3) | 5 (9.8) | 1 (4.2) | 76 (12.9) | 7 (2.0) |
| Nucleo‐some | 44 (29.1) | 3 (3.4) | 0 (0.0) | 6 (5.9) | 6 (4.4) | 1 (8.3) | 2 (3.9) | 1 (4.2) | 63 (10.7) | 4 (1.1) |
| dsDNA | 38 (25.2) | 3 (3.4) | 1 (4.0) | 8 (7.9) | 9 (6.6) | 2 (16.7) | 5 (9.8) | 2 (8.3) | 68 (11.5) | 12 (3.4) |
| PCNA | 3 (2.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 3 (0.5) | 0 (0.0) |
| CENP B | 9 (6.0) | 8 (9.2) | 7 (28.0) | 7 (6.9) | 12 (8.8) | 0 (0.0) | 1 (2.0) | 0 (0.0) | 44 (7.5) | 9 (2.5) |
| Jo‐1 | 0 (0.0) | 1 (1.1) | 0 (0.0) | 0 (0.0) | 5 (3.7) | 1 (8.3) | 0 (0.0) | 0 (0.0) | 7 (1.2) | 1 (0.3) |
| PM/Scl | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) |
| Scl‐70 | 5 (3.3) | 0 (0.0) | 2 (8.0) | 4 (4.0) | 4 (2.9) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 15 (2.5) | 3 (0.8) |
| SS‐B | 20 (13.2) | 22 (25.3) | 2 (8.0) | 9 (8.9) | 26 (19.1) | 2 (16.7) | 3 (5.9) | 0 (0.0) | 84 (14.2) | 6 (1.7) |
| Ro52 | 55 (36.4) | 42 (48.3) | 6 (24.0) | 22 (21.8) | 64 (47.1) | 3 (25.0) | 11 (21.6) | 1 (4.2) | 204 (34.6) | 18 (5.0) |
| SS‐A | 71 (47.0) | 46 (52.9) | 5 (20.0) | 24 (23.8) | 58 (42.6) | 2 (16.7) | 10 (19.6) | 0 (0.0) | 216 (36.6) | 17 (4.7) |
| Sm | 17 (11.3) | 0 (0.0) | 2 (8.0) | 0 (0.0) | 6 (4.4) | 0 (0.0) | 1 (2.0) | 0 (0.0) | 26 (4.4) | 1 (0.3) |
| nRNP | 41 (27.2) | 5 (5.7) | 4 (16.0) | 11 (10.9) | 25 (18.4) | 0 (0.0) | 1 (2.0) | 2 (8.3) | 89 (15.1) | 1 (0.3) |
Comparison Between FANA and sp‐ANAs Test Results
FANA positivity with confirmation by the sp‐ANAs test showed higher specificity of 89.4% compared to the 52.5% specificity of the results that were only found positive by FANA (Table 1). The overall agreement between FANA and sp‐ANAs results was 69.2% with a Kappa coefficient of 0.404, indicating moderate agreement (Table 3). According to the clinical diagnosis, the levels of agreement were variable, ranging from 33.3 to 83.1% (k value; –0.090 to 0.579). Although the number of cases was small, the lower concordance between FANA and sp‐ANAs test results was observed among patients with PM/DM (33.3%), RA (56.9%), and non‐SRD (58.1%). The positive and negative predictive value (PPV and NPV) of the FANA test for the presence of sp‐ANAs was 60.5% (374/618) and 85.5% (282/330), respectively.
Table 3.
Results of the FANA and sp‐ANAs Tests According to Clinical Diagnoses
| Diagnosis (no. of patients) | Results of the FANA and sp‐ANAs tests | Agreementa | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| N/N | N/P | P/N | P/P | % | k value | |||||
| SLE (n = 154) | 9 | (5.8)† | 2 | (1.3) | 28 | (18.2) | 115 | (74.7) | 80.5 | 0.298 |
| Sjogren (n = 87) | 20 | (23.0) | 7 | (8.0) | 9 | (10.3) | 51 | (58.6) | 81.6 | 0.579 |
| SS (n = 25) | 6 | (24.0) | 0 | (0.0) | 5 | (20.0) | 14 | (56.0) | 80.0 | 0.573 |
| MCTD (n = 101) | 26 | (25.7) | 7 | (6.9) | 25 | (24.8) | 43 | (42.6) | 68.3 | 0.369 |
| Overlap syndrome (n = 136) | 22 | (16.2) | 8 | (5.9) | 15 | (11.0) | 91 | (66.9) | 83.1 | 0.570 |
| PM/DM (n = 12) | 1 | (8.3) | 1 | (8.3) | 7 | (58.3) | 3 | (25.0) | 33.3 | ‐0.090 |
| RA (n = 51) | 13 | (25.5) | 5 | (9.8) | 17 | (33.3) | 16 | (31.4) | 56.9 | 0.180 |
| Vasculitis (n = 24) | 15 | (62.5) | 0 | (0.0) | 6 | (25.0) | 3 | (12.5) | 75.0 | 0.385 |
| Non‐SRD (n = 358) | 170 | (47.5) | 18 | (5.0) | 132 | (36.9) | 38 | (10.6) | 58.1 | 0.132 |
| Total (n = 948) | 282 | (29.7) | 48 | (5.1) | 244 | (25.7) | 374 | (39.5) | 69.2 | 0.404 |
Agreement between results of the FANA and sp‐ANAs test, †number of patients (%). N, negative; P, positive.
