Abstract
Background
Approximately 6% of sera positive in a dengue virus IgM‐capture enzyme immunoassay (EIA) represent false‐positives due to interaction between IgM and horseradish peroxidase (HRP)‐labeled monoclonal antibody (MAb) 6B6C1 (IgG2a). To better understand this interaction, we assessed the reactivity of captured IgM from these sera with other HRP‐labeled MAbs. J. Clin. Lab. Anal. 27:27–30, 2013. © 2012 Wiley Periodicals, Inc.
Methods
Fifty dengue IgM false‐positive sera (recognizing 6B6C1) were evaluated for IgM reactivity with the HRP‐labeled MAbs H3A4 (IgG2a), 53.8 (IgG2b), and IL‐A2 (IgG1). The sera were also tested in an EIA for human anti‐mouse antibody (HAMA).
Results
Forty‐three sera (86%) reacted with IgG2a MAb (H3A4). Most (31/43 = 72%) of these sera recognizing 6B6C1 and H3A4 also recognized the IgG2 MAb and/or the IgG1 MAb. In contrast, HAMA was increased in only 9 of 50 (18%) sera reacting with 6B6C1.
Conclusions
IgM from most sera‐binding IgG2a MAb 6B6C1 also binds another IgG2a MAb, suggesting that IgM‐6B6C1 reactivity is not idiotype specific. In many cases, IgM recognizing 6B6C1 also binds MAbs of other IgG subclasses, but is negative in a HAMA assay. These findings indicate that samples positive in IgM‐capture EIAs utilizing conjugated MAbs should always be retested in the absence of antigen to identify false‐positive reactivity caused by direct IgM‐MAb interaction. J. Clin. Lab. Anal. 00:1‐4, 2012. © 2012 Wiley Periodicals, Inc.
Keywords: human anti‐mouse antibodies, IgM‐capture EIA, false positivity, monoclonal IgG subclasses
INTRODUCTION
Our earlier studies demonstrated that approximately 6% of sera tested for dengue virus IgM using a screening mu‐capture enzyme immunoassay (EIA) give false‐positive results 1. Identification of this false‐positive reactivity requires that sera positive in the screening assay be retested using a background subtraction modification of the assay, in which reactivity in the presence and absence of dengue antigen is evaluated. This background subtraction assay has clearly demonstrated that the false‐positive screening assay reactivity reflects direct interaction between captured IgM and the horseradish peroxidase (HRP) conjugated anti‐flavivirus murine IgG2a monoclonal antibody (MAb) 6B6C1 1. As has been described for similar assays 2, 3, 4, inclusion of murine polyclonal IgG or another IgG2a MAb in the assay system inhibited the reactivity of some, but not all, false‐positive sera (unpublished observations), leaving the background subtraction method as the most sensitive option to identify such false‐positive sera. In an effort to better understand the reactivity between captured IgM and MAb 6B6C1, we assessed the reactivity of captured IgM from dengue IgM false‐positive sera with another HRP‐conjugated IgG2a MAb, as well as HRP‐conjugated IgG2b and IgG1 MAbs.
MATERIALS AND METHODS
The study employed 50 consecutive sera giving false‐positive reactivity and 40 blood donor sera giving negative results in the dengue virus IgM screening EIA 1.
The HRP‐conjugated MAbs and dilutions utilized were as follows: (a) 6B6C1 anti‐flavivirus (IgG2a) 1:9,000, (b) H3A4 anti‐hepatitis B core antigen (IgG2a) 1:500, (c) 53.8 anti‐talin‐2 (IgG2b) 1:3,000, and (d) IL‐A2 anti‐bovine IgG (IgG1) 1:300. A commercially available HRP‐conjugated IgG3 MAb could not be identified. Conjugated 6B6C1 was purchased from Jackson ImmunoResearch (Westgrove, PA); all other MAb conjugates were purchased from AbD Serotec (Raleigh, NC). All conjugates were diluted in Eagle's minimum essential medium (Irvine Scientific, Santa Ana, CA) containing 5% fetal bovine serum (Hyclone, Logan, UT), 1% Ficoll (Sigma Aldrich, St. Louis, MO), and 0.1% tween 20 (BioRad, Hercules, CA). The MAb dilutions selected for use were those giving absorbance values of 0.100–0.300 in preliminary studies using sera from two healthy blood donors.
Sera were diluted and added to microtiter wells coated with rabbit anti‐human IgM as described 1. After an hour at room temperature and washing, wells received 0.1 mL of diluted conjugate. After 30 min at room temperature and washing, enzyme substrate and stop solution were utilized as described for the routine dengue IgM EIA procedure 1. Absorbance was measured at 450 nM. Absorbance values greater than the 97.5th percentile absorbance value for the 40 blood donor sera were considered positive; these cutoff absorbance values were 0.241 for 6B6C1 (IgG2a), 0.158 for H3A4 (IgG2a), 0.148 for 53.8 (IgG2b), and 0.357 for IL‐A2 (IgG1). Because results were expressed as absorbance values generated on different days, interassay precision studies were conducted to ensure assay consistency.
