Abstract
Background
RHD alleles leading to a reduced expression of D antigen of the red blood cell (RBC) surface may be erroneously typed as D− by serology and may cause anti‐D immunizations when transfused to recipients.
Methods
To determine the occurrence of such alleles among apparent D− blood donors, molecular typing was implemented as a routine test using a pool of DNA. A total of 2,450 pretyped D− samples were tested in pools of 10 for the RHD‐specific polymorphism in intron 4 and exon 7. Samples in polymer chain reaction (PCR) positive pools were individually reevaluated by exon‐specific PCRs, sequencing, and serologic methods.
Results
Among 2,450 serologically D− blood donor samples tested, 101 (4.1%) carried the RHD gene. Nonfunctional RHD (RHDψ, RHD*CE(2–9)‐D, and RHD*CE(3–7)‐D), different weak D alleles such as RHD*weak D type 1, RHD*weak D type 4.3, RHD*weak D type 5, RHD*weak D type 38, and RHD*DEL were identified.
Conclusion
We employed a PCR‐based assay for RHD as a routine test using pools of ten DNA blood donor samples. The integration of RHD genotyping into the routine screening program using pools of DNA samples was straightforward. As a consequence, 19 (0.8%) blood donors carrying a weak D and Del phenotypes with the potential of causing anti‐D immunizations in recipients were reclassified as D+. For each population, it would be necessary to adapt the RHD genotyping strategy to the spectrum of prevalent alleles.
Keywords: pool of DNA, RHD alleles, D antigen, blood donors, PCR
INTRODUCTION
The Rh blood group system is clinically important, because antibodies against Rh antigens are involved in hemolytic disease of the newborn, hemolytic transfusion reactions, and autoimmune hemolytic anemia 1. The Rh blood group system belongs to one of the most diverse antigen systems presently known in humans and the D antigen is the most important blood group antigen determined by a protein because D‐negative individuals can be easily anti‐D immunized 1, 2.
Rh antigens are carried by two highly homologous proteins, the RhD and RhCE, which are encoded by two genes, RHD and RHCE, located at chromosomal position 1p34–1p36. Both genes encompass ten exons, and their structures are highly homologous 2. In previous studies, using pools of DNA, it has demonstrated that different genetic forms of D negativity occur with high incidences of unexpressed RHD alleles among D‐negative individuals with the presence of C and/or E antigens 3, 4, 5, 6. In general, Caucasians with the RhD‐negative phenotype have the absence of the RHD gene due to a deletion of RHD between the upstream and downstream Rhesus boxes 7. However, other D‐negative haplotypes exist. In African populations, some individuals who are D‐negative can harbor nonfunctional RHD alleles, the RHDψ gene and RHD‐CE(3–7)‐D hybrid 8, 9, 10. In Asians, a very weak expression of the D antigen, the DEL phenotype, was found in individuals who were apparently D‐negative and represents 10–33% of the Japanese and Chinese serologically typed as D‐negative 11. Less common D‐negative alleles are caused by hybrid RHD‐CE‐D alleles or nonsense and frame shift mutations 3, 4, 5, 6.
Among Europeans, 0.6–1% carries RHD alleles producing weak D and partial D 12. Up to 80 weak RHD alleles have been described and are due to missense mutations exclusively located in the transmembranous or intracellular parts of the RhD protein 13. A small fraction of weak D samples are explained by qualitatively altered RhD proteins, called partial D and frequently caused by a RHD/RHCE hybrid allele 13, 14. A weak D expression is also caused by the suppressive effects of Cde haplotypes placed in trans position 15, 16.
RHD alleles leading to a reduced expression of D antigen on the red blood cell (RBC) surface may be erroneously typed as D− by standard serologic methods, including the indirect antiglobulin test (IAT) and may cause anti‐D immunizations when transfused to D− recipients 17. The adsorption/elution technique considered more sensitive is not suitable for routine diagnostic use. D variants detectable by this technique are known as Del and have also caused the generation of anti‐D when transfused to D‐negative patients 18.
The molecular typing overcomes the limitations of serologic methods in order to solve discrepancies and to identify RHD variants. The RHD alleles are currently determined by performing PCR assays in individual samples. However, molecular typing of each sample can be time consuming, expensive, and labor intensive, representing a challenge to be performed as a routine in the clinical setting. One of the major concerns in daily practice transfusions is to prevent anti‐D alloimmunization without adding much to the costs of antigen D typing and without incurring in wasting D– units unnecessarily. Therefore, a method for RHD genotyping that is less expensive and straightforward is highly desirable.