Two hundred and forty‐four (25.7%) sera samples were FANA(+)/sp‐ANAs(−), and 48 (5.1%) sera samples were FANA(−)/sp‐ANAs(+). Out of the 48 FANA(−)/sp‐ANAs(+) sera, 18 (37.5%) were from the patients with non‐SRD (Table 3, Fig. 3). Out of 154 samples from SLE patients, two (1.3%) were FANA(−)/sp‐ANAs(+) and SS‐A‐positive. Among patients with SRD except for individuals with SLE, 5.9–8.3% of the sera were FANA(−)/sp‐ANAs(+) (Fig. 3).
Figure 3.
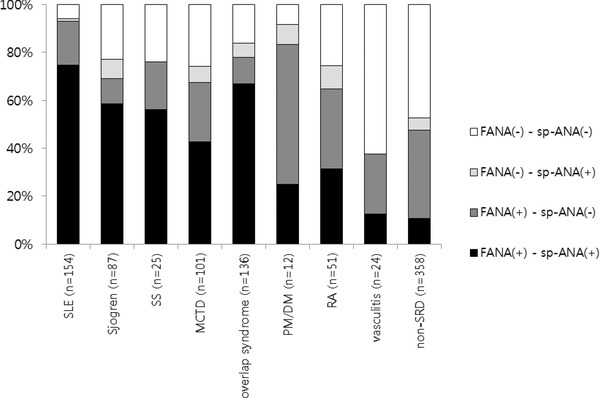
Results of the FANA and sp‐ANAs tests according to the clinical diagnoses.
Table 4 summarizes the positive rates of sp‐ANAs for each FANA dilution in FANA‐positive sera. At a 1:100 dilution, 71.3% of FANA‐positive samples were sp‐ANAs‐negative. There was a tendency for higher dilutions of FANA‐positive sera to increase the sp‐ANAs positivity rate. The positive rates for each sp‐ANA are shown in Table 5 according to the FANA pattern. The PPVs of each FANA pattern for the detection of sp‐ANAs were less than 50% except for the DS pattern for which the PPV was 91.7% (44/48) of them showed CNEP B‐positive. In sera showing a homogeneous pattern, sp‐ANAs against SS‐A, Ro52, histones, nucleosomes, or DNA were frequently detected. In sera showing a speckled pattern, antibodies were specific for SS‐A, Ro52, RNP, or SS‐B. Among 48 FANA(−)/Sp‐ANAs(+) sera, sp‐ANAs were mostly directed against SS‐A, Ro52, dsDNA, and histones.
Table 4.
