Abstract
Background
Current methods used to diagnose the thalassemia minor (TM) patients require high‐cost assays, while broader screening based on routine blood count has limited specificity and sensitivity. This study developed a new screening technique for TM patients’ diagnosis.
Methods
The study enrolled 526 patients database that included 185 verified α and β TM cases, and control group consisted of iron‐deficiency anemia (IDA), myelodysplastic syndrome (MDS), and healthy patients. More than 1,500 artificial neural networks (ANNs) models were created and the networks that gave high accuracy were selected for the study. TM patients were identified from the general database using the best‐optimized ANNs.
Results
Comparison between three or six routine blood count parameters determined a slightly higher accuracy of the model with the three‐parameter scheme, including mean corpuscular volume, red blood cell distribution width, and red blood cell. Based on these parameters, we were able to separate TM patients from the control group and MDS group, with specificity of 0.967 and sensitivity of 1. Including IDA patients into comparison gave lower but, still, very good values of specificity of 0.968 and sensitivity of 0.9.
Conclusion
ANN‐based TM diagnostics should be used for broad automatic screening of general population prior diagnosis with high‐cost tests.
Keywords: artificial neural networks, blood count, differential diagnosis, thalassemia minor, thalassemia screening
INTRODUCTION
The thalassemias represent the most common monogenic defect worldwide and are particularly prevalent in the Mediterranean region, Middle East, Southeast Asia, and some regions of Africa, representing a major public health problem in these areas 1. There are several thalassemia defects, all affecting the genes controlling globin production, and among them α and β thalassemia are the most common. Heterozygous thalassemia condition, called thalassemia minor (TM), is often unrecognized and undiagnosed. Currently, the best methods to determine the existence of hemoglobin (HB) defects are high‐performance liquid chromatography, protein electrophoresis, and mutation screening by PCR and DNA sequencing 2. All these techniques require special instrumentation, trained technicians and are of relatively high cost.
The early recognition of the TM condition in a patient will prevent unnecessary treatment and costly tests, and impede birth of thalassemia major children. Therefore, several attempts were previously made to identify the condition by a simple blood count with varying degrees of success. The use of complete blood count (CBC) parameters to screen for thalassemia can be divided into three major approaches: index based on a single parameter 3, 4, 5, 6, index based on several parameters 7, 8, 9, 10, 11, 12, 13, 14, 15, 16, 17, and index based on a nonlinear approach 18, 19, 20, 21, 22.
Analysis of an index based on a single parameter 3, 4, 5, 6 provides some insight into the variables that might assist in determining whether a patient is carrying thalassemia, however, the sensitivity and specificity seems low, and fails to identify thalassemia in the general population, apparently due to the influence of unconcerned parameters and large variance in blood sample characteristics. The indexes based on several parameters 7, 8, 9, 10, 11, 12, 13, 14, 15, 16, 17 are proven to be more successful, but aggregating the set of parameters to a single index reduces separation ability of this approach. Thus, the combination of sensitivity and specificity values of these indexes is insufficient for broad and, still, accurate screening of general population.
The use of the nonlinear approach requires a more complicated mathematical treatment; however, it may significantly improve the separation ability, specificity, and sensitivity. Artificial neural networks (ANNs) are one of the nonlinear tools, which are successfully applied to various fields in order to find patterns in ambiguous and cloudy data. Basically, ANNs are the mathematical algorithms generated by computers and resembling the biological cluster of neurons structurally and functionally. ANN consists of an interconnected group of artificial neurons that learn through experience, not from programming. Like a biological neuron network, the ANN is designed to obtain an input, process and amplify the signal through neuron connections, and supply the output, which in most cases would be some prediction about the patterns and behavior of the studied system. The power of neural computations comes from connecting neurons in a network like in a human brain and their greatest advantage is the ability to detect complex relationships in data where real human brain may fail to detect. In medicine, ANNs are widely used in diagnosis, medical image analysis, modeling, and medical data mining 23.
There are previous studies that are concerned with the problem of identifying TM patients from a healthy population using ANNs 18, 19, 20, 21, 22. These studies proved the usefulness of the ANNs method and were able to separate various thalassemia types among general datasets of CBC. Despite the fact that most of these studies used large number (up to 20) of CBC parameters, the results showed insufficient values of sensitivity and specificity. Adding chromatography HB typing improves the results but requires additional costly analysis 18. The only study that applied the ANN approach for thalassemia and had been based on only four blood parameters achieved a sensitivity value of 0.92 and a specificity value of 0.95 20.