We developed a method herein for RHD typing that applies a pool of 10 DNA samples from apparently D‐negative blood donors. Our lower‐cost method has shown to have great sensibility/sensitivity and to be a great alternative to be used in transfusions centers.
MATERIAL AND METHODS
Blood Samples
A total of 2,450 blood donor samples from Southeast (São Paulo, SP) and Northeast (Recife, PE) Brazil labeled D− were investigated. Blood samples used for screening purposes were the by‐product of blood donation and were collected according to the approved protocol of the blood banks. Whole units of blood were taken with informed consent.
Study Design and Validation of DNA Pool
The study design used to analyze the samples is shown in Figure 1.
Figure 1.
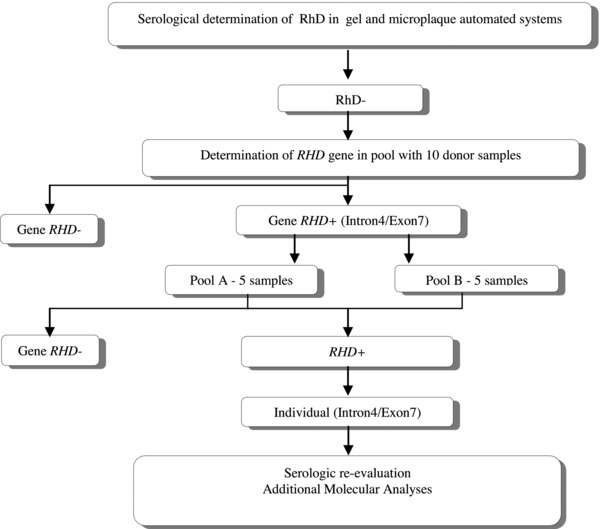
Study design.
Validation of DNA pool typing was performed by mixing nine pretyped negative DNA samples with one reference DNA sample genotyped for RHD variants. The sensitivity and specificity of the assay were verified by the presence of the specific amplification product, at the DNA concentration of 10ng/μL per sample.
Serologic Studies
D antigen reactivity was analyzed by agglutination in duplicate screening on an automated gel system (Wadiana®, Grifols, Derio, Spain) using two different anti‐D MoAbs, one IgM (clone P3×61), and oligoclonal antibodies (clones P3×290 (IgG), P3×35 (IgG), P3×61(IgM), P3×21223B10 (IgM)) (Griffols, Derio, Spain) and on an automated microplate system (PK7200, Olympus Diagnostic Systems) using anti‐D oligoclonal antibodies (clones MS26(IgG)/MS201(IgM)) (Fresenius‐Kabi, São Paulo, Brazil). Nonreactive samples were tested with anti‐D blend (clones MS26/MS201) using the IAT in tube. C, E, c, e status of all RBCs were determined by hemagglutination in gel neutral cards (Diamed AG, Morat, Switzerland) using routine anti‐C, anti‐E, anti‐c, anti‐e MoAbs (Fresenius Kabi, São Paulo, Brazil).
PCR Assays
Genomic DNA was individually isolated using the QIAmp DNA Blood Mini‐Kit (Qiagen, Valencia, CA), according to the manufacturer's recommendations.
Pools of ten samples of serologically D‐negative were tested with a PCR assay used to determine the presence or absence of RHD‐specific amplified products from sequences in the intron 4 and exon 7 10. We used 1μL of genomic DNA at the optimal DNA concentration of 10 ng/μL per sample in a final volume of 20 μL of reaction mix. When positive results were found, the pool was first divided in two and the PCR repeated in each pool of five samples.
When a five sample pool generated a PCR‐positive result, each sample of the pool was tested individually in a PCR system using sequence specific primers that detect the common weak D types 19, a multiplex PCR that detects the RHD gene hybrid alleles 20. RHD zygosity was determined by checking for the presence of the hybrid Rhesus box, by an allele‐specific PCR 21 and a Pst I‐RFLP 22.
Sequence Analysis
Sequence analysis was performed on PCR products amplified from genomic DNA to determine the specific allele present using RHD specific primers as previously reported 23. PCR products were purified by elution from 1% agarose gels using a Qiaex II gel extraction kit (Qiagen), and sequenced directly, without subcloning, on an ABI 373XL Perkin Elmer Biosystems (PEB) sequencer, and the PEB Big Dye reagent BD Half‐term (GenPak, Perkin Elmer Biosystems, Foster City, CA).
