Abstract
Background
Treatment failure of antiretroviral therapy in HIV‐1 infection is increasing due to development of viral resistance. Trends of resistance‐associated mutation lead to the ineffective treatment in HIV‐infected individuals.
Methods
Extracted viral RNA from HIV‐infected subjects in 2009 to 2010 was performed. The genotypic resistance testing was investigated for HIV‐1 drug resistance in RT and PR genes. Frequencies of mutation were compared by a Fischer's exact test.
Results
Three hundred and sixty‐nine samples (147 in 2009 and 222 in 2010) were genotyped. At least one mutation was found in 90.8% (335/369) in PR gene and 87.0% (321/369) in RT gene. Three sequences in PR gene, M36I, H69K, and L90M, were decreased significantly in 2010 when compared to 2009. Mutations associated with resistance to nucleoside analogue reverse transcriptase inhibitors (NRTI's) were found in 61.0% and 64.2% in nonnucleoside analogue reverse transcriptase inhibitors (NNRTI's). A total of 49.6% was found in combined NRTI and NNRTI. In 2010, M41L was increased significantly from 7.5% to 14.9%. However, there was a decrease in the frequency of the mutations at position 67, 70, and 184 between 2009 and 2010.
Conclusions
In 2010, three mutations in PR gene, M36I, H69K, and L90M, were decreased significantly. However, only one mutation in RT gene, M41L was significantly increased.
Keywords: HIV, prevalence, drug resistance
INTRODUCTION
The rapid development of resistance to single drug led to the use of combination therapies of three or more drugs. These combinations usually consist of a backbone of two complementary NRTI and either an NNRTI or one or two PIs. These combination therapies have been called highly active antiretroviral therapy (HAART). In terms of preventing the onset of resistance, this strategy has a number of advantages. First, the combination of drugs results in much greater levels of viral suppression; the reduction in viral turnover reduces the rate at which mutants are produced. Second, the development of resistance is much more complex as the virus must acquire mutations that induce resistance to a range of drugs, raising the genetic barrier 1, 2. However, virological failure of these regimens, due to development of viral resistance is becoming increasingly common 3, 4, 5, 6, 7. This indicates that current therapeutic regimens may not suppress virus sufficiently in the clinical situation to prevent development of resistance with selection of resistant quasi‐species 2, 8, 9.
Factors leading to treatment failure in HIV‐1 infection are numerous and complex. These are related to drug factors (limited potency of regimen, low genetic barrier, sub‐inhibitory plasma levels, pharmacological factors), host‐related factors (poor adherence, limited recovery capacity of the immune system, prior drug experience), and viral‐related factors (viral kinetics, error‐prone reverse transcription, presence of resistant variants) 6, 10, 11, 12, 13, 14, 15, 16, 17, 18, 19, 20, 21.
The capacity of HIV to develop drug resistance mutations is a major obstacle to effective long‐term therapy 22, 23, 24, 25, 26, 27. The increased use of combination antiretroviral therapies for HIV‐1 infection has led to the emergence of viral strains resistant to all licensed reverse transcriptase inhibitors (RTIs) and protease inhibitors (PIs) 27, 28, 29, 30, 31. Mutations selected by drug resistance are classified as primary and secondary mutations. Primary mutations are generally selected early in the process of resistance mutation accumulation and tend to be specific for each compound. They have a marked effect on virus drug susceptibility in phenotypic assays. Secondary mutations by themselves have little or no discernible effect on resistance but may be selected because they compensate for reduced viral fitness induced by the primary mutation. Secondary mutations usually occur in viral genomes already containing one or more primary mutations. High prevalence of drug‐resistant virus among HIV‐1 infected patients on therapy has increased the probability of transmission of virus resistant to one or more classes of antiretroviral drugs 27, 28, 29, 30, 31. Complete suppression of HIV‐1 could be compromised if therapy‐naïve patients already harbor virus with mutations conferring resistance to antiretroviral drugs used for initial therapy 29, 31, 32. Furthermore, incomplete suppression of viral replication promotes the development of broader drug resistance, compromising subsequent treatment regimens. A number of studies have evaluated rates of resistance mutations in recently transmitted virus by looking at subjects with primary infection 28, 29, 32, 33. Reported rates of drug resistance in these studies vary from 1.4 to 37.0% with most of the resistance mutations seen in the RT gene 23, 27, 31, 34, 35, 36, 37, 38, 39, 40. In this study, rates of genotypic resistance in both PR and RT genes were determined in HIV infection from 2009 to 2010 in Thailand.
MATERIALS AND METHODS
Patients and Blood Samples
The study was commenced in January 2009. All patients were identified with HIV infection and received antiretroviral therapy prior at the first plasma sample being collected. A sterile plasma sample stored at −70°C was included in the study.