Here, we created more than 1,500 ANNs and located the networks that gave an optimized accuracy result. This broad approach allowed us to separate thalassemia‐carrying patients from the general population, with specificity above 0.96 and sensitivity of 1.
MATERIALS AND METHODS
Patients
The patient database consisted of 526 patients of various ages and both genders living in Israel. The patients were divided into four groups as detailed in Table 1. For each patient, the following parameters were recorded: HB, mean corpuscular volume (MCV), mean corpuscular HB (MCH), red blood cell distribution width (RDW), red blood cell (RBC), and platelet count (PLT).
Table 1.
Patients Composition Inside the Database
| Group name | Number of patients |
|---|---|
| Control | 229 |
| Myelodysplastic syndrome (MDS) | 58 |
| Iron‐deficiency anemia (IDA) | 54 |
| α and β thalassemia minor | 185 |
The patients' data were collected in the database over a duration of 10 years. The data acquisition was conducted in accordance with Helsinki declaration. The database does not contain identifiable information such as age, gender, or name.
System Design
The motivation for dividing the system's design into several stages was to differentiate between a group of patients known to carry the thalassemia trait and other diseases known to change the blood parameters. Iron‐deficient anemia is known to have symptoms that are close to the symptoms exhibited by TM and we felt it important to examine our method's ability to separate these two groups. The group of patients diagnosed with myelodysplastic syndrome (MDS) was used as an additional control, in order to test our system's ability to differentiate a disease that changes the CBC parameters nonspecifically.
At each stage of the study, we compared the group of TM patients and the group of patients with different subgroups (control: healthy individuals, MDS, and iron‐deficiency anemia (IDA)).
Another important aspect of these stages was the choice of CBC parameters. In each comparison, first three parameters were used (stages 1–3) and then six (stages 4– 6). Table 2 outlines the different stages deployed in our study.
Table 2.
System Design Regarding Stages and CBC Parameters
| Stage | Groups | CBC parameters |
|---|---|---|
| 1 | Thalassemia vs.control group | MCV, RDW, RBC |
| 2 | Thalassemia vs. control and MDS group | |
| 3 | Thalassemia vs. control, MDS, and IDA | |
| 4 | Thalassemia vs. control group | MCV, RDW, RBC, HB, MCH, PLT |
| 5 | Thalassemia vs. control and MDS group | |
| 6 | Thalassemia vs. control, MDS, and IDA |
ANN Optimization
Above 1,500 ANNs were created and set to the task of recognizing patterns in the data using MATLAB programming environment 24. The constructed networks were designed to contain up to 16 fully connected neurons.
Selection of the best or optimal network was based on our selection of appropriate optimization of the various metrics. The scheme we decided to employ was designed from the viewpoint of the best solution in a medical sense. We wanted a minimal false negative (FN)—percentile of patients wrongly identified as healthy, and a maximal true positive (TP)—percentile of patients correctly identified with TM, while maintaining the simplest ANN possible. Optimizing for a minimal FN measure ensures that we will not miss any patients (under the risk of falsely identifying others) carrying thalassemia. Such system design allows very flexible and accurate ANN optimization. Thus, the accuracy metric was defined as the difference between the FN and the TP values, while keeping the network as simple as possible. This relationship is described as accuracy value η and calculated as in equation (1):
| (1) |
While optimizing ANNs, the error in output prediction was minimized across many learning cycles until the networks reached the requested level of accuracy, and then a single ANN for each stage of the study was selected.
Stage Analysis
The metrics we employed were sensitivity, specificity, positive predictive value (PPV), and negative predictive value (NPV). In addition, the following metrics were also recorded: TP,the percentile of patients correctly identified with TM; false positive (FP), the percentile of patients wrongly identified with TM; true negative (TN), percentile of patients correctly identified as healthy; and FN, percentile of patients wrongly identified as healthy. All values were calculated as described in equations (2), (3), (4), (5):
| (2) |
| (3) |
| (4) |
| (5) |
RESULTS
Accuracy Level Comparison for Different Number of Parameters
In order to check the impact of the number of parameters on the accuracy of the model, above 1,500 different ANNs were constructed and their accuracy level was calculated with three (MCV, RDW, RBC) or six (MCV, RDW, RBC, HB, MCH, PLT) CBC parameters. Figure 1 illustrates accuracy vs. total number of ANNs for the six stages of comparison, as described in the Materials and Methods section.