Serological Followup
Blood samples with positive results in the PCR screening were reevaluated by an IAT in two gel matrix techniques (Diamed and Grifols) using a MoAb anti‐D IgG (clone ESD1). Adsorption/elution test was also performed on all nonreactive samples in IAT but positive in the PCR screening using a MoAb anti‐D IgG (clone MS‐26) (Fresenius‐Kabi, São Paulo, Brazil). For anti‐D adsorption to RBCs, 500 μL of RBCs were incubated for 1 hr at 37°C with 500 μL of anti‐D diluted 1:2. Elution was performed by using the ELU–KIT II (Immucor Gamma, Houston, TX).
RESULTS
Molecular Analyses
We found 101 donors (4.1%) who carried the RHD gene. All 101 samples genotyped as D+ were RHD heterozygous. Table 1 summarizes the distribution of the RHD alleles found in this D− donor population, their molecular alteration detected, and the associated haplotypes.
Table 1.
Distribution of the RHD Alleles Identified Among D− Blood Donors, the Polymorphisms Detected, and the Associated Haplotypes
| RHD allele | N (%) | Polymorphisms detected | Associated haplotype |
|---|---|---|---|
| RHDψ | 78 (3.2) | 37‐bp insertion | ce |
| RHD*CE(2‐9)‐D | 1 (0.04) | CE exons 2–9 | Ce trans |
| RHD*CE(3‐7)‐D | 3 (0.1) | CE exons 3–7 | (C)ces |
| RHD*DEL | 5 (0.2) | 1227G>A | Ce |
| RHD*weak D type 1 | 1 (0.04) | 809T>C | Ce |
| RHD*weak D type 4.3 | 5 (0.2) | 602 C>G, 667 T>G, 819 G>A, 872 C>G | ce |
| RHD*weak D type 5 | 3 (0.1) | 446C>A | cE |
| RHD*weak D type 38 | 5 (0.2) | 833 G>A | Ce |
Regional differences in the composition of the group of donors studied were encountered and reflected the diverse Brazilian genetic background. The five DEL carriers identified in this study were from Asian descent, the five weak D type 38 were from Portuguese descent, the weak D types 1 and 5 were from Caucasian descent, and the 5 weak D type 4.3 were from African descent. The RhCE phenotypes associated with weak D and DEL alleles were consistent with published data 24.
Serological Followup
Nine different weak D phenotypes such as weak D type 1, weak D type 5, and the weak D type 38 were identified by serologic reevaluation showing positive IAT results. Weak D type 4.3 and Del phenotypes were only detected by adsorption and elution. Table 2 shows the RHD alleles identified among D− blood donors and their serologic results.
Table 2.
RHD Alleles Identified Among D− Blood Donors and Serologic Results
| RHD allele | IAT/gela | Adsorption/elutionb |
|---|---|---|
| RHDψ | − | − |
| RHD*CE(3‐7)‐D | − | − |
| RHD*DEL | − | + |
| RHD*weak D type 1 | + | + |
| RHD*weak D type 4.3 | − | + |
| RHD*weak D type 5 | + | + |
| RHD*weak D type 38 | + | + |
IAT/gel (gel cards from Diamed and Grifols) using a MoAb anti‐D IgG (clone ESD1).
Adsorption/elution test was performed using a MoAb anti‐D IgG (clone MS‐26) and the ELU–KIT II (Immucor Gamma).
DISCUSSION
Any blood bank must ensure that D‐negative products are appropriately labeled, such that a recipient of a D‐negative product does not form anti‐D in response to transfused RBCs 1, 17. In this study, we screened serologically D‐ donors for the presence of the RHD gene and presented a fast and reliable method for RHD genotyping using DNA pools. We established this practice in our blood bank because current serologic practice has failed to determine weak D types 16, and to discriminate between the Del and the true D–phenotypes.
This approach revealed that 19 (0.8%) blood donors carrying weak D and Del phenotypes were mistyped as D‐negative by serology, higher than the incidence of 0.4% previously reported in Caucasians 4 and comparable to the relative occurrence of 0.6 and 1% described for Germany and Canada 5, 25, 26, 27.
The presence of the nonfunctional RHD, RHD‐CE(2–9)‐D, RHD‐CE(3–7)‐D and the high prevalence of RHDψ (3.2%) found in this donor population reinforce that the ethnic background of the population may govern which RH alleles are prevalent. Our population, in fact, has ethnic characteristics related to the marriage among Caucasians, Amerindians, and Africans.
One of the major concerns in daily transfusion practice is to prevent anti‐D alloimmunization, as anti‐D may induce hemolytic transfusion reactions and remains the major cause of hemolytic disease of the fetus and newborn 1, 7.