Genotypic Resistance Analysis
Viral RNA and genotypic resistance testing from plasma was successfully determined on all subjects using standard methodologies. Plasma viral RNA was sequenced using commercially available kits. Briefly, the procedure was as follows: viral RNA was isolated from 140 μl plasma samples using the QIAamp Viral Extraction Kit (Qiagen Inc., Chatsworth, CA). The extracted viral RNA was reversed transcribed to cDNA using the RT‐PCR Trugene kit. A 1.3 kb amplicon covering the entire polymerase gene, produced by single‐tube PCR, was subjected to bidirectional DNA sequencing employing three primer pairs, and fluorescent dye primer chemistry on both forward and reverse strands (TruGene, Siemen). The sequencing reactions were loaded into an automated DNA sequencing system (CLIP, Siemen). This assay allows the sequencing of the amino acids 1–99 of PR and 1–247 of RT. Sequences were assembled and aligned with a lymphadenopathy‐associated virus type 1 (LAV‐1) consensus sequence using Trugene Gene Librarian Software. The software incorporates a rules based algorithm by comparing the derived patient sequence against the consensus to determine the presence of primary and secondary mutations. Genotypic resistance mutations were defined according to the International AIDS Society‐USA recommendations 41.
Statistical Analysis
Comparison between frequencies of wild‐type and mutant sequences were compared by a Fischer's exact test using Statistical Package for the Social Sciences (SPSS).
RESULTS
Between 1st January 2009 and 31st December 2010, 369 patients with HIV infection and appropriately stored plasma samples were identified. The demographic characteristics of this group are summarized in Table 1. Mean age was 39 years and the most common risk factor for acquisition of HIV‐1 was sexual contact. These patients had high viral load (Table 1). Most of all viral sequences were A/E subtype (359/369) as determined using the Program Manual for the Wisconsin Package, Version 8 (Genetics Computer Group, Madison, Wisconsin) software. Ten of 369 were B subtype. Of these 369 patients, 147 presented in 2009 and 222 presented in 2010. PR and RT sequences were obtained on all individuals. Table 2 summarizes the primary and secondary mutations detected in PR and RT genes in this population. At least one mutation associated with resistance to antiretroviral drugs was detected in 90.8% (335/369) in PR and 87.0% (321/369) in RT.
Table 1.
Baseline Demographic Characteristics of 369 Subjects With HIV Infection
| Age, mean (range) | 39 (20–75) |
| Sex, number (%) | |
| Male | 110 (29.8) |
| Female | 101 (27.4) |
| MSM | 158 (42.8) |
| Initial plasma HIV RNA level, (copies/ml) Mean | 6350 |
| (Range) | (2,200–10,500) |
Table 2.
Frequency of Multiple Resistance Mutations in Single Isolate in 2009 and 2010
| Protease | Reverse transcriptase | |||
|---|---|---|---|---|
| Number of mutations | 2009 | 2010 | 2009 | 2010 |
| 1 | 22.0% | 39.1% | 19.5% | 19.0% |
| 2 | 29.3% | 16.3% | 9.8% | 2.3% |
| 3 | 4.9% | 5.8% | 2.4% | 1.2% |
| 4 | 0 | 1.2% | 2.4% | 0.4% |
| 5 | 0 | 0.4% | 0 | 0.4% |
Resistance Mutations in PR Gene
Two hundred and sixty five (72.9%) sequences carried a primary protease inhibitor resistance mutation. Primary mutations observed were M46N/L, V82I, and L90M. All other resistance mutations were secondary mutations (Table 2). Among these primary mutations, L90M was found in 64.8%. In decreasing order of frequency, they were M36I, H69K, L10I/V, L63P/Q/N/T/A/S, K20R, V77I, and A71T/V. Figure 1 shows the number of primary and secondary mutations seen in PR each year. L90M was associated with H69K, while M46N/L and V82I were not accompanied by any secondary mutations. The most frequency of secondary mutations was found at position 36 (82.1%). If the data are considered, the most common polymorphism was M36I that occurred in 96.7% (142 of 147) in 2009 and 72.5% (161 of 222) in 2010. Three sequences in PR gene, M36I, H69K, and L90M, were decreased significantly in 2010 when compared to 2009 (P < 0.05). However, there was not significant increase in the proportion of PR resistance mutations represented by L101/V pre‐ and post‐2009 from 23.13 to 26.1% (Fig. 1). If the data set is divided into sequences from patients presenting between 2009 and 2010 then 46.3% (68/147) and 48.2% (107/222), respectively of these groups carried sequences with at least one secondary mutation in the PR gene (Table 2).
Figure 1.
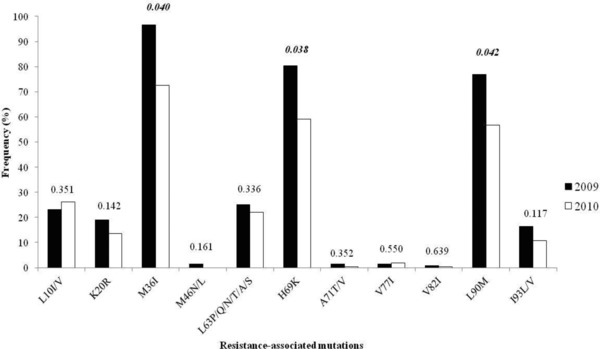
Frequency histogram showing the rates of primary and secondary resistance mutations in the PR gene 2009 and 2010. P values are determined from a Fischer's exact test.