Figure 1.
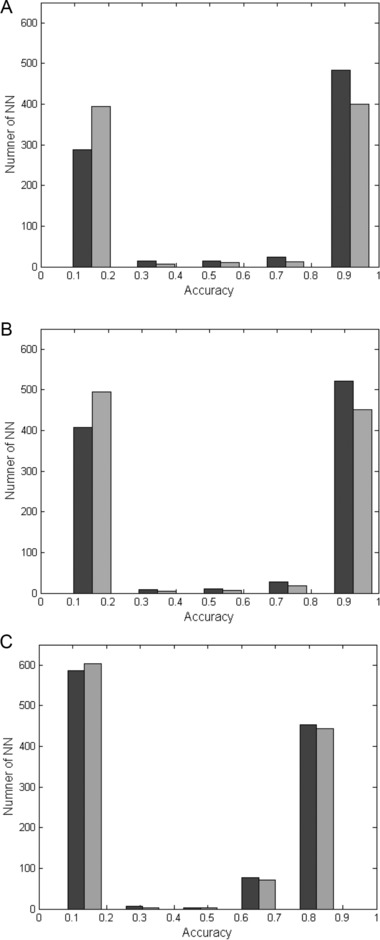
Accuracy value for three or six CBC parameters. The accuracy is presented against the number of ANNs for the three (black columns) or the six (gray columns) blood parameters as detailed in the Materials and Methods section. (A) TM vs. control group—stages 1 and 4; (B) TM vs. control and MDS groups—stages 2 and 5; (C) TM vs. control, MDS, and IDA groups—stages 3 and 6.
These results illustrate that slightly higher accuracy of ANNs is achieved with the three parameters than with the six parameters scheme. The effect is more prominent for the classification of TM from the healthy population and the MDS patients (Fig. 1A and B); including IDA into the comparison reduces the positive effect of limiting the model for the three CBC parameters: MCV, RDW, and RBC.
The Most Successful Neural Network for Each Stage of the Study
From the collection of 1,500 ANNs constructed, we optimized the best ANN for each stage of analysis based on the accuracy metric. Then we evaluated the values of FN, TP, NPV, PPV, specificity, and sensitivity for the best ANN at each stage (Table 3).
Table 3.
Results of the Best ANN at Each Stage of the Study
| Stage | FN | TP | NPV | PPV | Specificity | Sensitivity |
|---|---|---|---|---|---|---|
| 1 | 0 | 0.957 | 1 | 0.957 | 0.958 | 1 |
| 2 | 0 | 0.966 | 1 | 0.966 | 0.967 | 1 |
| 3 | 0.105 | 0.971 | 0.895 | 0.971 | 0.968 | 0.902 |
| 4 | 0 | 0.957 | 1 | 0.957 | 0.958 | 1 |
| 5 | 0 | 0.966 | 1 | 0.966 | 0.967 | 1 |
| 6 | 0.111 | 0.971 | 0.889 | 0.971 | 0.968 | 0.897 |
The results for the separation of the TM group from the control group (stages 1 and 4) achieve a sensitivity of 1 and specificity of 0.958. The separation of the TM group from the control group and the nonrelevant blood disease, MDS (stages 2 and 5), achieve a sensitivity of 1 and specificity of 0.967. The separation of the TM group from the control group, MDS group, and IDA group (stages 3 and 6) achieved a sensitivity of around 0.9 and specificity of 0.968. Our accuracy metric is based on the assumption that the model should not miss any TM patients. Thus, the best ANN for the separation of TM from the healthy and the control group show 0% of FN results and NPV of 1.
Accuracy Level Comparison for Different Patient Group Size
In order to examine the performance of the best ANN selected, accuracy for different patient groups was evaluated. We varied the amount of data given to the best ANN achieved (a specific ANN) for stage 2 in order to understand the effect of different patient group sizes on the accuracy of the optimized ANN.
Figure 2 shows that the accuracy of the ANN is high for patient groups over 300. In our study, each ANN was optimized for the complete patients’ database. Some accuracy fluctuations seen under 500 patients can be attributed to the fact that the optimization of the ANN was done for the maximal patient group (526 patients) and not for the smaller groups. This result shows that the best TM classification can be achieved when ANN is optimized on the database of at least 300 patients.
