Abstract
Aberrant expression of CD5 has been reported in 5–10% of diffuse large B-cell lymphomas (DLBCLs). CD5+ DLBCL had been recognized as an aggressive immunophenotypic subgroup of DLBCL in the 2008 WHO classification of haematolymphoid neoplasm; however, it was eliminated from the list of subgroups of DLBCLs in the revised 2016 classification. Nevertheless, there is much controversy regarding the clinical significance of CD5 expression, and many researchers still assert that this subgroup exhibits an extremely unfavorable prognosis with frequent treatment failure. We retrospectively investigated 405 DLBCLs recruited from three university hospitals in Korea from 1997 to 2013. The clinical profile, immunophenotype, and chromosomal structural alterations of the BCL2 and MYC genes were compared according to CD5 expression. A total of 29 cases of de novo CD5+ DLBCL were identified out of 405 in our series (7.4%). Clinicopathologic correlation was performed in all 29 CD5+ DLBCLs and 166 CD5- DLBCLs which were eligible for full clinical review and further pathologic examination. Compared with CD5- counterparts, CD5+ DLBCLs showed female preponderance, frequent bone marrow involvement, higher lactate dehydrogenase level, advanced Ann Arbor stages and poorer prognosis (all p<0.05). Pathologically, the expression of CD5 positively correlated with that of BCL2, MYC and Ki-67 (all p<0.05). Coexpression of BCL2 and MYC, which is referred to as a double-expressor, was relatively more common in CD5+ DLBCL, whereas translocation or amplification of these genes was very rare. in conclusion, the expression of CD5 is an independent poor prognostic factor of DLBCLs, and this subgroup displays unique clinicopathologic features. Although the exact mechanism remains uncertain, consistent activation of BCL2 and MYC by alternative pathways other than chromosomal translocation may contribute to the pathogenesis.
Introduction
Pathologic diagnosis of malignant lymphoma is based on the application of immunohistochemistry (IHC) using lineage-specific surface markers such as CD3 or CD20. However, aberrant expression of some T-cell markers, of which the most representative is CD5, has been well documented in a subset of B-cell neoplasms [1–3]. In fact, CD5 is an important diagnostic marker of chronic lymphocytic leukemia/small lymphocytic lymphoma (CLL/SLL) and mantle cell lymphoma [4].
The expression of CD5 in diffuse large B-cell lymphoma (DLBCL) can be observed in Richter transformation of CLL but can also be found in de novo DLBCLs. Since it was first recognized in 1995 [2], many de novo CD5+ DLBCL cases have been documented, and the overall incidence comprises 5–10% of all DLBCLs [1, 5, 6]. The CD5+ DLBCL had been introduced as an immunophenotypic subgroup of DLBCL in the 2008 WHO classification of haematolymphoid neoplasms, however, the revised 2016 version has omitted designation of the CD5+ subtype. Nevertheless, accumulating evidences suggest that CD5+ DLBCL is a distinctive subgroup which typically presents aggressive clinical features and adverse outcomes [1, 4, 7–11]. Previous studies have confirmed that the prognosis of CD5+ DLBCL is still poor regardless of Rituximab based chemotherapy [1, 9, 10], and even with the salvage stem cell transplantation [12]. To achieve optimal therapeutic responses, better understanding of pathogenic mechanisms and risk stratification are crucial.
To date, most large-scale studies of de novo CD5+ DLBCL have been performed in Japan, and there are only few reports from other Asian countries or Western areas [1, 8, 9, 12–15]. We performed a retrospective study to review detailed characteristics of CD5+ DLBCL among Korean patients, particularly focusing on the relationship to other constitutional prognostic factors, such as cell of origin by IHC, and BCL2 and MYC status.
Materials and methods
Case selection and analysis of the clinical characteristics
Cases diagnosed as DLBCL, not otherwise specified (NOS), were retrieved from three university hospitals (Seoul National University Hospital, Seoul National University Bundang Hospital and Seoul National University Boramae Hospital) in Korea from January 1996 to January 2016. The diagnosis was confirmed by two experienced hematopathologists (HYN and JEK), based on the 2017 WHO classification of Tumours of Haematopoietic and Lymphoid Tissues [4]. Clinical profiles and follow-up data were obtained from electronic medical records.