Resistance Mutations in RT Gene
Mutations associated with resistance to nucleoside analogue reverse transcriptase inhibitors (NRTI's) were found in 61.0%, while those associated with resistance to nonnucleoside analogue reverse transcriptase inhibitors (NNRTI's) were found in 64.2%. Patients had combined NRTI and NNRTI mutations were found in 49.6%. Eleven different primary RT mutations were found: M184V (58.5%), Y181C/I (42.9%), K103N (21.0%), D67N (19.7%), G109S/A (19.7%), K70R (15.6%), K219Q/E (9.5%), M41L (7.5%), L210W (6.8%), T215Y/E (6.8%), and P225H (4.1%) in 2009 and M184V (53.1%), Y181C/I (35.1%), K103N (22.5%), G109S/A (19.4%), D67N (15.3%), M41L (14.9%), K70R (11.7%), K219Q/E (11.3%), L210W (6.8%), T215Y/E (9.9%), and P225H (2.7%) in 2010. Secondary mutations in the RT gene were observed at positions 50 and 67. A total of 50.6% of subjects had one or two RT primary or secondary resistance mutations, 6.8% carried more than two mutations (Table 2). The frequency of primary and secondary mutations in the RT gene is shown in Figures 2 and 3. There was a decrease in the frequency of the following mutations: D67N (19.7 to 15.3%), K70R (15.6 to 11.7%), and M184V (58.5 to 53.2%) between 2009 and 2010 (Fig. 2) whereas the primary mutations at position 41, 215, and 219 were increased. In 2010, primary mutation in the RT gene at position 41 was increased from 7.5 to 14.9%. There was a statistically significant difference (P < 0.05). Of those, M184V was the most common (55.3%) that confers FTC or 3TC resistance. Combinations of this mutation are usually associated with high‐level zidovudine resistance. Combined M184V and T215Y mutations were detected in 2.4% (9/369) and 12.7% (47/369) in 2009 and 2010, respectively. The resistance mutations of NNRTIs were detected in 64.2%. The most frequency of NNRTI resistance mutations were Y181C/I (38.2%), K103N (22.0%), and G190S/A (19.5%). The mutations at position 181, 188, 190, and 225 were decreased in 2010 when compared to 2009. In contrast to the mutation at position 103, there was an increase in 2010. However, it was not statistically significant difference (P < 0.05).
Figure 2.
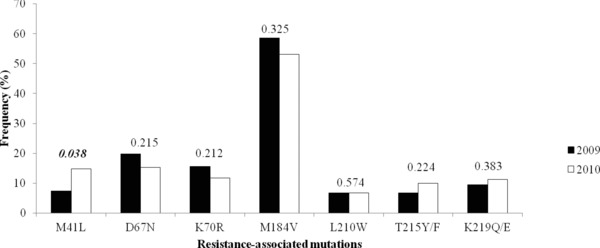
There was AZT‐related resistance mutation in RT pre‐ and post‐2009. P values are determined from a Fischer's exact test.
Figure 3.
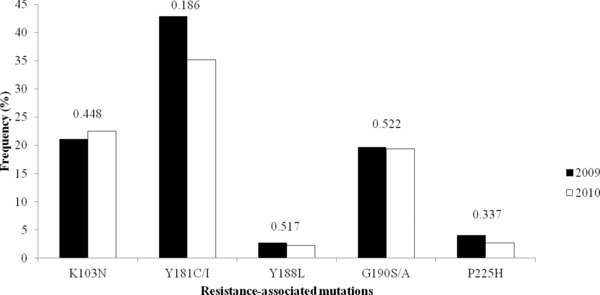
There was NNRTI‐related resistance mutation in RT pre‐ and post‐2009. P values are determined from a Fischer exact test.
DISCUSSION
The development of resistance mutations in patients receiving combination therapy has resulted in resistant virus becoming more common within the HIV‐infected population. It has raised concerns that such mutants may be transmitted more frequently, compromising not only the effectiveness of treatment of an individual but also the effectiveness of therapy on a population basis. Some reports have suggested that in excess of a quarter of subjects with recent acquisition of HIV have potentially drug‐resistant virus 42, 43, 44, 45, 46.
The increasing prevalence of drug‐resistant mutations among HIV‐1 infected patients on treatment has increased the probability of transmission of virus resistant to one or more classes of antiretroviral drugs 47, 48, 49, 50, 51, 52. Incomplete suppression in turn promotes the development of broader drug resistance, compromising subsequent treatment regimens 1, 2, 6, 13.
Rates of new infection numbers caused by resistant virus vary markedly from study to study. Most studies report that at least 10% of new primary HIV‐1 infected people carry virus resistant to at least one of the antiretroviral drugs while they are still therapy naive, suggesting that they have been infected with drug‐resistant virus 53, 54, 55, 56, 57, 58, 59, 60, 61, 62, 63, 64, 65, 66. However, some recent studies have suggested significant increases in the rates of transmission of drug‐resistant HIV‐1 within excess of 25% of newly infected individuals carrying virus with at least one primary mutation. 55, 67, 68, 69, 70, 71, 72. By contrast, another European study sampling genotypes from 369 patients with recently acquired infection in Greece between 2000 and 2007 found that only 7.6% of viral sequences had mutations associated with NRTI resistance, 5.4% had mutations associated with NNRTI resistance, and only 3.3% had mutations associated with PI resistance 31.