Figure 2.
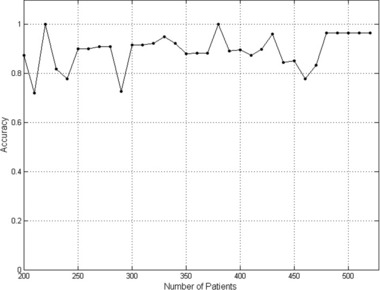
Accuracy of the best ANN. The accuracy value was calculated for the best ANN of stage 2 of the study and is presented against the number of patients tested.
DISCUSSION
Identification of TM patients is important to prevent misdiagnosis and a birth of thalassemia major children. Current methods for identification of thalassemia carriers are high‐performance liquid chromatography and protein electrophoresis for the β TM, and mutation screening by PCR and DNA sequencing for the α type 2. However, the need for specific instrumentation and the relatively high cost of these methods prevent their application from a high‐throughput screening of wide population. A low‐cost and fast screening that can be performed directly and together with routine blood count is required to achieve the effective recognition of TM patients. In the present study, we used patients' database of 526 patients, which was gathered over a period of 10 years. TM patients in the database were diagnosed and verified prior to the study. Our database included both α and β types of TM as well as non‐TM patients.
From the CBC, three of the parameters (MCV, RBC, and RDW) were chosen for the analysis because of their high relevance for the diagnosis of TM 25, 26. To study the contribution of additional parameters, we added three more parameters: HB, MCH, and PLT, which we thought to be associated with TM. The first two parameters, HB level and MCH, are broadly used for TM detection 11, 13, 14, 15, while the latter (PLT) was reported to differentiate between thalassemia trait and IDA 27. We found that the first three parameters were enough and even gave improved results for TM differentiation than the six parameters. Thus, we conclude that at least in our model, the HB level, MCH, and PLT are of lesser importance than MCV, RBC count, and RDW. Analysis of the best ANN at each stage showed that the only differences between the three and the six parameters scheme is observed for the stages 3 and 6, when the IDA patients are added to the comparison. The FN value, that is missed TM patients, is slightly higher with the six parameters, while together with a slightly lower sensitivity value, agrees with the suggestion that three parameters perform better than six. As noted earlier, precise identification of the parameters that contribute the most to thalassemia identification was not conducted here. A more rigorous examination of the parameters included in the routine CBC analysis is possible and will require additional mathematical method, such as principal component analysis 28.
After we found the best ANN, we used it to study the significance of the number of patients included in the study. We have shown that at least 300 patients are needed to gain stable accuracy results. Some fluctuations in accuracy measure could arise from different devices used to get CBC, as our database was collected from different clinics over 10 years. Our study shows that the use of smaller scale databases can significantly reduce the accuracy of the study. It is likely that as more samples are added in a future study, the performance of the ANN would increase.
Our results show that effective TM differentiation can be achieved using the ANNs analysis based on a standard CBC test. Differentiation between TM and the nonthalassemia patients (control group and MDS) achieved zero FN results, NPV value of 1, and PPV value of 0.966. Calculation of specificity and sensitivity resulted in 0.967 and 1 values, respectively. Therefore, a positive result of our method yields a very limited number of false outcomes, while a negative result reliably excludes a patient for the presence of TM condition. The method we presented in this study is flexible and can be used to optimize results according to the scheme selected. In our study, we performed optimization toward higher sensitivity over lower specificity values. A different optimization scheme could have been chosen.
Previous studies using linear methods were not able to achieve satisfactory results and usually accomplished a rather low specificity and sensitivity values 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 13, 14, 15, 16, 17. Nonlinear approaches resulted in improved values. For example, nonlinear approach using support vector machines and K nearest neighbors achieved a specificity of 0.95 and sensitivity of 0.92 21. A different study conducted a comparison between decisions trees, evolved through genetic programming, and ANN. This study had the ANN outperform the decision tree, while also showing that a multilayer ANN performs better than a single‐layer ANN 18. A more recent study shows that a different method of implementing decision trees can increase performance to a specificity of 0.998 and sensitivity of 0.972 that outperforms multilayer ANN 19. This study included healthy population and other hemoglobinopathies and excluded patients with IDA. In the work presented here, we felt it important to include nonthalassemia disease, MDS (which changes blood parameters non‐specifically), α and β thalassemia types, and even IDA patients. Although IDA patients decreased the overall accuracy; specificity and sensitivity were 0.968 and 0.9, respectively. These results are good as we believe them to more accurately describe the real population in a clinic, because sometimes TM patient are misdiagnosed as IDA.