All 90 sera were tested for human antimouse antibody (HAMA) using a commercially available EIA kit (Immunomedics, Morris Plains, NJ); HAMA levels greater than the 97.5th percentile value for the group of 40 sera from blood donors (43 ng/mL) were considered increased.
RESULTS
Table 1 shows interassay precision results for a blood donor serum and a dengue IgM false‐positive serum that recognized all the MAb conjugates evaluated; coefficient of variation values were all ≤17%, indicating good between‐run reproducibility.
Table 1.
Interassay Variation over Six Runs for EIAs Employing HRP‐Labeled MAbs
| Sample | Parameter | IgG2a (6B6C1) | IgG2a (H3A4) | IgG2b (53.8) | IgG1 (IL‐A2) |
|---|---|---|---|---|---|
| Blood donor serum | Mean absorbance | 0.12 | 0.13 | 0.11 | 0.22 |
| Standard deviation | 0.01 | 0.01 | 0.01 | 0.01 | |
| Coefficient of variation | 8% | 8% | 9% | 5% | |
| Dengue IgM false‐positive serum | Mean absorbance | 1.33 | 0.63 | 0.40 | 0.57 |
| Standard deviation | 0.11 | 0.11 | 0.03 | 0.07 | |
| Coefficient of variation | 8% | 17% | 8% | 12% |
Figure 1 presents absorbance values for reactivity to the 4 MAb conjugates for the 50 false‐positive sera. By definition, all 50 sera recognized 6B6C1 (IgG2a); 43 of 50 sera (86%) recognized H3A4 (IgG2a), 29 of 50 (58%) recognized 53.8 (IgG2b), and 17 of 50 (34%) recognized IL‐A2 (IgG1). Table 2 presents the distribution of the 50 samples among all possible patterns of reactivity with H3A4, 53.8, and IL‐A2 MAbs. The 43 sera reactive with 6B6C1 (IgG2a) and H3A4 (IgG2a) segregated into three reactivity patterns with respect to 53.8 (IgG2b) and IL‐A2 (IgG1) binding; 12 of 43 (28%) recognized neither the IgG2b nor the IgG1 MAb, 19 of 43 (44%) recognized either the IgG2b or the IgG1 MAb, and 12 of 43 (28%) recognized both the IgG2b and IgG1 MAbs. Only two 6B6C1 (IgG2a)‐reactive sera recognized the IgG2b and/or the IgG1 MAb but not H3A4 (IgG2a). Five of 50 6B6C1‐reactive sera (10%) did not recognize any of the three additional MAbs. Only 9 of 50 (18%) dengue IgM false‐positive sera contained increased levels of HAMA (Table 2).
Figure 1.
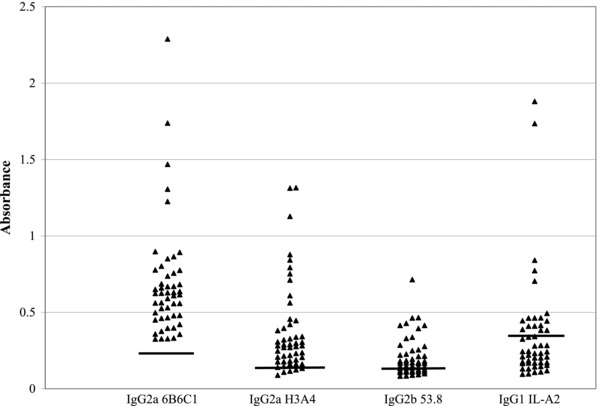
Reactivity of captured IgM from dengue virus IgM false‐positive sera with other MAbs. Results represent absorbance values. Horizontal lines represent the cutoff absorbance value, defined as the 97.5th percentile value for a group of 40 sera from blood donors.
Table 2.