Overall, most of the reported transmitted mutations occur in the RT rather than in the PR gene though relative frequencies vary markedly from study to study and may depend upon the dominant risk factors associated with transmission 31, 35, 45, 73, 74, 75, 76. In general, higher rates of PR resistance have been seen in populations with intravenous drug use as a major risk factor, however the sample sizes employed in these studies are too small to make any firm conclusions regarding whether the types of transmitted resistance mutations is related to the mode of transmission (e.g. mucosal vs. blood borne transmission) 77, 78, 79.
While resistance mutations in RT are more commonly seen than mutations in PR; the RT mutations associated with AZT are the most commonly described 27, 29, 35, 80, 81. Relative rates of drug use are unlikely to explain the preferential finding of certain resistance mutations among resistant virus. It may be that the mutations most commonly reported in transmitted virus are those that have less implications in terms of viral fitness, especially in regards to transmission and initiation of infection 69, 82. There is a growing body of evidence supporting the phenomenon of reversion of mutations in the absence of drug pressure in transmitted virus, but reversion may not be to wild‐type but to an intermediate strain. The best evidence for this phenomenon comes from the study of mutations at codon 215 in RT. The mutation T215Y is a common and well‐described primary mutation for AZT resistance. In combination with M41L, it can induce high‐level resistance to AZT. Although both these mutations are seen in transmitted virus, either alone or in combination, another set of mutations at 215 coding for one of the amino acids, D, C, or S have been found in a significant minority of samples (approximately 3–4%) in three independent studies 60, 69, 83, 84.
In our findings, frequency of mutations in PR gene has decreased in 2010. One of the reasons for the lack of increase in protease resistance mutations may be that the introduction of these drugs coincided with the institution of combination therapy. The resulting reductions in viral load and reduced rates of viral turnover resulting from these therapeutic strategies may have played a role in the lack of generation and transmission of protease inhibitor resistance.
Not only has the rate of mutations found in transmitted virus decreased but also the type of mutations seen also has changed, again reflecting changes in drug usage. The number of individual mutations associated with AZT resistance decreased, as did the number of combined AZT resistance mutations with decreased rates of AZT usage within the treated population. Notably, there was no significant increase in the rates of mutations associated with other RT inhibitors used within this population, which may reflect the increased use of these drugs in combination therapies during this period. This is especially noteworthy in the case of 3TC which is the most common NRTI included in the regimens of greater than 60% of those on treatment.
These data provide insight into how treatment strategies impact upon drug resistance virus. While the rates of transmission of resistance virus during the last decade are notable, they are much lower than those seen prior to the introduction of combination therapy. The change in rates of resistance seen in this study suggests that data derived from cross sectional studies should be interpreted with caution.
In this population, primary PR resistance mutations decreased and the rate of secondary resistance mutations was high since the introduction of protease inhibitors into this population. The prevalence of RT resistance mutations appears to be decreased. Only one mutation in this gene, M41L was significantly increased.
ACKNOWLEDGEMENT
This paper is supported by the 90th Anniversary of Chulalongkorn University Fund (Ratchadaphiseksomphot Endowment Fund).
REFERENCES
- 1. Hirsch MS, Conway B, D'Aquila RT, et al. Antiretroviral drug resistance testing in adults with HIV infection: Implications for clinical management. International AIDS Society–USA Panel. Jama 1998;279(24):1984–1991. [DOI] [PubMed] [Google Scholar]
- 2. Wegner SA, Brodine SK, Mascola JR, et al. Prevalence of genotypic and phenotypic resistance to anti‐retroviral drugs in a cohort of therapy‐naive HIV‐1 infected US military personnel. Aids 2000;14(8):1009–1015. [DOI] [PubMed] [Google Scholar]
- 3. Blower SM, Aschenbach AN, Gershengorn HB, Kahn JO. Predicting the unpredictable: Transmission of drug‐resistant HIV. Nat Med 2001;7(9):1016–1020. [DOI] [PubMed] [Google Scholar]