The method we propose here is novel in some aspects, as unlike other studies implementing ANN, in this study a more broad approach was conducted. Usually designing ANN for a specific purpose requires intensive optimization of network configuration (e.g., number of neurons and their connections) and currently, the trial and error is the best method for this task. Therefore, most studies with medical ANN applications are limited to a small number of ANN configurations, attempting to optimize only several best working networks. In the present work, we designed more than 1,500 networks that differed by their configurations and selected the best ANN for each task. This novel approach has saved most of the trial‐and‐error optimization steps and allowed us a broad screening from the large ANN repository. The selected networks are highly optimized for the TM screening task.
In comparison with other ANN‐based attempts of TM screening, our method gives the best results based on only three input parameters and does not require any additional data besides the standard CBC. The input parameters are a part of a routine CBC and our approach imposes no further test or calculation other than the ones already performed by laboratories. Thus, the ANN model we present here can be used to review current medical databases in order to locate potential TM patients by a simple one‐step computational screening.
ANNs bear a strong resemblance to many well‐known statistical methods, for example, linear regression, but are usually easier to apply. The networks are implemented as a computer program obtaining the input of specified CBC parameters and providing the prediction output as a simple yes or no answer. Such software can be designed in a user‐friendly manner and operated by a nontrained laboratory technician. There is no need to re‐optimize the ANNs constructed in this study, our algorithms can be implemented directly in various scenarios. We propose that our method will be applied as a screening before diagnosis with complicated and high‐cost testing. This screening will be simple, rapid, and indicate the possibility of having TM. The ANN software attached to routine blood count devices or applied by a technician on specific samples will lead to low cost and high sample throughput analysis. Positive result will need further confirmation by conventional methods. Due to high NPV, the number of samples forwarded to this extensive analysis would be significantly reduced. The low cost of our model will be of special importance in the developing countries with high frequency of thalassemia alleles. Moreover, the model we propose here can be applied for the screening of other diseases that change any CBC parameters, such as early diagnosis of nonsymptomatic patients with myelo‐ and lympho‐proliferative disorders. Furthermore, our model is not limited to CBC parameters and can be used with any countable parameters available.
REFERENCES
- 1. Rachmilewitz EA, Giardina PJ. How I treat thalassemia. Blood 2011;118:3479–3488. [DOI] [PubMed] [Google Scholar]
- 2. Hartwell SK, Srisawang B, Kongtawelert P, Christian D, Grudpan K. Review on screening and analysis techniques for hemoglobin variants and thalassemia. Talanta 2005;65:1149–1161. [DOI] [PubMed] [Google Scholar]
- 3. Lafferty JD, Crowther MA, Ali MA, Levine M. The evaluation of various mathematical RBC indices and their efficacy in discriminating between thalassemic and non‐thalassemic microcytosis. Am J Clin Pathol 1996;106:201–205. [DOI] [PubMed] [Google Scholar]
- 4. Karimi M, Rasekhi AR. Efficiency of premarital screening of beta‐thalassemia trait using MCH rather than MCV in the population of Fars Province, Iran. Haematologia (Budap) 2002;32:129–133. [DOI] [PubMed] [Google Scholar]
- 5. Okan V, Cigiloglu A, Cifci S, Yilmaz M, Pehlivan M. Red cell indices and functions differentiating patients with the β‐thalassaemia trait from those with iron deficiency anaemia. J Int Med Res 2009;37:25–30. [DOI] [PubMed] [Google Scholar]
- 6. Bessman JD, Gilmer PR, Gardner FH. Improved classification of anemias by MCV and RDW. J Int Med Res 1983;80:322–326. [DOI] [PubMed] [Google Scholar]