Positive Rates for sp‐ANAs According to the Different Dilutions of FANA
| Sp‐ANA | FANA dilution | ||||||
|---|---|---|---|---|---|---|---|
| 0 | 1:100 | 1:200 | 1:400 | 1:800 | ≥ 1:1600 | Total | |
| n = 330 | n = 230 | n = 110 | n = 40 | n = 93 | n = 145 | n = 948 | |
| Negative | 282 (85.5)a | 164 (71.3) | 50 (45.5) | 9 (22.5) | 14 (15.1) | 7 (4.8) | 526 (55.5) |
| Positive | 48 (14.5) | 66 (28.7) | 60 (54.5) | 31 (77.5) | 79 (84.9) | 138 (95.2) | 422 (44.5) |
| M2 | 2 (0.6) | 1 (0.4) | 0 (0.0) | 1 (2.5) | 5 (5.4) | 5 (3.4) | 14 (1.5) |
| Rib‐P | 0 (0.0) | 1 (0.4) | 1 (0.9) | 0 (0.0) | 3 (3.2) | 9 (6.2) | 14 (1.5) |
| Histone | 8 (2.4) | 8 (3.5) | 11 (10.0) | 2 (5.0) | 19 (20.4) | 35 (24.1) | 83 (8.8) |
| Nucleosome | 5 (1.5) | 2 (0.9) | 8 (7.3) | 2 (5.0) | 14 (15.1) | 36 (24.8) | 67 (7.1) |
| DsDNA | 10 (3.0) | 12 (5.2) | 10 (9.1) | 4 (10.0) | 13 (14.0) | 31 (21.4) | 80 (8.4) |
| PCNA | 0 (0.0) | 1 (0.4) | 0 (0.0) | 1 (2.5) | 0 (0.0) | 1 (0.7) | 3 (0.3) |
| CENP B | 2 (0.6) | 2 (0.9) | 4 (3.6) | 4 (10.0) | 14 (15.1) | 27 (18.6) | 53 (5.6) |
| Jo‐1 | 5 (1.5) | 1 (0.4) | 2 (1.8) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 8 (0.8) |
| PM/Scl | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) |
| Scl‐70 | 1 (0.3) | 2 (0.9) | 0 (0.0) | 4 (10.0) | 2 (2.2) | 9 (6.2) | 18 (1.9) |
| SS‐B | 3 (0.9) | 9 (3.9) | 13 (11.8) | 6 (15.0) | 25 (26.9) | 34 (23.4) | 90 (9.5) |
| Ro52 | 14 (4.2) | 28 (12.2) | 35 (31.8) | 19 (47.5) | 49 (52.7) | 77 (53.1) | 222 (23.4) |
| SS‐A | 15 (4.5) | 31 (13.5) | 42 (38.2) | 18 (45.0) | 53 (57.0) | 74 (51.0) | 233 (24.6) |
| Sm | 3 (0.9) | 5 (2.2) | 0 (0.0) | 0 (0.0) | 3 (3.2) | 16 (11.0) | 27 (2.8) |
| nRNP | 1 (0.3) | 11 (4.8) | 3 (2.7) | 3 (7.5) | 12 (12.9) | 60 (41.4) | 90 (9.5) |
Number (%) of samples with positive sp‐ANA results for each FANA dilution.
Table 5.
Numbers of Samples With Positive sp‐ANA Results According to the Different FANA Patterns
| Sp‐ANA | FANA pattern | |||||||
|---|---|---|---|---|---|---|---|---|
| H | Sp | DS | Nu | H+Sp | H+Nu | Sp+Nu | Others | |
| n = 153 | n = 174 | n = 48 | n = 81 | n = 62 | n = 22 | n = 25 | n = 53 | |
| Negative | 80 (52.3)a | 47 (27.0) | 4 (8.3) | 49 (60.5) | 20 (32.3) | 8 (36.4) | 4 (16.0) | 32 (60.4) |
| Positive | 73 (47.7) | 127 (73.0) | 44 (91.7) | 32 (39.5) | 42 (67.7) | 14 (63.6) | 21 (84.0) | 21 (39.6) |
| M2 | 1 (0.7)* | 4 (2.3) | 1 (2.1) | 1 (1.2) | 2 (3.2) | 0 (0.0) | 0 (0.0) | 3 (5.7) |
| Rib‐P | 3 (2.0) | 5 (2.9) | 0 (0.0) | 3 (3.7) | 1 (1.6) | 1 (4.5) | 1 (4.0) | 0 (0.0) |
| Histone | 37 (24.2) | 16 (9.2) | 2 (4.2) | 4 (4.9) | 11 (17.7) | 4 (18.2) | 1 (4.0) | 0 (0.0) |
| Nucleosome | 31 (20.3) | 10 (5.7) | 1 (2.1) | 5 (6.2) | 11 (17.7) | 4 (18.2) | 0 (0.0) | 0 (0.0) |
| dsDNA | 33 (21.6) | 9 (5.2) | 3 (6.3) | 6 (7.4) | 12 (19.4) | 5 (22.7) | 2 (8.0) | 0 (0.0) |
| PCNA | 1 (0.7) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 1 (1.6) | 1 (4.5) | 0 (0.0) | 0 (0.0) |
| CENP B | 2 (1.3) | 3 (1.7) | 44 (91.7) | 0 (0.0) | 2 (3.2) | 0 (0.0) | 0 (0.0) | 0 (0.0) |
| Jo‐1 | 0 (0.0) | 1 (0.6) | 0 (0.0) | 1 (1.2) | 0 (0.0) | 0 (0.0) | 1 (4.0) | 0 (0.0) |
| PM/Scl | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) |
| Scl‐70 | 3 (2.0) | 4 (2.3) | 0 (0.0) | 5 (6.2) | 1 (1.6) | 3 (13.6) | 1 (4.0) | 0 (0.0) |
| SS‐B | 12 (7.8) | 33 (19.0) | 4 (8.3) | 7 (8.6) | 12 (19.4) | 5 (22.7) | 12 (48.0) | 2 (3.8) |