- 4. Brenner BG, Wainberg MA. The role of antiretrovirals and drug resistance in vertical transmission of HIV‐1 infection. Ann N Y Acad Sci 2000;918:9–15. [DOI] [PubMed] [Google Scholar]
- 5. Opravil M, Hirschel B, Lazzarin A, et al. A randomized trial of simplified maintenance therapy with abacavir, lamivudine, and zidovudine in human immunodeficiency virus infection. J Infect Dis 2002;185(9):1251–1260. [DOI] [PubMed] [Google Scholar]
- 6. Hirsch MS, Brun‐Vezinet F, Clotet B, et al. Antiretroviral drug resistance testing in adults infected with human immunodeficiency virus type 1: 2003 recommendations of an International AIDS Society‐USA Panel. Clin Infect Dis 2003;37(1):113–128. [DOI] [PubMed] [Google Scholar]
- 7. D'Aquila RT, Schapiro JM, Brun‐Vezinet F, et al. Drug resistance mutations in HIV‐1. Top HIV Med 2003;11(3):92–96. [PubMed] [Google Scholar]
- 8. Frost SD, McLean AR. Quasispecies dynamics and the emergence of drug resistance during zidovudine therapy of HIV infection. Aids 1994;8(3):323–332. [DOI] [PubMed] [Google Scholar]
- 9. Kellam P, Boucher CA, Tijnagel JM, Larder BA. Zidovudine treatment results in the selection of human immunodeficiency virus type 1 variants whose genotypes confer increasing levels of drug resistance. J Gen Virol 1994;75(Pt 2):341–351. [DOI] [PubMed] [Google Scholar]
- 10. Shafer RW, Winters MA, Palmer S, Merigan TC. Multiple concurrent reverse transcriptase and protease mutations and multidrug resistance of HIV‐1 isolates from heavily treated patients. Ann Intern Med 1998;128(11):906–911. [DOI] [PubMed] [Google Scholar]
- 11. Cabana M, Clotet B, Martinez MA. Emergence and genetic evolution of HIV‐1 variants with mutations conferring resistance to multiple reverse transcriptase and protease inhibitors. J Med Virol 1999;59(4):480–490. [DOI] [PubMed] [Google Scholar]
- 12. Bangsberg DR, Perry S, Charlebois ED, et al. Non‐adherence to highly active antiretroviral therapy predicts progression to AIDS. Aids 2001;15(9):1181–1183. [DOI] [PubMed] [Google Scholar]
- 13. Taylor S, Cane P, Hue S, et al. Identification of a transmission chain of HIV type 1 containing drug resistance‐associated mutations. AIDS Res Hum Retroviruses 2003;19(5):353–361. [DOI] [PubMed] [Google Scholar]
- 14. Paterson DL, Swindells S, Mohr J, et al. Adherence to protease inhibitor therapy and outcomes in patients with HIV infection. Ann Intern Med 2000;133(1):21–30. [DOI] [PubMed] [Google Scholar]
- 15. Louie M, Hogan C, Di Mascio M, et al. Determining the relative efficacy of highly active antiretroviral therapy. J Infect Dis 2003;187(6):896–900. [DOI] [PubMed] [Google Scholar]
- 16. Kirk O, Pedersen C, Law M, et al. Analysis of virological efficacy in trials of antiretroviral regimens: drawbacks of not including viral load measurements after premature discontinuation of therapy. Antivir Ther 2002;7(4):271–281. [PubMed] [Google Scholar]
- 17. Volberding P. Adherence, resistance, and timing: current issues in the use of new therapies. AIDS Read 2002;12(8):349–350, 356–357, 368. [PubMed] [Google Scholar]
- 18. Hogg RS, Heath K, Bangsberg D, et al. Intermittent use of triple‐combination therapy is predictive of mortality at baseline and after 1 year of follow‐up. Aids 2002;16(7):1051–1058. [DOI] [PubMed] [Google Scholar]
- 19. Walsh JC, Pozniak AL, Nelson MR, Mandalia S, Gazzard BG. Virologic rebound on HAART in the context of low treatment adherence is associated with a low prevalence of antiretroviral drug resistance. J Acquir Immune Defic Syndr 2002;30(3):278–287. [DOI] [PubMed] [Google Scholar]
- 20. Masuhr A, Mueller M, Simon V, et al. Predictors of treatment failure during highly active antiretroviral therapy (racing trial). Eur J Med Res 2002;7(8):341–346. [PubMed] [Google Scholar]
- 21. Rambaut A, Posada D, Crandall KA, Holmes EC. The causes and consequences of HIV evolution. Nat Rev Genet 2004;5(1):52–61. [DOI] [PubMed] [Google Scholar]
- 22. Deeks SG, Gange SJ, Kitahata MM, et al. Trends in multidrug treatment failure and subsequent mortality among antiretroviral therapy‐experienced patients with HIV infection in North America. Clin Inf Dis 2009;49(10):1582–1590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Jakobsen MR, Tolstrup M, S gaard OS, et al. Transmission of HIV‐1 drug‐resistant variants: Prevalence and effect on treatment outcome. Clin Inf Dis 2010;50(4):566–573. [DOI] [PubMed] [Google Scholar]
- 24. Moreno S, L pez Aldeguer J, Arribas JR et al. The future of antiretroviral therapy: Challenges and needs. J Antimicrob Chemoth 2010;65(5):827–835. [DOI] [PubMed] [Google Scholar]