- 7. Ehsani MA, Shahgholi E, Rahiminejad MS, Seiqhali F, Rashidi A. A new index for discrimination between iron deficiency anemia and beta‐thalassemia minor: Results in 284 patients. Pak J Biol Sci 2009;12:473–475. [DOI] [PubMed] [Google Scholar]
- 8. Johnson CS, Tegos C, Beutler E. Thalassemia minor: Routine erythrocyte measurements and differentiation from iron deficiency. Am J Clin Pathol 1983;80:31–36. [DOI] [PubMed] [Google Scholar]
- 9. Kneifati‐Hayek J, Fleischman W, Bernstein LH, Riccioli A, Bellevue R. A model for automated screening of thalassemia in hematology (math study). Lab Hematol 2007;13:119–123. [DOI] [PubMed] [Google Scholar]
- 10. Sirdah M, Tarazi I, Al Najjar E, Al Haddad R. Evaluation of diagnostic reliability of different RBC indices and formulas in the differentiation of the β thalassemia minor from iron deficiency in Palestinian population. Int J Lab Hematol 2007;30:324–330. [DOI] [PubMed] [Google Scholar]
- 11. Green R, King R. A new red cell discriminant incorporating volume dispersion for differentiating iron deficiency anemia from thalassemia minor. Blood Cells 1989;15:481–495. [PubMed] [Google Scholar]
- 12. Mentzer WC, Jr . Differentiation of iron deficiency from thalassemia trait. Lancet 1973;1:882. [DOI] [PubMed] [Google Scholar]
- 13. England JM, Fraser PM. Differentiation of iron deficiency from thalassemia trait by routine blood count. Lancet 1973;1:449–452. [DOI] [PubMed] [Google Scholar]
- 14. Srivastava PC, Bevington JM. Iron deficiency and/or thalassemia trait. Lancet 1973;1:832. [DOI] [PubMed] [Google Scholar]
- 15. Shine I, Lal S. A strategy to detect beta‐thalassemia minor. Lancet 1977;1:692–694. [DOI] [PubMed] [Google Scholar]
- 16. Ricerca BM, Storti S, d’Onofrio G, et al. Differentiation of iron deficiency from thalassemia trait: A new approach. Haematologica 1987;72:409–413. [PubMed] [Google Scholar]
- 17. Urrechaga E, Borque L, Escanero JF. The role of automated measurement of RBC subpopulations in differential diagnosis of microcytic anemia and β‐thalassemia screening. Am J Clin Pathol 2011;135:374–379. [DOI] [PubMed] [Google Scholar]
- 18. Wongseree W, Chaiyaratana N, Vichittumaros K, Winichagoon P, Fucharoen S. Thalassemia classification by neural networks and genetic programming. Inform Sci 2007;177:771–786. [Google Scholar]
- 19. Piroonratana T, Wongseree W, Assawamakin A, et al. Classification of haemoglobin typing chromatograms by neural networks and decision trees for thalassaemia screening. Chemom Intell Lab Syst 2009;99:101–110. [Google Scholar]
- 20. Amendolia SR, Brunetti A, Carta P, et al. A real‐time classification system of thalassemic pathologies based on artificial neural networks. Med Decis Making 2002;22:18–26. [DOI] [PubMed] [Google Scholar]
- 21. Amendolia SR, Cossu G, Ganadu ML, Golosio B, Masala GL, Mura GM. A comparative study of k‐nearest neighbour, support vector machine and multi‐layer perceptron for thalassemia screening. Chemom Intell Lab Syst 2003;69:13–20. [Google Scholar]
- 22. Yeh J, Cheng C. Using hierarchical soft computing method to discriminate microcyte anemia. Expert Syst Appl 2005;29:515–524. [Google Scholar]
- 23. Patel JL, Goyal RK. Applications of artificial neural networks in medical science. Curr Clin Pharmacol 2007;2:217–226. [DOI] [PubMed] [Google Scholar]
- 24. Sivanandam SN, Sumathi S, Deepa SN. Introduction to Neural Networks Using Matlab 6.0. Tata McGraw‐Hill, Noida, UP, India; 2006. [Google Scholar]
- 25. Muncie HL, Campbell JS. Alpha and beta thalassemia. Am Fam Physician 2009;15:339–344. [PubMed] [Google Scholar]
- 26. Urrechaga E, Borkue L, Escanero JF. Erythrocyte and reticulocyte parameters in iron deficiency and thalassemia. J Clin Lab Anal 2011;25:223–228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Timuragaoglu A, Coban E, Erbasan F. The importance of platelet indexes in discriminating between β‐thalassemia trait and iron deficiency anemia. Acta Haematol 2004;111:235–236. [DOI] [PubMed] [Google Scholar]
- 28. Abdi H, Williams LJ. Principal component analysis. Wiley Interdiscip Rev Comput Stat; 2010;2:433–459. [Google Scholar]