A total of nine weak D phenotypes such as weak D type 1, weak D type 5, and the weak D type 38 were identified by serologic reevaluation showing positive IAT results in gel, reinforcing that the sensitivity of the method and the anti‐D reagent used may influence on D donor typing result. To avoid further mistyping in our blood bank we started to use molecular techniques, as a way to avoid potential alloimmunization in the patients. However, the use of molecular techniques may represent higher costs. For this reason we investigated an alternative technique in which pools of 10 DNA samples were combined in a single reaction to decrease labor and costs of PCR reactions. By performing this pool, we were able to decrease the costs of PCR typing from approximately US$21 to US$13, which represents a reduction of almost 50%. Previous studies employing pools of DNA to genotype D‐negative donors also showed that this strategy can contribute significantly to the total costs what is considered an acceptable strategy in terms of cost saving 5, 6. Another benefit of using PCR over serological analysis is the fact that PCR genotyping usually does not need to be repeated. Few laboratories, however, are using molecular workup on a daily basis to type D antigen.
In an editorial of Transfusion, Garratty 28 stressed the caveat that not every blood donor in the D− donor pool who is carrying an RHD gene necessarily expresses the D antigen. Donors with the RHD pseudogene, for instance will not express the D antigen; however D− donor carrying aberrant RHD alleles leading to a reduced expression of D antigen on the RBC surface may cause anti‐D immunizations when transfused to D− recipients. The overall appearance of 0.8% antigen D+ samples among D− blood donors indicates that RHD genotyping of D− donors could improve the RBC unit safety by permanently removing D+ donors from the D− pool and transferring their RBC units to the D+ pool as already discussed by Flegel et al. 5. According these authors, it also has the advantage that it obviates the need for a sensitive D antigen testing at the serologic level: a very strict quality assurance for sensitivity may not be required anymore, which may return some cost savings 5.
Another important benefit of using PCR is that the genotyping information can be transferred from institution to institution nationally and internationally, which would eliminate the need for repeat testing of blood donors. The method described herein also opens the opportunity to extend DNA typing to other antigens using pools, especially when we look for rare donors. It is not unreasonable to think that donor testing in pools of DNA may also be used for typing other highly polymorphic systems, such as platelets and leukocytes.
Grant sponsor: FAPESP; Grant number: 2009/05924‐0
REFERENCES
- 1. Westhoff CM. The Rh blood group system in review: a new face for the next decade. Transfusion 2004;44:1663–1673. [DOI] [PubMed] [Google Scholar]
- 2. Wagner FF, Flegel WA. Review: the molecular basis of the Rh blood group phenotypes. Immunohematology 2004; 20:23–36. [PMC free article] [PubMed] [Google Scholar]
- 3. Wagner FF, Frohmajer A, Flegel WA. RHD positive haplotypes in D negative Europeans. BMC Genet 2001;2–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Polin H, Danzer M, Gaszner W, Broda D, St‐Louis M, Proll J, Hofer K, Gabriel C. 2009. Identification of RHD alleles with the potential of anti‐D immunization among seemingly D‐ blood donors in Upper Austria. Transfusion 49:676–681. [DOI] [PubMed] [Google Scholar]
- 5. Flegel WA, von Zabern I, Wagner FF. Six years’ experience performing RHD genotyping to confirm D‐ red blood cell units in Germany for preventing anti‐D immunizations. Transfusion 2009;49:465–471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Gassner C, Doescher A, Drnovsek TD, Rozman P, Eiche RNI, Legler TJ, Lukin S, Garritsen H, Kleinrath T, Egger B, Ehling R, Kormoczi GF, Kilga‐Nogler S, Schoenitzer D, Petershofen EK. Presence of RHD in serologically D‐, C/E+ individuals: a European multicenter study. Transfusion 2005;45:527–538. [DOI] [PubMed] [Google Scholar]
