Abstract
Multidrug-resistant Staphylococcus aureus is a leading concern worldwide. Coagulase-Negative Staphylococci are claimed to be the reservoir and source of important resistant elements in S. aureus. However, the origin and evolutionary route of resistant genes in S. aureus are still remaining unknown. Here, we performed a detailed phylogenomic analysis of 152 completely sequenced S. aureus strains in comparison with 7,529 non-Staphylococcus aureus reference bacterial genomes. Our results reveal that S. aureus has a large open pan-genome where 97 (55%) of its known resistant-related genes belonging to its accessory genome. Among these genes, 47 (27%) were located within the Staphylococcal Cassette Chromosome mec (SCCmec), a transposable element responsible for resistance against major classes of antibiotics including beta-lactams, macrolides, and aminoglycosides. However, the physically linked mec-box genes (MecA–MecR–MecI) that are responsible for the maintenance of SCCmec elements is not unique to S. aureus, instead it is widely distributed within Staphylococcaceae family. The phyletic patterns of SCCmec-encoded resistant genes in Staphylococcus species are significantly different from that of its core genes indicating frequent exchange of these genes between Staphylococcus species. Our in-depth analysis of SCCmec-resistant gene phylogenies reveals that genes such as blaZ, ble, kmA, and tetK that are responsible for beta-lactam, bleomycin, kanamycin, and tetracycline resistance in S. aureus were laterally transferred from non-Staphylococcus sources. In addition, at least 11 non-SCCmec-encoded resistant genes in S. aureus, were laterally acquired from distantly related species. Our study evidently shows that gene transfers played a crucial role in shaping the evolution of antibiotic resistance in S. aureus.
Keywords: microbial genome evolution, pan-genome, antibiotic resistance, SCCmec, lateral/horizontal gene transfer, S. aureus
Introduction
Antibiotic-resistant bacteria are causing serious global threat (Center for Disease Control 2013; World Health Organization 2016). Large population size and short generation time of bacteria are beneficial for the evolution of resistance within 2–4 years of introducing a new antibiotic (Harkins 2017). Clinically relevant opportunistic pathogens like Staphylococcusaureus have acquired resistance against multiple antibiotics such as penicillin, methicillin, and vancomycin. Methicillin-resistant S. aureus (MRSA) has become a major problem worldwide and is increasingly being detected in both hospitals and communities (Chambers and DeLeo 2009). The lateral component of microbial genome evolution (LGT/HGT) has played an essential role in shaping the bacterial genome content as well as their metabolic capabilities (Nelson-Sathi et al. 2015; Soucy et al. 2015). For example, S. aureus metabolic capabilities are connected to the acquisition of both resistant and virulent traits (McCarthy et al. 2014; Bosi et al. 2016). About 15–20% of the S. aureus genome includes parts of bacteriophage genomes, pathogenicity islands, plasmids, transposons, and cassette chromosomes (Lindsay 2010; Alibayov et al. 2014). The gene MecA is responsible for beta-lactam antibiotic resistance along with its regulatory proteins being embedded within the unique mobile genetic element called Staphylococcal Cassette Chromosome mec (SCCmec) (Ito et al. 2001). So far, 12 SCCmec types, designated as I to XII and different subtypes have been identified and the resistance capacity of the species varies based on its SCCmec types (Chongtrakool et al. 2006; Deurenberg and Stobberingh 2008). All SCCmec elements reported to date carry the following common gene complexes: 1) orfX (for the easy integration of SCCmec), 2) five classes of mec-box genes (include MecA/MecC/MecB and its regulatory proteins MecR and MecI located upstream of MecA), 3) flanking Insertion-Sequence elements (IS431 and IS1272), 4) three allotypes of recombinases (ccrA, ccrB, and ccrC; sequence similarity between ccr allotypes is <50%), which are responsible for the easy excision and integration of the SCCmec cassette, 5) transposons (tnp and Tn554, which are required for transposition), and 6) other genes include heavy metal and antibiotic-resistant genes such as ars and blaZ (IWG-SCC 2009; Ito et al. 2012).
Resistant genes encoded from different types and subtypes of highly variant SCCmec elements and other non-SCCmec regions; are the main reason behind the fast adaptation of S. aureus to different environments (IWG-SCC 2009). Previous studies on SCCmec elements have suggested that the MecA genes are distributed in multiple Staphylococcus species such as Staphylococcusepidermidis, Staphylococcushaemolyticus, and Staphylococcussaprophyticus (Grundmann et al. 2006). A recent study by Subakishita et al. (2010) found the presence of plasmid-encoded mec-gene complex (mecAm–mecRm–mecIm–blaZm) in a Macrococcus caseolyticus JSCS5402 strain, suggest that human pathogenic staphylococci acquired mec-box genes from Macrococcus lineage. Few other studies indicated that SCCmec elements assembled within Coagulase-Negative Staphylococci (CoNS), such as S. epidermidis, S. saprophyticus, Staphylococcusschleiferi and then transferred to S. aureus (IWG-SCC 2009; Ito et al. 2012; Otto 2013). In addition, study on MecA gene distribution pattern, Rolo et al. (2017) claimed that the most primitive staphylococcal species such as Staphylococcusfleurettii, Staphylococcussciuri, and Staphylococcusvitulinus were involved in the stepwise assembly of SCCmec elements and it is later transferred to S. aureus.
However, there are limitations in the existing studies; mainly due to the lack of diverse data used. Most of these studies focused on the evolution of SCCmec-encoded elements, which contribute only 27% of resistance genes in S. aureus. Many efforts have been taken to understand the evolution of antibiotic resistance in S. aureus and ended up with multiple observations, yet an overall picture is still lacking. Since most of the resistant genes in S. aureus located on mobile genetic elements and plasmids, we strongly hypothesize that gene exchange among closely as well as distantly related species, might have shaped the current form of resistant genes distribution in S. aureus.