| Ro52 | 41 (26.8) | 78 (44.8) | 11 (22.9) | 14 (17.3) | 21 (33.9) | 6 (27.3) | 19 (76.0) | 18 (34.0) |
| SS‐A | 48 (31.4) | 79 (45.4) | 16 (33.3) | 17 (21.0) | 22 (35.5) | 9 (40.9) | 19 (76.0) | 8 (15.1) |
| Sm | 4 (2.6) | 14 (8.0) | 0 (0.0) | 1 (1.2) | 4 (6.5) | 1 (4.5) | 0 (0.0) | 0 (0.0) |
| nRNP | 9 (5.9) | 58 (33.3) | 2 (4.2) | 4 (4.9) | 12 (19.4) | 0 (0.0) | 0 (0.0) | 4 (7.5) |
Number (%) of samples with positive sp‐ANAs findings for FANA pattern. H, homogeneous; Sp, speckled; DS, discrete speckled; Nu, nucleolar.
DISCUSSION
The aim of this study was to investigate and compare the clinical performance of FANA and LIA detecting 15 sp‐ANAs for identifying cases of variable SRD. As criteria for determining positive and negative results, we used clinical diagnoses obtained from the medical records of patients for whom both FANA and sp‐ANAs tests were simultaneously ordered.
Autoimmune serology is an important tool for the diagnosis of SRD and related disorders. Discrepancies among the results for autoantibodies evaluated with different immunoassay pose a problem for clinicians. Although FANA is the most routine method used to screen for SRD, lower specificity of this assay is associated with sp‐ANAs reactivity in a low percentage of FANA‐positive sera 3, 9. In this study, the prevalence of positive FANA was 65.2% at 1:100 titer cut‐off values, but a previous study for adult Korean reported FANA positivity as 13.3% (cut‐off 1:40) in referred FANA screening 10. This difference may be due to a possible bias associated with requested samples as the present study only included sera requested for both FANA and sp‐ANAs tests.
The sensitivity and specificity of the present FANA test for detecting SRD was 75.9% and 52.5%, respectively; these were similar to the results of the previous report 9. However, the level of overall agreement between the FANA and sp‐ANAs results was moderate with a Kappa coefficient of 0.404, and the sensitivity of FANA test varied according to clinical diagnosis with the highest sensitivity found among patients with SLE. Since the fluorescent intensity with 1:100 was observed in about half of the FANA‐positive samples from the patients with RA, vasculitis, and non‐SRD, FANA positivity with a 1:100 titer may potentially decrease the overall specificity 2, 11.
HEp‐2 cells have been used as a substrate because the results offer the detecting a fluorescent pattern that suggests clinical associations with certain types of SRD. However, the DS pattern only was associated with a PPV of more than 50% for the clinical diagnosis of SRD as previously reported 12. In the FANA test, a mixed fluorescence pattern was frequently observed in clinical samples and each fluorescence pattern could not orient ANA specificities in a considerable number of specimens similar to a prior report 13. It is also possible that a dominant pattern can mask other combined fluorescence patterns and the FANA test with HEp‐2 cells has less sensitive cut‐off dilution than sp‐ANAs test. Overall false‐positive rate of the speckled or mixed speckled pattern (≥1:100) was 26.5% in patients with non‐SRD. Although the dense fine speckled pattern was not described in this study, this finding supports a previous report showing that the dense fine speckled pattern may not be disease specific 14.