- 25. Hoffmann CJ, Charalambous S, Sim J, et al. Viremia, resuppression, and time to resistance in human immunodeficiency virus (HIV) subtype C during first‐line antiretroviral therapy. Clin Inf Dis 2009;49(12):1928–1935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Volberding PA, Deeks SG. Antiretroviral therapy and management of HIV infection. The Lancet 2010;376(9734):49–62. [DOI] [PubMed] [Google Scholar]
- 27. Berm dez‐Aza EH, Kerr LRFS, Kendall C, et al. Antiretroviral drug resistance in a respondent‐driven sample of HIV‐infected men who have sex with men in Brazil. JAIDS J Acq Immun Def Synd 2011;57:S186–S192. [DOI] [PubMed] [Google Scholar]
- 28. Myers JE, Taylor BS, Rojas Ferm n RA, et al. Transmitted drug resistance among antiretroviral‐naive patients with established HIV type 1 infection in Santo Domingo, Dominican Republic and Review of the Latin American and Caribbean Literature. AIDS Res Hum Retroviruses 2011;28(7):667–674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Lai A, Violin M, Ebranati E, et al. Transmission of resistant HIV‐1 variants and epidemiological chains in italian newly diagnosed individuals. AIDS Res Hum Retroviruses 2011;28(8):857–865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Jones LR, Moretti F, Calvo AY, et al. Drug resistance mutations in HIV pol sequences from Argentinean patients under antiretroviral treatment: Subtype, gender and age issues. AIDS Res Hum Retroviruses 2011;28(8):949–955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Skoura L, Metallidis S, Buckton AJ, et al. Molecular and epidemiological characterization of HIV‐1 infection networks involving transmitted drug resistance mutations in Northern Greece. J Antimicrob Chemother 2011;66(12):2831–2837. [DOI] [PubMed] [Google Scholar]
- 32. Ndembi N, Abraha A, Pilch H, et al. Molecular characterization of human immunodeficiency virus type 1 (HIV‐1) and HIV‐2 in Yaounde, Cameroon: Evidence of major drug resistance mutations in newly diagnosed patients infected with subtypes other than subtype B. J Clin Microbiol 2008;46(1):177–184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Turriziani O, Russo G, Lichtner M, et al. Study of the genotypic resistant pattern in HIV‐infected women and children from rural west Cameroon. AIDS Res Hum Retroviruses 2008;24(6):781–785. [DOI] [PubMed] [Google Scholar]
- 34. Booth CL, Geretti AM. Prevalence and determinants of transmitted antiretroviral drug resistance in HIV‐1 infection. J Antimicrob Chemother 2007;59(6):1047–1056. [DOI] [PubMed] [Google Scholar]
- 35. Vercauteren J, Wensing AMJ, van de Vijver DAMC, et al. Transmission of drug‐resistant HIV‐1 is stabilizing in Europe. J Inf Dis 2009;200(10):1503–1508. [DOI] [PubMed] [Google Scholar]
- 36. Brindeiro RM, Diaz RS, Sabino EC, et al. Brazilian network for HIV drug resistance surveillance (HIV‐BResNet): A survey of chronically infected individuals. Aids 2003;17(7):1063–1069. [DOI] [PubMed] [Google Scholar]
- 37. Pedroso C, Queiroz ATL, Alc ntara LC, et al. High prevalence of primary antiretroviral resistance among HIV‐1‐infected adults and children in Bahia, a northeast state of Brazil. J Acq Immun Def Synd 2007;45(2):251–253. [DOI] [PubMed] [Google Scholar]
- 38. Barreto CC, Nishyia A, Ara jo LV, Ferreira JE, Busch MP, Sabino EC. Trends in antiretroviral drug resistance and clade distributions among HIV‐1–infected blood donors in Sao Paulo, Brazil. J Acq Immun Def Synd 2006;41(3):388–341. [DOI] [PubMed] [Google Scholar]
- 39. Sucupira MCA, Caseiro MM, Alves K, et al. High levels of primary antiretroviral resistance genotypic mutations and B/F recombinants in Santos, Brazil. Aids Patient Care STDs 2007;21(2):116–128. [DOI] [PubMed] [Google Scholar]
- 40. Soria J, Bull M, Mitchell C, et al. Transmitted HIV resistance to first‐line antiretroviral therapy in Lima, Peru. AIDS Res Hum Retroviruses 2011;28(4):333–338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Hirsch MS, Brun‐Vezinet F, D'Aquila RT, et al. Antiretroviral drug resistance testing in adult HIV‐1 infection: Recommendations of an International AIDS Society‐USA Panel. Jama 2000;283(18):2417–2426. [DOI] [PubMed] [Google Scholar]
- 42. Cozzi‐Lepri A, Dunn D, Pillay D, et al. Long‐term probability of detecting drug‐resistant HIV in treatment‐naive patients initiating combination antiretroviral therapy. Clin Infect Dis 2010;50(9):1275–1285. [DOI] [PubMed] [Google Scholar]
- 43. Koyalta D, Charpentier C, Beassamda J, et al. High frequency of antiretroviral drug resistance among HIV‐infected adults receiving first‐line highly active antiretroviral therapy in N'Djamena, Chad. Clin Infect Dis 2009;49(1):155–159. [DOI] [PubMed] [Google Scholar]
- 44. Hamers RL, Derdelinckx I, van Vugt M, et al. The status of HIV‐1 resistance to antiretroviral drugs in sub‐Saharan Africa. Antivir Ther 2008;13(5):625–639. [PubMed] [Google Scholar]