In the present study, we performed a detailed phylogenomic analysis of all the resistance encoding genes in 152 completely sequenced S. aureus strains and compared them with 7,529 reference bacterial genomes to construct a better picture of resistance evolution in S. aureus. Our findings revealed that the gene distribution and phyletic pattern of SCCmec elements in Staphylococcus species are significantly different from its core genes indicating that they are evolved in a different fashion. Our results showed that LGT (Lateral Gene Transfers) plays a crucial role in shaping the evolution of SCCmec and non-SCCmec-encoded resistant genes in S. aureus.
Materials and Methods
Data
Completely sequenced 7,681 bacterial genomes (protein sequences) and its feature tables were downloaded from National Center for Bioinformatics Information-GenBank database (July 2017) (ftp://ftp.ncbi.nlm.nih.gov/genomes/genbank/bacteria/; last accessed October 10, 2019). This includes 152 S. aureus strains from different hosts (Human, Bovin, Chicken, Swine) isolated from different geographic regions and 7,529 bacterial reference genomes (supplementary table 1, Supplementary Material online).
Pan-Genome Reconstruction and Functional Annotation of Genes
Homologous genes were obtained from a total of 424,469 genes encoded in the chromosomes and plasmids of 152 S. aureus genomes. An all-against-all genomes, BLASTp (Altschul et al. 1997) was performed with e-value <1e-05, ≥30% amino acid BLAST identity and ≥70% query coverage followed by reciprocal best BLAST hits (rBBH) (Tatusov et al. 1997). Homolog protein pairs were further globally aligned using Needleman–Wunsch algorithm with needle program of EMBOSS package (Rice et al. 2000) and homologs were filtered out with ≥25% global identity threshold. The resulting 428,070 protein pairs were clustered using the Markov Cluster algorithm (MCL) (Enright et al. 2002) with default parameters yielded a total of 4,562 protein families. There were 1,764 genes, which do not have any significant homolog in 152 strains were classified as unique cases. 192 paralog genes, which skipped the MCL clustering pipeline, were manually added back to the existing clusters using a best cluster match criteria. Altogether 4,562 protein families and 1,764 unique genes were converted into a binary matrix of presence/absence patterns (PAPs). Based on the distribution pattern of clusters within S. aureus strains, its pan-genome is classified into three categories (genes present in ≥90% strains as core genome, gene presence in >1% to <90% of strains as auxiliary genome and presence of a gene in a single strain as unique). The S. aureus pan-genome is summarized as a presence absence matrix, where x axis represents protein families and unique elements and y axis represents 152 S. aureus strains. If a protein family or unique gene is present in any of the S. aureus strain, its corresponding entry is represented with “1”; otherwise, a “0.” The presence absence matrix was sorted in ascending order of its distribution and visualized using MATLAB R2015a. Representative genes for each MCL cluster were functionally annotated using Clusters of Orthologus Groups (COG) database (Tatusov et al. 2003). The functional assignment of protein families and unique genes was done by mapping against the COG database using BLAST. If a COG ID mapped to more than one category, the category R (general function prediction only) was assigned. The COG categories for each protein are given in supplementary table 2, Supplementary Material online. Curve fitting of S. aureus pan-genome was performed using a power-law regression based on Heaps’ law n = k×N−α, as described in Tettelin et al. (2005, 2008) and Rasko et al. (2008). Fitting was conducted with the PanGP software (Zhao et al. 2014), where n is the expected number of genes for a given number of genomes, N is the number of genomes (i.e., 152), and k and α (α = 1−γ) are free parameters that are determined empirically. According to Heaps’ law, when α > 1 (γ < 0), the pan-genome is considered to be closed and the addition of new genomes will not increase the number of new genes significantly. On the other hand, when α < 1 (0 < γ < 1), the pan-genome is open, and for each newly added genome, the number of new genes will increase significantly (Tettelin et al. 2008).
Identification of Resistance-Related Clusters (SCCmec Encoded and Non-SCCmec Encoded)
Based on the classification proposed by International Working Group on Staphylococcal Cassette Chromosome (IWG-SCC 2009), 104 SCCmec-encoded elements from different SCCmec typed reference strains and previously reported 173 plasmid/chromosome-encoded resistant genes of S. aureus were downloaded from NCBI-GenBank database. Each reference element was BLASTed against 4,562 protein families and 1,764 unique genes and the best-hit entries (with an e-value <1e-05, query coverage ≥70 and BLAST identity ≥30) were considered as SCCmec elements and other resistant homologs. This yielded 47 SCCmec-encoded and 130 non-SCCmec-encoded multidrug-resistant gene families in S. aureus. To identify the homologs of these resistant genes in non-Staphylococcus aureus species BLASTp was performed using previously described thresholds. Coverage matrix was constructed by calculating the proportion of genomes containing SCCmec homologs within a non-Staphylococcus taxon. Distance between each genomic locations of SCCmec homolog genes within bacterial species was used to calculate the length of SCCmec elements.
Classification of MRSA, MSSA, and SCCmec Typing
Reference MecA sequence was BLASTed against 152 S. aureus strains and based on the presence or absence of homolog (e-value <1e-05, query coverage ≥70 and BLAST identity ≥90) strains were classified as resistant and sensitive. Similarly, ccr allotypes of known SCCmec reference strains were BLASTed and based on best hits (BLAST identity ≥80% and query coverage of ≥70%, e-value <1e-05), strains were classified into 12 SCCmec types (I to XII).