MAb Reactivity Patterns of Captured IgM from 50 Sera Giving Dengue Virus IgM False‐Positive Results due to Binding of Captured IgM to MAb 6B6C1 (IgG2a)
| Reactivity with indicated MAb | ||||
|---|---|---|---|---|
| IgG2a (H3A4) | IgG2b (53.8) | IgG1 (IL‐A2) | No. of samples (% of total) | No. with detectable HAMA |
| + | − | − | 12 (24) | 4 |
| + | + | − | 15 (30) | 2 |
| + | − | + | 4 (8) | 1 |
| + | + | + | 12 (24) | 2 |
| − | + | − | 1 (2) | 0 |
| − | − | + | 0 (0) | 0 |
| − | + | + | 1 (2) | 0 |
| − | − | − | 5 (10) | 0 |
DISCUSSION
Our findings demonstrate that the IgM from 86% of sera reactive with MAb 6B6C1 (IgG2a) also reacts with another IgG2a MAb, indicating that most IgM‐6B6C1 reactivity reflects IgM binding to subclass determinants, rather than IgM recognition of idiotypic determinants of 6B6C1. Although this finding was not unexpected, we did not anticipate the heterogeneity in recognition of other murine IgG subclasses by IgM recognizing murine IgG2a MAbs. Sera containing IgM that bound to both IgG2a MAbs (6B6C1 and H3A4) segregated into three subsets with respect to binding to IgG2b and IgG1 MAbs; one subset (28%) recognized neither the IgG2b nor IgG1 MAb, another subset (44%) recognized either the IgG2b or IgG1 MAb, and a third subset (28%) recognized both IgG2b and IgG1 MAbs. Koshida et al. 5 performed a similar analysis, and found a similar distribution of 22, 55, and 22%, respectively.
Another unexpected finding from the study was that only 18% of sera containing IgM recognizing murine MAbs exhibited increased levels of HAMA. One would expect the vast majority of sera reactive with murine MAbs to also bind to murine polyclonal IgG, the target antigen used in the EIA we employed to measure HAMA 3. Further, this EIA is a bridging assay, and should thus detect IgM as well as IgG HAMAs 5, 6. However, a positive HAMA assay result requires binding to both immobilized murine IgG and liquid‐phase murine IgG 3, whereas a positive result in our assay for IgM recognition of murine MAbs requires only binding to liquid‐phase murine IgG. Thus, a possible explanation for the low concordance between HAMA EIA and our EIA results is that the MAb epitopes recognized by captured IgM in our assay are not available on immobilized polyclonal murine IgG.
A limitation to our study is lack of information on possible stimuli triggering the production of human IgM capable of binding to murine MAbs, as detected by our EIA. Known stimuli include therapeutic murine MAbs and dietary exposure to murine proteins 2, 3, 4, 7; other stimuli, as yet unidentified, may exist. The reactive sera we identified were submitted for dengue virus antibody testing, making it unlikely that the reactive IgM detected was induced by MAb therapy.
The findings presented here have important practical implications for patient care; sera giving false‐positive results in IgM‐capture assays that employ a murine IgG2a MAb conjugate may also give false‐positive results in IgM‐capture assays that employ conjugates of other murine MAb IgG subclasses. Laboratorians should consider using a background subtraction modification of all IgM‐capture EIAs employing murine MAb conjugates, regardless of MAb IgG subclass, in order to identify false‐positive results due to direct interactions between captured IgM and conjugated MAb. Indeed, we have implemented background subtraction retesting of all screen‐positive sera identified using FDA‐cleared IgM‐capture EIA kits for rubella virus IgM, cytomegalovirus IgM, and Toxoplasma gondii IgM, even though the package inserts do not call for such retesting. Through this process, we have identified sera exhibiting false‐positive reactivity in each of these three assays.
REFERENCES
- 1. Prince HE, Yeh C, Lapé‐Nixon M. Development of a more efficient algorithm for identifying false‐positive reactivity results in a dengue virus immunoglobulin M screening assay. Clin Vaccine Immuno 2008;15:1304–1306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Reinsberg J. Interferences with two‐site immunoassays by human anti‐mouse antibodies formed by patients treated with monoclonal antibodies: comparison of different block reagents. Clin Chem 1998;44:1742–1744. [PubMed] [Google Scholar]
- 3. Kricka LJ. Human anti‐animal antibody interferences in immunological assays. Clin Chem 1999;45:942–956. [PubMed] [Google Scholar]
- 4. Levinson SS, Miller JJ. Towards a better understanding of heterophile (and the like) antibody interference with modern immunoassays. Clin Chim Acta 2002;325:1–15. [DOI] [PubMed] [Google Scholar]
- 5. Koshida S, Asanuma K, Kuribayashi K, et al. Prevalence of human anti‐mouse antibodies (HAMAs) in routine examinations. Clin Chim Acta 2010;411:391–394. [DOI] [PubMed] [Google Scholar]
- 6. Itaka M, Kitahama S, Fukasawa N, et al. Incidence of anti‐mouse IgG in normal subjects and patients with autoimmune thyroid disease. Intern Med 1992;31:984–988. [DOI] [PubMed] [Google Scholar]
- 7. Schroff RW, Foon KA, Beatty SM, Oldham RK, Morgan AC. Human anti‐murine immunoglobulin responses in patients receiving monoclonal antibody therapy. Cancer Res 1985:45:879–885. [PubMed] [Google Scholar]