- 45. Price MA, Wallis CL, Lakhi S, et al. Transmitted HIV type 1 drug resistance among individuals with recent HIV infection in East and Southern Africa. AIDS Res Hum Retroviruses 2011;27(1):5–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Tanuma J, Hachiya A, Ishigaki K, et al. Impact of CRF01_AE‐specific polymorphic mutations G335D and A371V in the connection subdomain of human immunodeficiency virus type 1 (HIV‐1) reverse transcriptase (RT) on susceptibility to nucleoside RT inhibitors. Microbes Infect 2010;12(14–15):1170–1177. [DOI] [PubMed] [Google Scholar]
- 47. Vaishnav YN, Wong‐Staal F. The biochemistry of AIDS. Annu. Rev Biochem 1991;60(1):577–630. [DOI] [PubMed] [Google Scholar]
- 48. Wilson W, Braddock M, Adams SE, Rathjen PD, Kingsman SM, Kingsman AJ. HIV expression strategies: Ribosomal frameshifting is directed by a short sequence in both mammalian and yeast systems. Cell 1988;55(6):1159–1169. [DOI] [PubMed] [Google Scholar]
- 49. Jacks T, Power MD, Masiarz FR, Luciw PA, Barr PJ, Varmus HE. Characterization of ribosomal frameshifting in HIV‐1 gag‐pol expression. 1988;331(6153):280–283. [DOI] [PubMed] [Google Scholar]
- 50. Brierley I. Ribosomal frameshifting on viral RNAs. J Gen Virol 1995;76(8):1885–1892. [DOI] [PubMed] [Google Scholar]
- 51. Kollmus H, Honigman A, Panet A, Hauser H. The sequences of and distance between two cis‐acting signals determine the efficiency of ribosomal frameshifting in human immunodeficiency virus type 1 and human T‐cell leukemia virus type II in vivo. J Virol 1994;68(9):6087–6091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Reil H, Kollmus H, Weidle U, Hauser H. A heptanucleotide sequence mediates ribosomal frameshifting in mammalian cells. J Virol 1993;67(9):5579–5584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Balotta C, Berlusconi A, Pan A, et al. Prevalence of transmitted nucleoside analogue‐resistant HIV‐1 strains and pre‐existing mutations in pol reverse transcriptase and protease region: Outcome after treatment in recently infected individuals. Antivir Ther 2000;5(1):7–14. [PubMed] [Google Scholar]
- 54. Boden D, Hurley A, Zhang L, et al. HIV‐1 drug resistance in newly infected individuals. Jama 1999;282(12):1135–1141. [DOI] [PubMed] [Google Scholar]
- 55. Briones C, Perez‐Olmeda M, Rodriguez C, del Romero J, Hertogs K, Soriano V. Primary genotypic and phenotypic HIV‐1 drug resistance in recent seroconverters in Madrid. J Acquir Immune Defic Syndr 2001;26(2):145–150. [DOI] [PubMed] [Google Scholar]
- 56. Duwe S, Brunn M, Altmann D, et al. Frequency of genotypic and phenotypic drug‐resistant HIV‐1 among therapy‐naive patients of the German Seroconverter Study. J Acquir Immune Defic Syndr 2001;26(3):266–273. [DOI] [PubMed] [Google Scholar]
- 57. Little SJ, Daar ES, D'Aquila RT, et al. Reduced antiretroviral drug susceptibility among patients with primary HIV infection. Jama 1999;282(12):1142–1149. [DOI] [PubMed] [Google Scholar]
- 58. Salomon H, Wainberg MA, Brenner B, et al. Prevalence of HIV‐1 resistant to antiretroviral drugs in 81 individuals newly infected by sexual contact or injecting drug use. Investigators of the Quebec Primary Infection Study. Aids 2000;14(2):F17–F23. [DOI] [PubMed] [Google Scholar]
- 59. Tamalet C, Pasquier C, Yahi N, et al. Prevalence of drug resistant mutants and virological response to combination therapy in patients with primary HIV‐1 infection. J Med Virol 2000;61(2):181–186. [DOI] [PubMed] [Google Scholar]
- 60. Yerly S, Kaiser L, Race E ea. Transmission of antiretroviral drug resistant HIV‐1 variants. Lancet 1999;354:729–733. [DOI] [PubMed] [Google Scholar]
- 61. Little SJ, Holte S, Routy JP, et al. Antiretroviral‐drug resistance among patients recently infected with HIV. N Engl J Med 2002;347(6):385–394. [DOI] [PubMed] [Google Scholar]
- 62. Barbour JD, Hecht FM, Wrin T, et al. Persistence of primary drug resistance among recently HIV‐1 infected adults. Aids 2004;18(12):1683–1689. [DOI] [PubMed] [Google Scholar]
- 63. Hanna GJ, Balaguera HU, Freedberg KA, et al. Drug‐selected resistance mutations and non‐B subtypes in antiretroviral‐naive adults with established human immunodeficiency virus infection. J Infect Dis 2003;188(7):986–991. [DOI] [PubMed] [Google Scholar]
- 64. Vercauteren J, Wensing AMJ, van de Vijver DAMC, et al. Transmission of drug‐resistant HIV‐1 is stabilizing in Europe. J Infect Dis 2009;200(10):1503–1508. [DOI] [PubMed] [Google Scholar]
- 65. Rockstroh J, Bhagani S, Benhamou Y, et al. European AIDS Clinical Society (EACS) guidelines for the clinical management and treatment of chronic hepatitis B and C coinfection in HIV‐infected adults. HIV Med 2008;9(2):82–88. [DOI] [PubMed] [Google Scholar]