Phylogenetic Tree Reconstruction and Comparison
16S rRNAs, SCCmec, and antibiotic-resistant genes with their homologs were aligned separately using MAFFT (Katoh et al. 2002) with the options –localpair, –maxiterate = 1,000 and the alignment confidence score is evaluated using GUIDANCE2 server (Sela et al. 2015). Maximum likelihood trees were reconstructed using RAxML (Stamatakis 2014) under the PROTGAMMAWAG model; with statistical support at nodes obtained by bootstrapping on 100 resample data sets. Maximum likelihood trees were rerooted between Firmicutes and non-Firmicutes using the nw_reroot program of Newick Utilities (Junier and Zdobnov 2010) and visualized using FigTree version1.4.3 (Rambaut 2012). To compare the phyletic patterns of SCCmec, chromosome, and core genes, we subsampled trees that contain at least 12 OTUs common in each set of trees (supplementary table 3, Supplementary Material online). Further, the pairwise distances between trees were computed by Robinson–Foulds (RF) metric (Robinson and Foulds 1981) using Phangorn package (Schliep 2011) and the Kernel Density Estimate (KDE) of the distances were plotted in R. Pairs of distance distributions were compared using a two-sample Kolmogorov–Smirnov test (KS test). The distance comparisons were repeated by independently sampling a subset of trees in triplicates and RF distances were pooled together for the statistical analysis.
LGT Inference
Any topological discordance between gene trees and 16S rRNA reference tree at the species level is considered as potential LGTs. In 177 resistant gene trees, we condensed OTUs at species-level and compared the nearest neighbor relationship of each OTU with the reference tree using in-house Perl script (supplementary table 4, Supplementary Material online). A gene is considered as acquisition in S. aureus if it is absent in nearest neighbors and widely distributed in distantly related species, which are different from its reference phylogeny. Taxa that are branching within S. aureus clade are considered as exports from S. aureus. 16S rRNA phylogeny used for the comparison is provided in figure 3 and its newick tree (including all OTUs) is provided in supplementary table 4, Supplementary Material online.
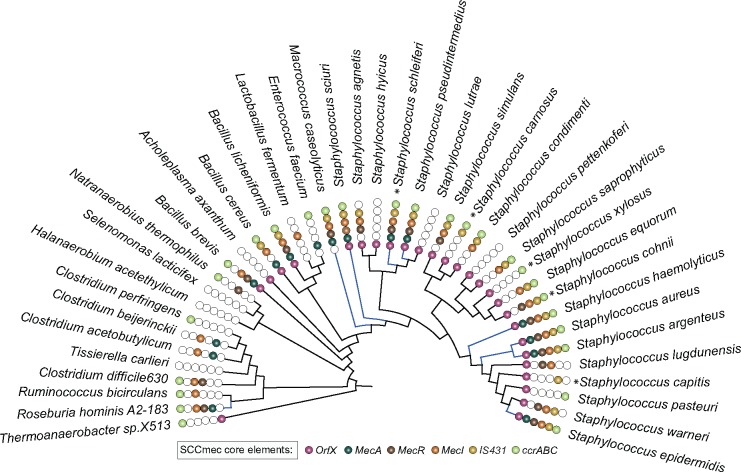
Fig. 3.
—16S rRNA reference phylogeny with the distribution of SCCmec-encoded core genes. Colored circles represent the presence of orfX-mec box-ccr box genes identified in different bacterial genera. Blue-colored branches in the phylogeny represent the species with a genomic organization of the core SCCmec elements (MecR–MecA–MecI) within 51 kb genomic regions. Most of the species in Staphylococcaceae family are pathogens, while the opportunistic pathogens are represented as asterisk (*).
Results
Characteristics of S. aureus Pan-Genome
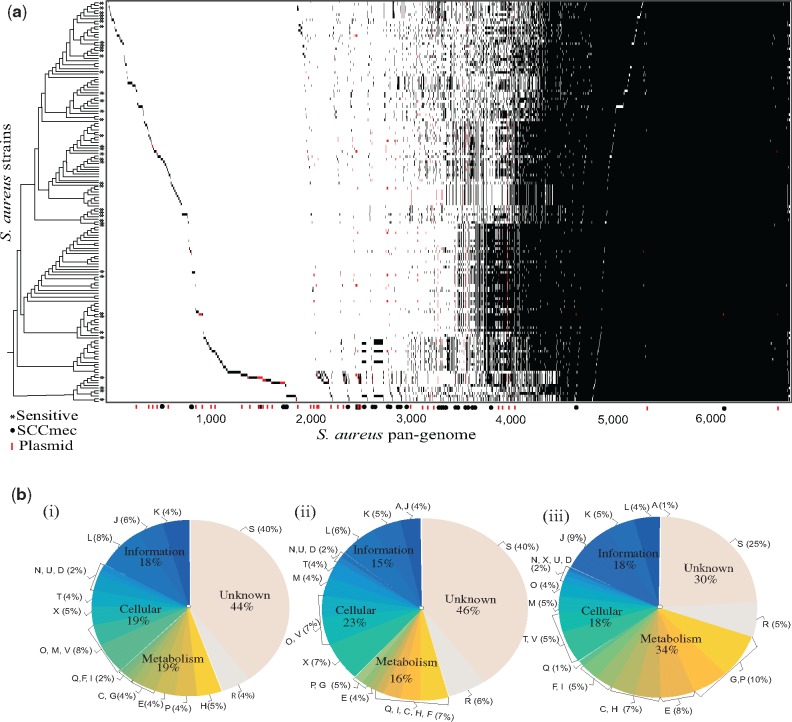
A total of 428,070 genes encoded in the genomes of S. aureus strains were clustered into 4,562 protein families and 1,764 unique genes using Markov Chain Clustering procedure (Enright et al. 2002). The S. aureus pan-genome is open (α < 1) and appears to be moderately expanding with the inclusion of new genomes (fig. 1 and supplementary fig. S1, Supplementary Material online). The openness of pan-genome and the presence of mobile genetic elements in S. aureus is similar to many other bacterial species, for example, Escherichiacoli that is known to have an essential capability of acquiring new genes into its genome via interspecies transfers (Touchon et al. 2009). Among 4,562 protein families and 1,764 unique genes in S. aureus pan-genome, 2,426 (38%) core genes are universally or nearly universally distributed (present in ≥90% strains) in 152 S. aureus strains (fig. 1 and supplementary table 2, Supplementary Material online). Majority of these core genes are encoded for the basic machinery of cells including essential aspects of transcription and translation, which is similar to that observed in Streptococcus and E. coli (Lefébure and Stanhope 2007; Touchon et al. 2009). The auxiliary (2,136) and unique genome (1,764) of S. aureus was found to have a different functional distribution as compared with that of the core genome. Unlike unique and core genomes, the auxiliary genome of S. aureus is more involved in cellular processing and signaling functions, which is similar to other bacterial species (Ozer et al. 2014). In addition, a large proportion (12%) of auxiliary and unique genes in S. aureus have been identified as mobile genetic elements like transposons and prophages genome (replication, recombination, and repair) (supplementary table 2, Supplementary Material online). S.aureus showed an extensive genetic diversity, with an average 27% of genes being specific for a single strain.