The laboratory tests should be requested based on the suspected clinical diagnoses 1, 2, 15. Antibodies against disease‐specific antigens can be detected with several highly specific methods. However, based on the currently proposed algorithms, the sp‐ANAs test can be performed only when ANA screening results are positive or if the patient has clear symptoms of SRD 5, 6, 7, 9, 15. For the sp‐ANAs test in the present study, the sensitivity for detecting SRD was less but the specificity was greater than the FANA test, similar to a previous report 9. The combined use of sp‐ANAs test with the FANA test increased the specificity up to 89.8%. The previous data from studies comparing the costs of diagnostic tests showed that strategies based on screening followed by identification are more efficient than a direct identification strategy alone 6. However, this traditional algorithm has several disadvantages including a long processing time and delayed diagnosis. Many laboratories tend to use a single technique for detecting sp‐ANAs with one to six assayed in each sample. The evaluation of FANA for detecting more than ten sp‐ANAs is rare.
Multiplex technologies including LIA have the advantage of simultaneous ANA testing for multiple reactivities 15. Our study had the benefit of examining a large population with various types of SRD and gathering data on a wide group of autoantibodies including less‐common disease‐specific ones. In this study, sera from non‐SRD patients were associated with FANA(−)/sp‐ANAs(+) and monoreactivity and anti‐SS‐A(−)/Ro52(+) on the LIA. In agreement with a previous report 16 that showed that antibody against Ro52 potentially decreases the overall clinical specificity, sera with antibodies against Ro52 that lacked anti‐SS‐A antibodies were more frequently collected from non‐SRD patients. Antibodies specific for Rib‐P and PCNA are very specific for SLE as shown in a previous study 17, 18, and FANA test may not detect anti‐Rib‐P and anti‐PCNA antibodies in sera with other co‐existing autoantibodies. Therefore, LIA capable of detecting these antibodies may be a useful tool for accurately determining a diagnosis.
In this study, 5.1% of total samples were FANA(−)/sp‐ANAs(+); SS‐A/Ro was the most common specificity in these samples. Our finding is in agreement with a previous report showing that anti‐SS‐A/Ro, anti‐SS‐B/La, and anti‐Jo‐1 antibodies have been occasionally detected in patients with negative HEp‐2 cell test results 19. Although the simultaneous detection of FANA and 15 sp‐ANAs can increase the costs, the quality of the diagnostic process can be improved and may help detect SRD with low‐titer ANAs and uncommon sp‐ANA positivity. Therefore, the results from this study highlight the need for careful interpretation of FANA screening tests according to the clinical diagnosis and a rigorous algorithm to derive a consensus between IIF and LIA results.
Our study had a few potential limitations including the use of an LIA for detecting antibodies against dsDNA rather than the use of an ELISA, Farr, or Crithidia assays. Although LIA showed good agreement with ELISA for anti‐ENA in a previous study (≥80%), the detection of dsDNA antibodies with an LIA has been reported to be less sensitive compared to other assays 20, 21. In addition, clinical diagnoses obtained from the medical records may be subject to bias since diagnostic errors and the effect of past and current treatments could not be taken into account.
In conclusion, a FANA or sp‐ANAs test alone was not enough to detect specific antibodies or precise diagnosis of SRD and the identification of autoantibodies, including ones not generally tested, was also important for diagnosing SRD. Therefore, screening with both FANA and sp‐ANAs tests could improve the efficiency of diagnosing patients suspected to have an SRD.
None of the authors have any potential conflicts of interest relevant to this article.
ACKNOWLEDGMENT
This research was supported by a grant of the Korea Healthcare technology R&D Project, Ministry for Health, Welfare & Family Affairs, Republic of Korea (A092258).
Grant sponsor: Korea Healthcare technology R&D Project, Ministry for Health, Welfare & Family Affairs, Republic of Korea; Grant number: A092258.