- 7. Flegel WA. Molecular genetics and clinical applications for RH (review). Transfus Apher Sci 2011;44:81–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Wagner FF, Moulds JM, Tounkara A, Kouriba B, Flegel WA. RHD allele distribution in Africans of Mali. BMC Genet 2003;4–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Rodrigues A, Rios M, Pellegrino J Jr, Costa FF, Castilho L. Presence of RHD pseudogene and the hybrid RHD‐CE‐Ds gene in Brazilians with the D‐negative phenotype. Braz J Med Biol Res 2002;35(5):767–773. [DOI] [PubMed] [Google Scholar]
- 10. Singleton BK, Green CA, Avent ND, Martin PG, Smart E, Daka A, Narter‐Olaga EG, Hawthorne LM, Daniels G. The presence of an RHD pseudogene containing a 37 base pair duplication and a nonsense mutation in Africans with the RhD‐negative blood group phenotype. Blood 2000;95:12–18. [PubMed] [Google Scholar]
- 11. Qun X, Grootkerk‐TaxM GHM, Maaskant‐van Wijk PA, van der Schoot C E. Systemic analysis and zigosity determination of the RHD gene in a D negative Chinese Han population reveals a novel D‐ negative RHD gene. Vox Sang 2005;88:35–40. [DOI] [PubMed] [Google Scholar]
- 12. Flegel W A, Wagner FF. Molecular biology of partial D and weak D: implications for blood bank practice. Clin Lab 2002;48:53–59. [PubMed] [Google Scholar]
- 13. Wagner FF. 1998; The RhesusBase, Department of Transfusion Medicine, Ulm, Germany: University Hospital; http://www.uni-ulm.de/~fwagner/RH/RB. Accessed March 11, 2011. [Google Scholar]
- 14. Wagner FF, Gassner C, Muller TH, Schonitzer D, Schunter F, Flegel W. Molecular basis of weak D phenotypes. Blood 1999;93:385–393. [PubMed] [Google Scholar]
- 15. Wagner FF. Influence of Rh phenotype on the antigen density of C, c, and D. Flow cytometric study using a frozen standard red cell. Transfusion 1994;34:671–676. [DOI] [PubMed] [Google Scholar]
- 16. Mota M, Fonseca NL, Rodrigues A, Kutner JM, Castilho L. Anti‐D alloimmunization by weak D type 1 red blood cells with a very low antigen density. Vox Sang 2005;88:130–135. [DOI] [PubMed] [Google Scholar]
- 17. Denomme GA, Dake LR, Vilensky D, Ramyar L, Judd WJ. Rh discrepancies caused by variable reactivity of partial and weak D types with different serologic techniques. Transfusion 2008;48:473–478. [DOI] [PubMed] [Google Scholar]
- 18. Körmöczi GF, Gassner C, Shao CP, Uchikawa M, Legler TJ. A comprehensive analysis of DEL types: partial DEL individuals are prone to anti‐D alloimmunization. Transfusion 2005;45:1561–1567. [DOI] [PubMed] [Google Scholar]
- 19. Mueller TH, Wagner FF, Trockenbacher A, Eicher NI, Flegel WA, Schonitzer D, Schunter F, Gassner C. PCR screening for common weak D types shows different distributions in three Central European populations. Transfusion 2001;41:45–52. [DOI] [PubMed] [Google Scholar]
- 20. Maaskant‐van Wijk PA, Faas BHW, de Ruijter JAM, Overbeeke MAM, von dem Borne AEGKr, van Rhenen DJ, van der Schoot CE. Genotyping of RHD by multiplex polymerase chain reaction analysis of six RHD‐specific exons. Transfusion 1998;38:1015–1021. [DOI] [PubMed] [Google Scholar]
- 21. Chiu RWK, Murphy MF, Fidler C, Zee B CY, Wainscoat JS, Lo DYM. Determination of RhD zygosity: comparison of a double amplification refractory mutation system approach and a multiplex real‐time quantitative PCR approach. Clin Chem 2001;47:667–672. [PubMed] [Google Scholar]
- 22. Wagner FF, Flegel W. RHD gene deletion occurred in the Rhesus box. Blood 2000;95:3662–3668. [PubMed] [Google Scholar]
- 23. Legler TJ, Maas JH, Köhler M, Wagner T, Daniels GL, Perco P, Panzer S. RHD Sequencing: a new tool for decision making on transfusion therapy and provision of Rh prophylaxis. Transf Med 2001;11:383–388. [DOI] [PubMed] [Google Scholar]
- 24. Ansart‐Pirenne H, Asso‐Bonnet M, Le Pennec PY, Roussel M, Patereau C, Noizat‐Pirenne F. RhD variants in Caucasians: consequences for checking clinically relevant alleles. Transfusion 2004;44:1282–1286. [DOI] [PubMed] [Google Scholar]
- 25. Flegel WA. Blood group genotyping in Germany. Transfusion 2007;47:47S–53S. [DOI] [PubMed] [Google Scholar]
- 26. Denomme GA, Wagner FF, Fernandes BJ, Li W, Flegel WA. Partial D, weak D types, and novel RHD alleles among 33,864 multiethnic patients: implications for anti‐D alloimmunization and prevention. Transfusion 2005;45:1554–1560. [DOI] [PubMed] [Google Scholar]
- 27. Flegel WA. How I manage donors and patients with a weak D phenotype. Curr Opin Hematol 2006;13:476–483. [DOI] [PubMed] [Google Scholar]
- 28. Garratty G. Do we need to be more concerned about weak D antigens? Transfusion 2005;45:1547–1551. [DOI] [PubMed] [Google Scholar]