Fig. 1.
—Pan-genome of S. aureus. (a) For each protein families, a black tick indicates the presence and white tick indicates the absence of a gene in the corresponding genome in the left. S. aureus strains on the left side are sorted according to the 16S rRNA reference tree. Genes encoded from plasmid are indicated in red in the presence and absence pattern. Methicillin-susceptible S. aureus (MSSA) are marked with a * symbol. (b) Functional classification of S. aureus proteins using COG is as follows: (i) the unique genome (genes that are unique to a single strain), (ii) the auxiliary genome (genes present in >1%≤90% of strains), and (iii) the core genome (genes present in ≥90% of strains). COG functional categories are as follows: For cellular processes and signaling: cell cycle control, cell division, and chromosome partitioning (D); cell wall/membrane/envelope biogenesis (M); cell motility (N); mobilome, prophages, transposons (X); posttranslational modification, protein turnover, and chaperones (O); signal transduction mechanisms (T); intracellular trafficking, secretion, and vesicular transport (U); defense mechanisms (V); extracellular structures (W); nuclear structure (Y) and cytoskeleton (Z). For information storage and processing: RNA processing and modification (A); translation, ribosomal structure, and biogenesis (J); transcription (K) and replication, recombination, and repair (L). For metabolism: energy production and conversion (C); amino acid transport and metabolism (E); nucleotide transport and metabolism (F); carbohydrate transport and metabolism (G); coenzyme transport and metabolism (H); lipid transport and metabolism (I); inorganic ion transport and metabolism (P) and secondary metabolites biosynthesis, transport, and catabolism (Q). General function prediction (R).
Evolution of SCCmec Elements in S. aureus
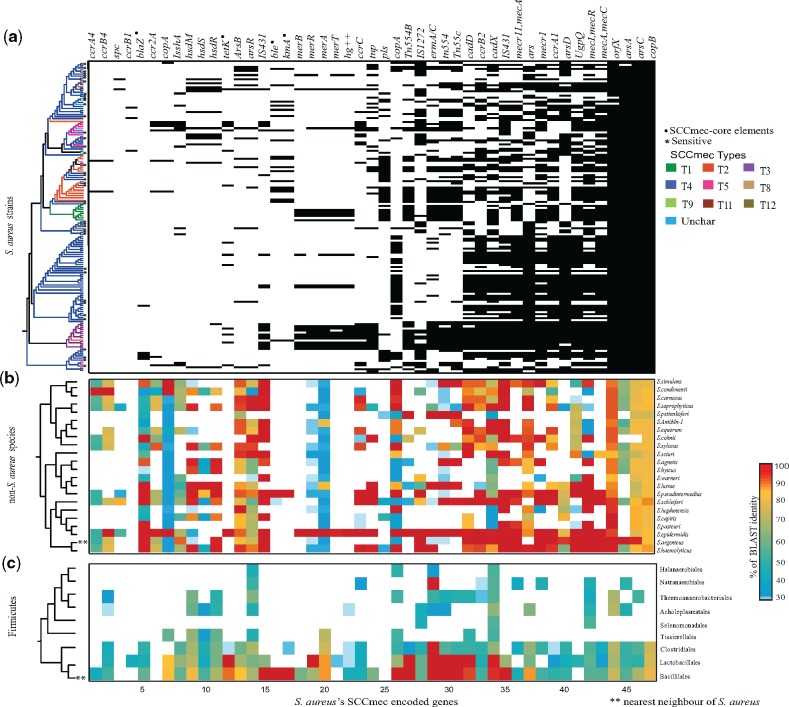
Among 4,562 protein families and 1,764 unique genes, 47 protein families pertain to SCCmec-encoded genes including MecA gene and its regulators (MecR and MecI), recombinases (ccrABC), insertion elements, transposons, heavy metal, and drug-resistant genes. Seventy-three percent of 152 S. aureus strains contains the MecA gene and were classified as “resistant,” while the remaining 27% strains were classified as susceptible. As expected, MecA was absent in most of the sensitive strains, but surprisingly 10% of sensitive S. aureus strains in our data set consist of SCCmec elements without MecA gene, this suggests that the presence of SCCmec is not limited to resistant strains. This distribution can also be due to the loss of MecA gene after the acquisition of the well-established SCCmec elements. Furthermore, based on the ccr gene allotypes classification, all the 152 strains were found to be in well-established I to XII SCCmec types; with community-associated S. aureus (Type IV) being the most abundant among them (63%) (supplementary table 1, Supplementary Material online). As suggested by Berglund et al. (2005), the observed wide distribution of Type IV S. aureus strains might be due to its small SCCmec size and the presence of the high number of functional recombinases. Among the 47 SCCmec-encoded genes, the crucial genes responsible for the maintenance and functioning of the SCCmec elements (MecA, MecR, MecI, orfX, IS431, Tn554A/B, tnp, and some ccr allotypes) were nearly universally distributed among the selected 152 S. aureus strains (fig. 2 and supplementary fig. S2, Supplementary Material online). The cassette flanking gene viz. orfX was found to be universally (99%) present in S. aureus strains, clearly indicating its importance in the integration of these elements. The mec-box genes (MecA–MecR–MecI), which are the decisive elements in SCCmec, are predominantly distributed among 152 S. aureus strains (73%, 73%, and 64% distribution, respectively). The ccr-box genes that code for recombinases are widely distributed within resistant strains of S. aureus (66%). Some of its allotypes such as ccrAB4 and ccrB1 occur at a lower frequency (1.3%) when compared with other ccr allotypes namely ccrA1 and ccrB2. In addition to the mec-box and ccr-box genes, various IS elements and genes associated with the mobility of SCCmec were also found to be widely distributed within S. aureus (fig. 2). Moreover, heavy metal-resistant genes such as ars (arsenate) and cop (copper) are also universally distributed in 152 S. aureus strains and their coexistence with the SCCmec elements hints the coselection under environmental pressure (Xue et al. 2015). Interestingly, most of the SCCmec core elements (orfX, MecA and its regulators, ccr genes, IS elements, transposons, and heavy metal-resistant genes such as ars and cop) are widely distributed within Staphylococcaceae with high sequence similarity (≥70% sequence similarity, fig. 2). While considering the entire Firmicutes phylum, the SCCmec homologs of S. aureus observed in many Bacillales, Clostridiales, and Lactobacillales species (supplementary fig. S2, Supplementary Material online).