Immunohistochemistry (IHC) and Epstein-Barr virus (EBV) detection
To determine the CD5+ subgroup, all DLBCL cases were reexamined and assessed by IHC. IHC was performed using 4 μm sections of paraffin-embedded tissue blocks using the following antibodies BCL2 (M0887, mouse, monoclonal, 1:100; Dako, Carpinteria, CA, USA), BCL6 (LN22, mouse, monoclonal, 1:100; Novocastra, Newcastle, UK), CD3 (M7254, mouse, monoclonal, 1:100; Dako), CD5 (M3641, mouse, monoclonal, 1:100; Dako), CD10 (PA0270, mouse, monoclonal, 1:100; Novocastra), CD20 (M0755, mouse, monoclonal, 1:400; Dako), IRF4/MUM1 (M7259, mouse, monoclonal, 1:100; Dako), Ki-67 (M7240, mouse, monoclonal, 1:100; Dako) and MYC (Y69, rabbit, monoclonal, 1:100; Epitomics, Burlingame, CA, USA). The IHC for antibodies were performed. First, sections were treated with Target Retrieval Solution (Dako, Glostrup, Denmark) at 115˚C for 15 min after inhibiting endogenous peroxidase activity for 30 min with 3% hydrogen peroxidase in methanol for antigen retrieval.Then, immune complexes were detected with the Envision Detection System (Dako) after overnight incubation. Finally, hematoxylin counterstaining was done.
For CD5 IHC, we used the cutoff of more than 50% tumor cells showing immunoreactivity for CD5 of any intensity as positive. Tumor cells with more than 30% staining were considered positive for BCL6, CD10 and IRF4/MUM1 [16]. For BCL2 and MYC, staining of more than 50%, and 40% was used as cutoff, according to the criteria suggested by Johnson et al [17]. EBV in situ hybridization (ISH) was performed using an EBV-encoded RNA (EBER) probe (INFORM EBER Probe; Ventana Medical Systems, Tucson, AZ, USA). The evaluation of IHC and EBV ISH were perfomed by the same two experienced hematopathologists (HYN and JEK), and discordant cases were discussed for consensus.
Detection of IgH/MYC translocation, MYC amplification and IgH/BCL2 translocation
Fluorescence in situ hybridization (FISH) was performed on paraffin embedded tissue block sections according to the manufacturer’s protocol. A Vysis LSI MYC dual-color, break-apart rearrangement probe (Abbott Molecular, Abbott Park, IL, USA) was used to detect MYC translocation, and a Vysis IgH/MYC/CEP 8 Tri-color DF probe (Abbott) was used for amplification. At least 100 cells from each case were assessed for split signals to identify MYC translocation and gene copy number alteration. FISH using a Vysis LSI BCL2 dual-color break-apart rearrangement probe (Abbott Molecular, Abbott Park, IL) was performed to identify BCL2 translocation. The result was considered positive for rearrangement and amplification when >20% of nuclei showed a break-apart signal or extra copies [18].
Statistical analysis
For the association analysis, the Mann–Whitney U-test, Fisher's exact test or Pearson’s chi-square test was performed. Spearman’s ρ was used to assess correlations between variables. Overall survival (OS) was measured from the date of diagnosis to the date of death. Progression-free survival (PFS) was estimated from the date of diagnosis to the date of disease progression, including relapse and death. Univariable Kaplan–Meier survival analysis with log-rank tests was conducted to compare the outcome based on the status of various parameters. Multivariable survival analysis using the Cox proportional hazards model was performed to identify independent prognostic markers. A two-tailed P-value of ≤0.05 was considered statistically significant. All data were analyzed with SPSS software, version 22.0 (SPSS Inc., IBM, Armonk, NY, USA).
Ethics
Ethical approval was obtained by the regional ethics committee in Seoul National University Boramae Hospital, Korea according to the declaration of Helsinki (2014/020, 2014/020/1, and 2014/233) (IRB No. 10-2018-19). Informed consent from participants was not required according to the ethics committee. All data were fully anonymized before accessed.