REFERENCES
- 1. Kavanaugh A, Tomar R, Reveille J, Solomon DH, Homburger Ha. Guidelines for clinical use of the antinuclear antibody test and tests for specific autoantibodies to nuclear antigens. American College of Pathologists. Arch Pathol Lab Med 2000;124:71–81. [DOI] [PubMed] [Google Scholar]
- 2. Tozzoli R, Bizzaro N, Tonutti E, et al. Italian society of laboratory medicine study group on the diagnosis of autoimmune diseases: Guidelines for the laboratory use of autoantibody tests in the diagnosis and monitoring of autoimmune rheumatic diseases. Am J Clin Pathol 2002;117:316–324. [DOI] [PubMed] [Google Scholar]
- 3. Peene I, Meheus L, Veys EM, De Keyser F. Detection and identification of antinuclear antibodies (ANA) in a large and consecutive cohort of serum samples referred for ANA testing. Ann Rheum Dis 2001;60:1131–1136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Malleson PN, Sailer M, Mackinnon MJ. Usefulness of antinuclear antibody testing to screen for rheumatic diseases. Arch Dis Child 1997;77:299–304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Bonaguri C, Melegari A, Dall'Aglio P, et al. An italian multicenter study for application of a diagnostic algorithm in autoantibody testing. Ann NY Acad Sci 2009;1173:124–129. [DOI] [PubMed] [Google Scholar]
- 6. Almeida González D, Cabrera de León A, Rodríguez Pérez Mdel C, et al. Efficiency of different strategies to detect autoantibodies to extractable nuclear antigens. J Immunol Methods 2010;360:89–95. [DOI] [PubMed] [Google Scholar]
- 7. Van Praet JT, Vander Cruyssen B, Bonroy C, Smith V, Delanghe J, De Keyser F. Validation of a new screening strategy for anti‐extractable nuclear antigen antibodies. Clin Exp Rheumatol 2009;27:971–976. [PubMed] [Google Scholar]
- 8. Parker JC, Bunn CC. Sensitivity of the Phadia EliA connective tissue disease screen for less common disease‐specific autoantibodies. J Clin Pathol 2011;64:631–633. [DOI] [PubMed] [Google Scholar]
- 9. Bizzaro N, Wiik A. Appropriateness in anti‐nuclear antibody testing: From clinical request to strategic laboratory practice. Clin Exp Rheumatol 2004;22:349–355. [PubMed] [Google Scholar]
- 10. Kang SY, Lee WI. Clinical significance of dense fine speckled pattern in anti‐nuclear antibody test using indirect immunofluorescence method. Korean J Lab Med 2009;29:145–151. [DOI] [PubMed] [Google Scholar]
- 11. Kang I, Siperstein R, Quan T, Breitenstein ML. Utility of age, gender, ANA titer and pattern as predictors of anti‐ENA and ‐dsDNA antibodies. Clin Rheumatol 2004;23:509–515. [DOI] [PubMed] [Google Scholar]
- 12. Ménard HA. Antinuclear antibodies: The medium is the message. Clin Exp Rheumatol 2000;18:429–430. [PubMed] [Google Scholar]
- 13. Muro Y. Antinuclear antibodies. Autoimmunity 2005;38:3–9. [DOI] [PubMed] [Google Scholar]
- 14. Mariz HA, Sato EI, Barbosa SH, Rodrigues SH, Dellavance A, Andrade Le. Pattern on the antinuclear antibody‐HEp‐2 test is a critical parameter for discriminating antinuclear antibody‐positive healthy individuals and patients with autoimmune rheumatic diseases. Arthritis Rheum 2011;63:191–200. [DOI] [PubMed] [Google Scholar]
- 15. Tampoia M, Brescia V, Fontana A, Zucano A, Morrone LF, Pansini N. Application of a combined protocol for rational request and utilization of antibody assays improves clinical diagnostic efficacy in autoimmune rheumatic disease. Arch Pathol Lab Med 2007;131:112–116. [DOI] [PubMed] [Google Scholar]
- 16. Parker JC, Burlingame RW, Bunn CC. Prevalence of antibodies to Ro‐52 in a serologically defined population of patients with systemic sclerosis. J Autoimmune Dis 2009;6:2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Bonfa E, Elkon KB. Clinical and serologic associations of the antiribosomal P protein antibody. Arthritis Rheum 1986;29:981–985. [DOI] [PubMed] [Google Scholar]
- 18. Fritzler MJ, McCarty GA, Ryan JP, Kinsella TD. Clinical features of patients with antibodies directed against proliferating cell nuclear antigen. Arthritis Rheum 1983;26:140–145. [DOI] [PubMed] [Google Scholar]
- 19. Hoffman IE, Peene I, Veys EM, De Keyser F. Detection of specific antinuclear reactivities in patients with negative anti‐nuclear antibody immunofluorescence screening tests. Clin Chem 2002;48:2171–2176. [PubMed] [Google Scholar]
- 20. Yang JY, Oh EJ, Kim Y, Park Yj. Evaluation of anti‐dsDNA antibody tests: Crithidia luciliae immunofluorescence test, immunoblot, enzyme‐linked immunosorbent assay, chemiluminescence immunoassay. Korean J Lab Med 2010;30:675–684. [DOI] [PubMed] [Google Scholar]
- 21. Kim JM, Ihm CH, Sin DH, Ihm MK, Sim Sc. Detection of anti‐ENA and anti‐dsDNA antibodies using line immunoassay in systemic autoimmune diseases. Korean J Lab Med 2008;28:353–361. [DOI] [PubMed] [Google Scholar]