Fig. 2.
—Distribution of 47 SCCmec-encoded protein families in Firmicutes phylum. x axis represents 47 SCCmec protein families and y axis represents 152 S. aureus strains, 22 non-Staphylococcus aureus species, and 9 orders within Firmicutes phylum. (a) The matrix indicates presence (black) or absence (white) of the SCCmec elements in the corresponding genome on the left. The 16S rRNA tree of 152 S. aureus strains is colored based on known 12 SCCmec types. Methicillin sensitive strains are indicated by an asterisk (*) symbol in the phylogenetic tree. LGT-influenced genes are represented as a black dot (.). The ccrA4 and ccrB4 are patchily distributed because of the uniqueness in their sequence. (b) The matrix depicts the presence and similarity of 47 S. aureus SCCmec-encoded genes in 22 non-Staphylococcus aureus species and the color bar on the right represents sequence similarity ranging from 30 (blue) to 100 (red). (c) The matrix represents the presence of S. aureus SCCmec gene homologs in 9 orders within the Firmicutes phylum.
The physically linked core mec-box genes (MecA–MecR–MecI) ubiquitously present within S. aureus is necessary for the proper functioning of SCCmec cassette. The genomic organization of SCCmec-core genes, MecA–MecR–MecI, is highly conserved within resistant strains of S. aureus as well as in some of the Staphylococcaceae species including S. sciuri, Staphylococcuspseudointermedius, S. epidermidis, Staphylococcusargenteus, S. haemolyticus, S. schleiferi, and M. caseolyticus. In these species, mec-box genes are located within 51 kb regions (fig. 3 and supplementary table 5, Supplementary Material online), which is similar to those of SCCmec carried by S. aureus (Ito et al. 2012). In terms of sequence similarity and linkage, all the species mentioned above seems to have genes that are comparable to the S. aureus mec-box genes. On the other hand, some Staphylococcus species including Staphylococcussimulans, Staphylococcuscohnii, Staphylococcuslugdunensis, Staphylococcuswarneri, Staphylococcusagnetis, S. saprophyticus, and Staphylococcusequorum have mec-box genes, but they are not located within the 51-kb range and do not seem to have any apparent genomic linkage.
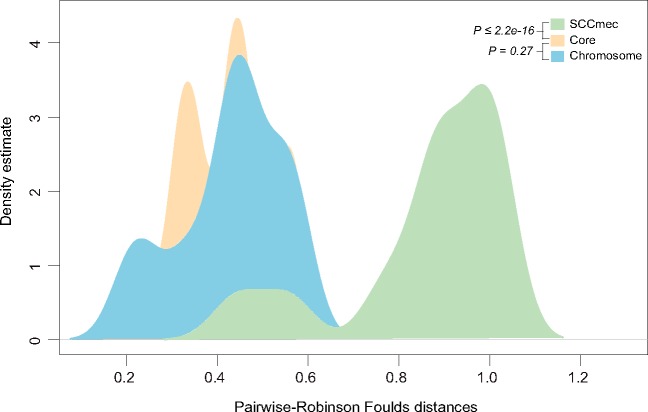
From gene distribution pattern and genomic linkage analysis, it is clear that SCCmec elements are widely present within Staphylococcus species. Moreover, the phylogenetic analysis of 47 genes encoded within the SCCmec cassette reveals that at least 42 (89%) of them are exhibiting significant topological discordances with respect to 16S rRNA reference phylogeny, while none of them are recovering individual species monophyly (supplementary table 4, Supplementary Material online). Tree dissimilarity distribution (Robinson–Foulds distances) of SCCmec and core genes in Staphylococcus species is significantly different from each other (P value <2.2e-16) (fig. 4) indicating frequent exchange of these genes within the Staphylococcus species. Lateral transfers of SCCmec genes were not limited within Staphylococcus species; rather distantly related lineages shared genes with Staphylococcus species (supplementary fig. S3 and table 4, Supplementary Material online). Resistant genes such as blaZ (beta-lactam resistance), kmA (kanamycin resistance), tetK (tetracycline resistance), and ble (bleomycin resistance) that embedded within the SCCmec elements of S. aureus are some of the good examples of laterally transferred genes from distantly related non-Staphylococcus species (supplementary fig. S4, Supplementary Material online). For instance, S. aureus might receive blaZ gene from Geobacillus stearothermophilus; a species where there is no evidence of SCCmec cassette but phylogenetically they are showing a close relationship with S. aureus. Another example is ble in S. aureus that might have been acquired from Paenibacillus beijingensis species. In case of tetK, S. aureus received it from one of its distantly related lineages such as Lactobacillales and Bacillales. Furthermore, kanamycin-resistant gene might have been acquired from Paenibacillus riograndensis species (supplementary fig. S4, Supplementary Material online). These precisely indicate that lateral influence of closely and distantly related species largely contributed to the current structure of SCCmec cassette of S. aureus. The MecA gene was subject to multiple lateral transfers within the Staphylococcaceae family members and M.caseolyticus species forming nearest neighbor to S. aureus (supplementary table 4, Supplementary Material online). In M.caseolyticus species, MecA gene is present in plasmid which increases the possibility that the MecA in S. aureus is originated from Macrococcus lineage. Moreover, species such as S. sciuri, S. pseudointermedius, S. epidermidis, S. argenteus, S. haemolyticus, and S. schleiferi are branching within S. aureus clade pointing exports of MecA from S. aureus.