Results
A total of 405 cases of DLBCL were investigated for CD5 expression on tumor cells, and 30 cases of CD5+ DLBCLs, consisting of one case transformed from CLL and 29 de novo cases (29 of 405, 7.2%), were identified. To compare the clinicopathologic findings between the CD5+ and CD5- groups, a total of 166 CD5-DLBCL, which had no evidence of transformation from low grade lymphoma, were selected, of which detailed clinical data and sufficient tissue for further pathologic studies were available. Clinicopathologic profiles of de novo CD5+ DLBCL (N = 29) and CD5-DLBCL patients are summarized in Table 1. Most CD5- DLBCL patients (21/29, 72.4%) were treated with CHOP (cyclophosphamide, doxorubicin, vincristine, and prednisone)-based chemotherapy with or without rituximab. Of these, 3 received etoposide in addition to the above combination (R-EPOCH). The remaining 4 patients were managed by supportive care only. Similarly, 129 (77.7%) CD5-DLBCL patients were treated with a CHOP-based regimen. The median follow-up time of de novo CD5+ DLBCL and CD5- DLBCL patients was 13 months (ranging from 1 to 163) and 43 months (ranging from 0 to 261), respectively.
Table 1. Comparison of clinicopathologic features of CD5+ and CD5- DLBCL patients.
| Clinicopathologic Feature | CD5+ DLBCL (n = 29) |
CD5- DLBCL (n = 166) |
P-value |
|---|---|---|---|
| Age over 65 yr | 15 (51.7%) | 59 (35.5%) | 0.098 |
| Sex (Male: female) | 9:20 | 103:63 | 0.002* |
| Initial diagnosis in lymph node | 16 (55.2%) | 60 (36.1%) | 0.039* |
| BM involvement | 11 (37.9%) | 20 (12.1%) | 0.010* |
| Ann Arbor Stage (I-II:III-IV) | 10:19 | 97:69 | 0.030* |
| ECOG PS (0–1:2–5) | 18:11 | 132:34 | 0.060 |
| Elevated LDH | 20 (69.0%) | 78 (47.0%) | 0.028* |
| >1 Extranodal sites | 13 (44.8%) | 30 (18.1%) | 0.002* |
| B symptoms | 8 (27.6%) | 41 (24.7%) | 0.689 |
| Bulky tumor | 4 (13.8%) | 23 (13.9%) | 0.980 |
| IPI (0–2:3–5) | 11:18 | 113:53 | 0.004* |
| Disease progression | 18 (62.1%) | 83 (50%) | 0.230 |
| CNS relapse | 1 (3.7%) | 8 (4.8%) | 0.745 |
| Treatment | 0.065 | ||
| R-CHOP | 18 (62.1%) | 78 (47.0%) | |
| CHOP | 3 (10.3%) | 51 (30.7%) | |
| Other chemotherapy | 3 (10.3%) | 18 (10.8%) | |
| Other treatment (e.g. Op, RT etc) | 0 (0%) | 9 (5.4%) | |
| No treatment | 4 (13.8%) | 2 (1.2%) | |
| PFS, median (range) | 10 (0–163) | 40 (0–192) | 0.003* |
| OS, median (range) | 13 (1–163) | 44 (0–261) | 0.008* |
| Hans system (GCB:Non-GCB) | 8:19 | 56:110 | 0.570 |
| CD10 | 5 (17.2%) | 26 (15.7%) | 0.830 |
| BCL6 | 14 (48.3%) | 76 (45.8%) | 0.804 |
| MUM1 | 16 (55.2%) | 58 (34.9%) | 0.038* |
| BCL2 | 17 (58.6%) | 50 (30.1%) | 0.003* |
| MYC | 9 (33.3%) | 7 (4.2%) | <0.001* |
| MYC/BCL2 double-expressor | 8 (27.6%) | 5 (3.0%) | <0.001* |
| Ki-67, median (range) (%) | 60 (20–100) | 50 (6.6–88.3) | 0.007* |
| MYC rearrangement | 2 (6.9%) | 6 (3.6%) | 0.614 |
DLBCL, diffuse large B-cell lymphoma; BM, bone marrow; IPI, ECOG, Eastern cooperative group; LDH, lactate dehydrogenase; IPI, international prognostic index; CNS, central nervous system; Op, operation; RT, radiation therapy; PFS, progression-free survival; OS, overall survival; Germinal center B-cell like subgroup.