- 66. Thompson MA, Aberg JA, Cahn P. Antiretroviral treatment of adult HIV infection: 2010 recommendations of the International AIDS Society‐USA panel. JAMA 2010;304(3):321–333. [DOI] [PubMed] [Google Scholar]
- 67. Little SJ. Transmission and prevalence of HIV resistance among treatment‐naive subjects. Antivir Ther 2000;5(1):33–40. [DOI] [PubMed] [Google Scholar]
- 68. UK Collaborative Group on Monitoring the Transmission of HIV Drug Resistance. Analysis of prevalence of HIV‐1 drug resistance in primary infections in the United Kingdom. BMJ 2001;322(7294):1087–1088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Garcia‐Lerma JG, Nidtha S, Blumoff K, Weinstock H, Heneine W. Increased ability for selection of zidovudine resistance in a distinct class of wild‐type HIV‐1 from drug‐naive persons. Proc Natl Acad Sci USA 2001;98(24):13907–13912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Brodine SK, Shaffer RA, Starkey MJ, et al. Drug resistance patterns, genetic subtypes, clinical features, and risk factors in military personnel with HIV‐1 seroconversion. Ann Intern Med 1999;131(7):502–506. [DOI] [PubMed] [Google Scholar]
- 71. Harrigan PR, Hogg RS, Dong WW, et al. Predictors of HIV drug‐resistance mutations in a large antiretroviral‐naive cohort initiating triple antiretroviral therapy. J Infect Dis 2005;191(3):339–347. [DOI] [PubMed] [Google Scholar]
- 72. Grossman Z, Lorber M, Maayan S, et al. Drug‐resistant HIV infection among drug‐naive patients in Israel. Clin Infect Dis 2005;40(2):294–302. [DOI] [PubMed] [Google Scholar]
- 73. Mosha F, Urassa W, Aboud S, et al. Prevalence of genotypic resistance to antiretroviral drugs in treatment‐naive youths infected with diverse HIV Type 1 subtypes and recombinant forms in Dar es Salaam, Tanzania. AIDS Res Hum Retroviruses 2011;27(4):377–382. [DOI] [PubMed] [Google Scholar]
- 74. Tebit DM, Sangaré L, Tiba F, et al. Analysis of the diversity of the HIV‐1 pol gene and drug resistance associated changes among drug‐naïve patients in Burkina Faso. J Med Virol 2009;81(10):1691–1701. [DOI] [PubMed] [Google Scholar]
- 75. Bártolo I, Casanovas J, Bastos R, et al. HIV‐1 genetic diversity and transmitted drug resistance in health care settings in Maputo, Mozambique. JAIDS 2009;51(3):323–331. [DOI] [PubMed] [Google Scholar]
- 76. Aghokeng AF, Vergne L, Mpoudi‐Ngole E, et al. Evaluation of transmitted HIV drug resistance among recently‐infected antenatal clinic attendees in four Central African countries. Antivir Ther 2009;14(3):401–411. [DOI] [PubMed] [Google Scholar]
- 77. Belyakov IM, Hel Z, Kelsall B, et al. Mucosal AIDS vaccine reduces disease and viral load in gut reservoir and blood after mucosal infection of macaques. Nat Med 2001;7(12):1320–1326. [DOI] [PubMed] [Google Scholar]
- 78. Beltrami EM, Williams IT, Shapiro CN, Chamberland ME. Risk and management of blood‐borne infections in health care workers. Clin Microbiol Rev 2000;13(3):385–407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Brenner B, Routy JP, Quan Y, et al. Persistence of multidrug‐resistant HIV‐1 in primary infection leading to superinfection. Aids 2004;18(12):1653–1660. [DOI] [PubMed] [Google Scholar]
- 80. Wensing A, Vercauteren J, Vijver D, et al. Transmission of drug‐resistant HIV‐1 in Europe remains limited to single classes. Aids 2008;22(5):625–635. [DOI] [PubMed] [Google Scholar]
- 81. Wensing AMJ, van de Vijver DA, Angarano G, et al. Prevalence of drug‐resistant HIV‐1 variants in untreated individuals in Europe: Implications for clinical management. J Infect Dis 2005;192(6):958–966. [DOI] [PubMed] [Google Scholar]
- 82. Garcia‐Lerma JG, MacInnes H, Bennett D, Weinstock H, Heneine W. Transmitted human immunodeficiency virus type 1 carrying the D67N or K219Q/E mutation evolves rapidly to zidovudine resistance in vitro and shows a high replicative fitness in the presence of zidovudine. J Virol 2004;78(14):7545–7552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Kuritzkes DR, Sevin A, Young B, et al. Effect of zidovudine resistance mutations on virologic response to treatment with zidovudine‐lamivudine‐ritonavir: Genotypic analysis of human immunodeficiency virus type 1 isolates from AIDS clinical trials group protocol 315. ACTG Protocol 315 Team. J Infect Dis 2000;181(2):491–497. [DOI] [PubMed] [Google Scholar]
- 84. Violin M, Cozzi‐Lepri A, Velleca R, et al. Risk of failure in patients with 215 HIV‐1 revertants starting their first thymidine analog‐containing highly active antiretroviral therapy. Aids 2004;18(2):227–235. [DOI] [PubMed] [Google Scholar]