Fig. 4.
—Distribution of pairwise tree distances of core and resistant genes. The pairwise tree distances of core, chromosome, and SCCmec-encoded resistant genes of Staphylococcus were calculated using Robinson–Foulds distance are shown. The x axis represents the distances between gene trees and reference tree and the y axis represent their densities. P values (two-tailed Kolmogorov–Smirnov test) from comparisons between core and SCCmec, core and chromosome genes were <2.2e-16 and 0.27, respectively.
Evolution of Non-SCCmec-Encoded Resistant Genes
Even though SCCmec elements are known to be the key factors responsible for antibiotic resistance in S. aureus, there are about 130 non-SCCmec-encoded resistance-related genes encoded from plasmids and the chromosome of S. aureus. The majority of these genes confer resistance to protein synthesis inhibitor class of antibiotics (33%) while a small number are showing resistance to folic acid synthesis inhibitors (0.38%). Similar to the SCCmec-encoded gene distribution, the non-SCCmec-encoded resistant genes are also widely distributed within most of the Staphylococcus species (≥70% sequence similarity, supplementary fig. S5, Supplementary Material online).
Among 130 non-SCCmec-encoded resistant genes in S. aureus 83 are encoded from chromosome while 47 are from plasmid and more than half of them showing topological resemblance with reference phylogeny. There are 53 genes showing S. aureus monophyly and nearest neighbor relationship as in 16S rRNA reference phylogeny (supplementary table 4, Supplementary Material online). Among these 53 genes, majority (87%) of them are chromosome-encoded, which suggest that a large fraction of vertically evolved genes retained within these species (supplementary fig. S6, Supplementary Material online). Robinson–Foulds distance metric computed for all pairwise comparisons revealed that the phylogenetic distributions of chromosome-encoded resistant genes are more similar to core genes (with P value <0.27) (fig. 4). Examples of vertically evolved resistant genes include mgrA, arlS, murA, sulA, and norB that are responsible for resistance against different classes of antibiotics such as fluoroquinolone, cephalosporin, sulfonamide, and fosfomycin. Even though a large proportion of chromosome-encoded resistant genes seems to be evolved vertically within Staphylococcus species, lateral gene transfers also influenced genes from this category. There are few genes in S. aureus including tetO, blmS, oppB, lnuA/linA, and OppC are examples of gene transfers from distinctly related linages (supplementary fig. S7a, Supplementary Material online).
As expected, frequent LGTs were detected on plasmid-encoded genes in S. aureus which includes important genes such as mupA, aadA5, tetL that are responsible for resistance against mupirocin, aminoglycoside, and tetracycline, respectively (supplementary fig. S7b, Supplementary Material online). Distantly related lineages such as Clostridiales, Proteobacteria, and Actinobacteria influenced most of the gene acquisitions in S. aureus. For example, aminoglycoside-resistant gene aadA5 phylogeny indicates that S. aureus might receive this gene from one of the distantly related lineages (Clostridiales, Bacteroidetes, and Virgibacillus); but in case of tetL, S. aureus might have acquired it from one of the species of Lactobacillales and Bacillales (supplementary fig. S7b, Supplementary Material online). Moreover, genes such as dfrA and mupA were acquired from some distantly related lineages in Bacillales.
Discussion
For better insights on the resistance evolution in S. aureus, we have considered completely sequenced bacterial genomes (all publicly available) in our analysis that is a limiting factor in the previous studies. The gene sharing patterns of S. aureus strains were not uniform between different resistant types; for example, Type IV strains are sharing more genes each other, but having less number of genes in its SCCmec element than others (supplementary fig. S1, Supplementary Material online). Interestingly, we noticed that the 16S rRNA reference phylogeny of S. aureus strains shows no monophyletic structure in terms of its resistant gene presence or SCCmec type, indicating that S. aureus lineages were not primarily evolved toward the resistant trait (fig. 2a) and LGTs might play a crucial role in shaping its resistant genes distribution. Some previous reports show that SCCmec element is originated in CoNS such as S. epidermidis, S. haemolyticus, S. schleiferi, and S. sciuri (Ito et al. 2012; Otto et al. 2013). However, we found that SCCmec elements are not limited to CoNS; rather mec-box and ccr-box gene homologs were observed in Coagulase-Positive Staphylococci such as S. argenteus and S. pseudointermedius. The physically linked mec-box genes (MecA–MecR–MecI) are widely distributed in Staphylococcus species, except in M.caseolyticus and Roseburia hominis A2-183, which are the only species having physically linked mec-box-like structure in non-Staphylococcus species (fig. 3). Tree-to-tree distances between SCCmec and core genes clearly indicate that SCCmec genes have undergone frequent exchange within the species. Notably, a couple of SCCmec core genes such as orfX and variant of mecR might have been less influenced by LGT because their dissimilarity indices were similar to that of core genes (fig. 4). Due to lack of availability of diverse set of complete genomes within each Staphylococcus species and frequent exchange of genes within the species, it is hard to polarize LGTs that happened between closely related species. Phylogenomic studies with additional diverse set of genomes of Staphylococcus species will provide further insights on the resistance evolution of S. aureus.