* P ≤0.05
Compared with CD5- DLBCL, the CD5+ group revealed female preponderance (p = 0.002), more than one extranodal site involvement (p = 0.002), and frequent bone marrow involvement (p = 0.01). In addition, elevated lactate dehydrogenase (LDH, p = 0.028) and a higher international prognostic index (p = 0.004) were more common in CD5+ DLBCL.
Both CD5+ and CD5- DLBCL showed non-germinal center B-cell type predominance according to the Hans algorithm [16]. Compared with CD5- cases, CD5+ DLBCL cases showed more frequent IRF4/MUM1 (p = 0.038), BCL2 (p = 0.003), and MYC (p<0.001) expression and a higher Ki-67 proliferation index (p = 0.007). Concurrent expression of BCL2 and MYC, which is known as double-expressor (DE), was significantly more common in CD5+ cases (8/29 vs. 5/166, p<0.001). Of note, only two (2 of 29 CD5+ DLBCLs, and 2 of 9 MYC overexpressed CD5+ DLBCLs) revealed MYC rearrangement, and no amplification was identified by FISH. BCL2 rearrangement or amplification was not found. (Figs 1 and 2).
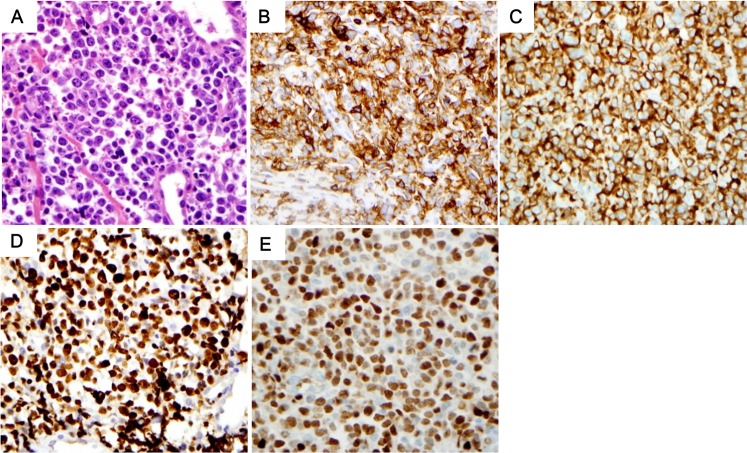
Fig 1. Pathologic characteristics of CD5+ DLBCL.
Pleomorphic large cells with many apoptotic features were shown in many cases (A). Tumor cells were positive for CD5 (B), BCL2 (C), IRF4/MUM1 (D). Note the diffuse strong pattern in CD5 IHC. Tumor cells also showed high Ki-67 proliferation index (E).
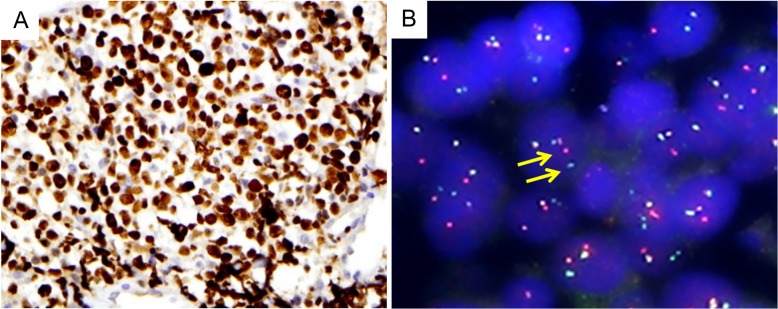
Fig 2. Representative case showing MYC overexpression and rearrangement.
High expression of MYC protein (A) and presence of MYC gene rearrangement examined by FISH for MYC, break apart probe (B, split signals indicated by arrows).
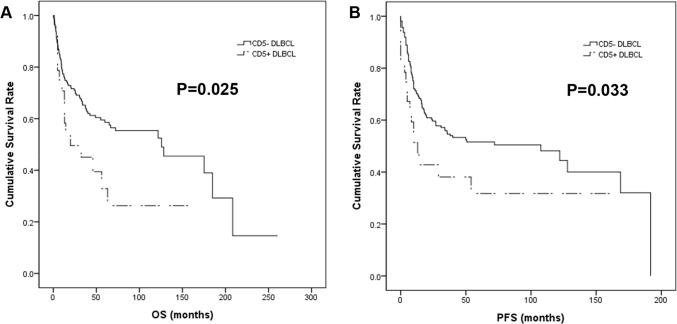
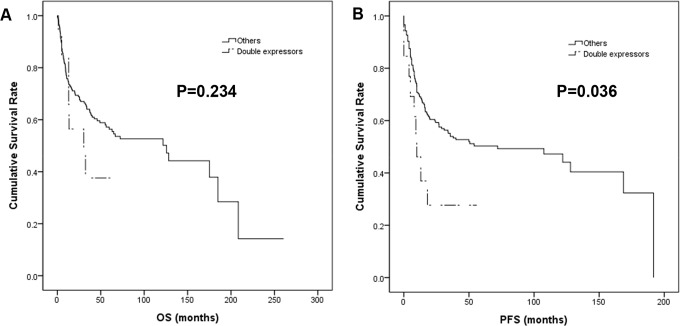
In the univariable survival analysis, patients with CD5+ DLBCL showed significantly shorter OS (p = 0.025) and PFS (p = 0.033) than those with CD5- tumors (Fig 3), whereas DE lymphomas revealed inferior PFS (p = 0.036) but not OS (p = 0.234) (Fig 4). Additionally, older age (>65 yrs), higher performance status (PS) by the Eastern Cooperative Oncology Group, elevated serum LDH, presence of B symptoms, more than one extranodal involvement and high Ann Arbor stages were all associated with inferior OS and PFS (all p<0.001) (Tables 2 and 3). In the multivariable Cox regression analysis, older age, higher PS, elevated serum LDH and the presence of B symptoms remained independent prognostic factors for OS and PFS, whereas CD5 positivity and DE did not (Tables 2 and 3).
Fig 3. Kaplan-Meier survival curves of CD5+ and CD5-DLBCL patients.
CD5 positivity was associated with significantly shorter OS (A) and PFS (B).
Fig 4. Kaplan-Meier survival curves of double-expressors compared with others.
DE was associated with inferior PFS (B) but not OS (A).
Table 2. Univariable and multivariable analysis for OS in DLBCL.
| Variables | Univariate analysis (OS) | Multivariate analysis (OS) | ||
|---|---|---|---|---|
| OR (95% CI) | P-value | OR (95% CI) | P-value | |
| All DLBCL | ||||
| Age > 65yr | 2.459 (1.622–3.728) | <0.001* | 2.111 (1.245–3.579) | 0.006* |
| Performance status | 3.473 (2.234–5.401) | <0.001* | 2.111 (1.280–3.481) | 0.003* |
| Elevated LDH | 2.362 (1.470–3.797) | <0.001* | 1.915 (1.080–3.395) | 0.026* |
| B symptom | 2.824 (1.828–4.363) | <0.001* | 2.206 (1.327–3.664) | 0.002* |
| > 1 Extranodal sites | 1.796 (1.113–2.900) | 0.017* | 1.037 (0.554–1.941) | 0.909 |
| Stage | 2.260 (1.475–3.464) | <0.001* | 1.308 (0.729–2.346) | 0.369 |
| Double-expressor | 1.465 (0.704–3.049) | 0.234 | 0.583 (0.222–1.531) | 0.273 |
| CD5 positivity | 1.825 (1.071–3.109) | 0.027* | 1.374 (0.723–2.612) | 0.331 |
| Non-GCB DLBCL | ||||
| Age > 65yr | 2.742 (1.642–4.579) | <0.001* | 2.429 (1.260–4.681) | 0.008* |
| Performance status | 3.559 (2.062–6.142) | <0.001* | 1.369 (0.704–2.664) | 0.355 |
| Elevated LDH | 2.687 (1.504–4.801) | 0.001 | 1.784 (0.866 = 3.676) | 0.116 |
| B symptom | 3.026 (1.791–5.115) | <0.001* | 2.456 (1.323–4.558) | 0.004* |
| > 1 Extranodal sites | 1.952 (1.108–3.438) | 0.021* | 0.867 (0.375–2.004) | 0.739 |
| Stage | 2.858 (1.675–4.876) | <0.001* | 2.831 (1.486–5.395) | 0.002* |
| Double-expressor | 2.189 (0.929–5.154) | 0.073 | 1.181 (0.384–3.635) | 0.772 |
| CD5 positivity | 2.446 (1.307–4.577) | 0.005* | 1.766 (0.821–3.798) | 0.145 |
DLBCL, diffuse large B-cell lymphoma; LDH, lactate dehydrogenase; OS, overall survival; OR, odds ratio; CI, confidence interval.