Conclusions
To better understand the evolution of antibiotic resistance-related genes in S. aureus, we have analyzed all known resistant genes present in 152 completely sequenced S. aureus genomes in comparison with 7,353 non-Staphylococcus. aureus reference bacterial genomes. The gene distribution pattern clearly shows that the SCCmec elements carry the antibiotic-resistant genes and its regulators are not unique to S. aureus. Moreover, they are widely distributed among other Staphylococcus species and also present in some distantly related non-Staphylococcus aureus species. Furthermore, many Staphylococcus species were observed to have a physically linked mec-box-like structure in their genomes with a similar structural organization and high amino acid sequence identity to the SCCmec element of S. aureus. Bacterial species having such SCCmec element is patchily distributed within Staphylococcus species. Tree dissimilarity distributions of SCCmec genes in Staphylococcus species with its core genes indicate frequent exchange of genes encoded from this element happened within the Staphylococcus species. The lateral gene transfers were not limited within Staphylococcus species; rather unique resistant genes such as blaZ, kmA, and ble are acquired from distantly related lineages. In 130 non-SCCmec-encoded resistant genes, most of the chromosome-encoded resistant genes seem to be vertically inherited, while plasmid genes are more prone to lateral gene transfers. Our phylogenomic analysis provides a better insight into the evolution of resistant genes in S. aureus strains and role of LGTs shaping its current distribution.
Supplementary Material
Supplementary data are available at Genome Biology and Evolution online.
Supplementary Material
Acknowledgments
We thank Prof. M. Radhakrishna Pillai, the Director, and Rajiv Gandhi Centre for Biotechnology (RGCB), for the strong support to setting up the computational facility. This work was supported by funding from Innovation in Science Pursuit for Inspired Research (INSPIRE) fellowship to S.N.-S. by Department of Science & Technology, Government of India (DST-INSPIRE) (DST/INSPIRE/04/2015/002935); and Kerala State Council for Science, Technology and Environment (KSCSTE) (043/YIPB/KBC/2017/KSCSTE); and Department of Science & Technology Early Career Award (DST-ECR) (ECR/2017/002980). The Council of Scientific & Industrial Research (CSIR), Government of India, funded J.J.
Author’s Contributions
S.N.-S. and J.J. designed and conceived the study. J.J. and S.M.G. coordinated the study and carried out the data analysis part. S.R.C.N. and J.J. performed the statistical analysis. S.N.-S. and J.J. drafted the article. All authors gave final approval for publication.
Literature Cited
- Alibayov B, Baba-Moussa L, Sina H, Zdeňková K, Demnerová K.. 2014. Staphylococcus aureus mobile genetic elements. Mol Biol Rep. 41(8):5005–5018. [DOI] [PubMed] [Google Scholar]
- Altschul SF, et al. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25(17):3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baba T, et al. 2009. Complete genome sequence of Macrococcus caseolyticus strain JSCS5402, reflecting the ancestral genome of the human-pathogenic staphylococci. J Bacteriol. 191(4):1180–1190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berglund C, Mölling P, Sjöberg L, Söderquist B.. 2005. Multilocus sequence typing of methicillin-resistant Staphylococcus aureus from an area of low endemicity by real-time PCR. J Clin Microbiol. 43(9):4448–4454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bosi E, et al. 2016. Comparative genome-scale modelling of Staphylococcus aureus strains identifies strain-specific metabolic capabilities linked to pathogenicity. Proc Natl Acad Sci U S A. 26:E3801–E3809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Center for Disease Control (CDC). 2013. Antibiotic resistance threats in the United States. United States: Centers for Disease Control and Prevention. Available from: https://www.cdc.gov/drugresistance/threat-report-2013/, last accessed October 10, 2019.
- Chambers HF, DeLeo FR.. 2009. Waves of resistance: Staphylococcus aureus in the antibiotic era. Nat Rev Microbiol. 9:629–641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan CX, Beiko RG, Ragan MA.. 2011. Lateral transfer of genes and gene fragments in Staphylococcus extends beyond mobile elements. J Bacteriol. 15:3964–3977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chongtrakool P, et al. 2006. Staphylococcal cassette chromosome mec (SCCmec) typing of methicillin-resistant Staphylococcus aureus strains isolated in 11 Asian countries: a proposal for a new nomenclature for SCCmec elements. Antimicrob Agents Chemother. 50(3):1001–1012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deurenberg RH, Stobberingh EE.. 2008. The evolution of Staphylococcus aureus. Infect Genet Evol. 8(6):747–763. [DOI] [PubMed] [Google Scholar]
- Enright AJ, Van Dongen S, Ouzounis CA.. 2002. An efficient algorithm for large-scale detection of protein families. Nucleic Acids Res. 30(7):1575–1584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grundmann H, Aires-de-Sousa M, Boyce J, Tiemersma E.. 2006. Emergence and resurgence of meticillin-resistant Staphylococcus aureus as a public-health threat. Lancet 368(9538):874–885. [DOI] [PubMed] [Google Scholar]
- Hanssen AM, Ericson Sollid JU.. 2006. SCCmec in staphylococci: genes on the move. FEMS Immunol Med Microbiol. 1:8–20. [DOI] [PubMed] [Google Scholar]
- Harkins CP, et al. 2017. Methicillin-resistant Staphylococcus aureus emerged long before the introduction of methicillin into clinical practice. Genome Biol. 18(1):13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- International Working Group on the Classification of Staphylococcal Cassette Chromosome Elements (IWG-SCC). 2009. Classification of staphylococcal cassette chromosome mec (SCCmec): guidelines for reporting novel SCCmec elements. Antimicrob Agents Chemother. 53:4961–4967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito T, et al. 2001. Structural comparison of three types of staphylococcal cassette chromosome mec integrated in the chromosome in methicillin-resistant Staphylococcus aureus. Antimicrob Agents Chemother. 45(5):1323–1336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito T, et al. 2012. Guidelines for reporting novel mecA gene homologues. Antimicrob Agents Chemother. 56(10):4997–4999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Junier T, Zdobnov EM.. 2010. The Newick utilities: high-throughput phylogenetic tree processing in the UNIX shell. Bioinformatics 26(13):1669–1670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh K, Misawa K, Kuma KI, Miyata T.. 2002. MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res. 30(14):3059–3066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lefébure T, Stanhope MJ.. 2007. Evolution of the core and pan-genome of Streptococcus: positive selection, recombination, and genome composition. Genome Biol. 8(5):R71.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindsay JA. 2010. Genomic variation and evolution of Staphylococcus aureus. Int J Med Microbiol. 300(2–3):98–103. [DOI] [PubMed] [Google Scholar]
- McCarthy AJ, et al. 2014. Extensive horizontal gene transfer during Staphylococcus aureus co-colonization in vivo. Genome Biol Evol. 10:2697–2708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Méric G, et al. 2015. Ecological overlap and horizontal gene transfer in Staphylococcus aureus and Staphylococcus epidermidis. Genome Biol Evol. 7(5):1313–1328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson-Sathi S, et al. 2015. Origins of major archaeal clades correspond to gene acquisitions from bacteria. Nature 517(7532):77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otto M. 2013. Coagulase-negative staphylococci as reservoirs of genes facilitating MRSA infection: staphylococcal commensal species such as Staphylococcus epidermidis are being recognized as important sources of genes promoting MRSA colonization and virulence. Bioessays 35(1):4–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozer EA, Allen JP, Hauser AR.. 2014. Characterization of the core and accessory genomes of Pseudomonas aeruginosa using bioinformatic tools Spine and AGEnt. BMC Genomics 15(1):737.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puigbo P, Wolf YI, Koonin EV.. 2010. The tree and net components of prokaryote evolution. Genome Biol Evol. 2:745–756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Core Team. 2018. R: a language and environment for statistical computing. Vienna (Austria: ): R Foundation for Statistical Computing; Available from: https://www.R-project.org/, last accessed October 10, 2019. [Google Scholar]
- Rambaut A. 2012. Tree figure drawing tool version 1.4. 2. Institute of Evolutionary Biology, Edinburgh: University of Edinburg; Available from: http://tree.bio.ed.ac.uk/software/figtree, last accessed October 10, 2019. [Google Scholar]
- Rasko DA, et al. 2008. The pangenome structure of Escherichia coli: comparative genomic analysis of E. coli commensal and pathogenic isolates. J Bacteriol. 190(20):6881–6893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rice P, Longden I, Bleasby A.. 2000. EMBOSS: the European molecular biology open software suite. Trends Genet. 16(6):276–277. [DOI] [PubMed] [Google Scholar]
- Robinson DF, Foulds LR.. 1981. Comparison of phylogenetic trees. Math Biosci. 53(1–2):131–147. [Google Scholar]
- Rolo J, et al. 2017. Evolutionary origin of the staphylococcal cassette chromosome mec (SCCmec). Antimicrob Agents Chemother. 61:02302–02316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schliep KP. 2011. phangorn: phylogenetic analysis in R. Bioinformatics 27(4):592–593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sela I, Ashkenazy H, Katoh K, Pupko T.. 2015. GUIDANCE2: accurate detection of unreliable alignment regions accounting for the uncertainty of multiple parameters. Nucleic Acids Res. 1:43.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soucy SM, Huang J, Gogarten JP.. 2015. Horizontal gene transfer: building the web of life. Nat Rev Genet. 16(8):472–482. [DOI] [PubMed] [Google Scholar]
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30(9):1312–1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subakishita T, Kuwahara-Arai K, Baba T, Hiramatsu K.. 2010. Staphylococcal cassette chromosome mec-like element in Macrococcus caseolyticus. Antimicrob Agents Chemother. 54:1469–1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tatusov RL, et al. 2003. The COG database: an updated version includes eukaryotes. BMC Bioinformatics 4:41.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tatusov RL, Koonin EV, Lipman DJ.. 1997. A genomic perspective on protein families. Science 278(5338):631–637. [DOI] [PubMed] [Google Scholar]
- Tettelin H, et al. 2005. Genome analysis of multiple pathogenic isolates of Streptococcus agalactiae: implications for the microbial “pan-genome”. Proc Natl Acad Sci U S A. 102(39):13950–13955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tettelin H, Riley D, Cattuto C, Medini D.. 2008. Comparative genomics: the bacterial pan-genome. Curr. Opin. Microbiol. 11(5):472–477. [DOI] [PubMed] [Google Scholar]
- Touchon M, et al. 2009. Organised genome dynamics in the Escherichia coli species results in highly diverse adaptive paths. PLoS Genet. 5(1):e1000344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- World Health Organization. 2016. Fact sheet – antibiotic resistance – media center. World Health Organization (WHO). Available from: https://www.who.int/en/news-room/fact-sheets/detail/antibiotic-resistance, last accessed October 10, 2019.
- Xue H, et al. 2015. Coexistence of heavy metal and antibiotic resistance within a novel composite staphylococcal cassette chromosome in a Staphylococcus haemolyticus isolate from bovine mastitis milk. Antimicrob Agents Chemother. 59(9):5788–5792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y, et al. 2014. PanGP: a tool for quickly analyzing bacterial pan-genome profile. Bioinformatics 9:1297–1299. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.