* P ≤0.05
Table 3. Univariable and multivariable ananlysis for PFS in DLBCL.
| Variables | Univariate analysis (PFS) | Multivariate analysis (PFS) | ||
|---|---|---|---|---|
| OR (95% CI) | P-value | OR (95% CI) | P-value | |
| All DLBCL | ||||
| Age > 65yr | 25884 (1.738–3.854) | <0.001* | 1.918 (1.158–3.176) | 0.011* |
| Performance status | 3.288 (2.155–5.018) | <0.001* | 2.266 (1.383–3.713) | 0.001* |
| Elevated LDH | 2.363 (1.497–3.729) | <0.001* | 1.917 (1.100–3.341) | 0.022* |
| B symptom | 2.686 (1.766–4.085) | <0.001* | 2.191 (1.346–3.569) | 0.002* |
| > 1 Extranodal sites | 1.702 (1.079–2.685) | 0.022* | 0.534 (0.508–1.771) | 0.972 |
| Stage | 2.134 (1.424–3.199) | <0.001* | 1.192 (0.684–2.079) | 0.535 |
| Double-expressor | 1.722 (0.892–3.325) | 0.036* | 0.676 (0.264–1.732) | 0.414 |
| CD5 positivity | 1.748 (1.304–2.954) | 0.037* | 1.681 (0.873–3.239) | 0.121 |
| Non-GCB DLBCL | ||||
| Age > 65yr | 3.186 (1.934–5.250) | <0.001* | 2.525 (1.402–4.548) | 0.002* |
| Performance status | 3.388 (2.003–5.731) | <0.001* | 1.570 (0.822–2.997) | 0.171 |
| Elevated LDH | 2.861 (1.613–5.075) | <0.001* | 1.752 (0.847–3.623) | 0.131 |
| B symptom | 2.665 (1.604–4.428) | <0.001* | 2.612 (1.443–4.728) | 0.002* |
| > 1 Extranodal sites | 1.949 (1.131–3.359) | 0.023* | 0.851 (0.365–1.983) | 0.708 |
| Stage | 2.598 (1.566–4.311) | <0.001* | 2.353 (1.264–4.380) | 0.007* |
| Double-expressor | 3.189 (1.500–6.782) | 0.003* | 1.297 (0.436–3.854) | 0.640 |
| CD5 positivity | 2.488 (1.347–4.598) | 0.004* | 2.840 (1.340–6.015) | 0.006* |
DLBCL, diffuse large B-cell lymphoma; LDH, lactate dehydrogenase; PFS, progression-free survival; OR, odds ratio; CI, confidence interval.
* P ≤0.05
Among non-GCB type DLBCL, CD5 expression was associated with inferior OS (p = 0.005) and PFS (p = 0.004), while DE was associated only with shorter PFS (p = 0.003) in univariable analysis. In multivariable analysis, CD5 positivity was an independent prognostic factor for poor PFS (p = 0.006), but not OS (p = 0.145) (Tables 2 and 3).
Discussion
In this study, we demonstrated that CD5 expression was associated with aggressive clinical features and poor survival in DLBCL. Pathologically, the expression of CD5 correlated with BCL2 and MYC positivity, and concurrent expression of these two proteins (DE) was more frequently found in CD5+ DLBCL, whereas chromosomal translocation or amplification was rarely noted. We suggest that CD5+ DLBCL is a distinct immunophenotypic subtype, and its pathogenesis and biologic nature are possibly related to alternative activation of MYC or BCL2 proteins other than gene rearrangement.
The overall incidence of CD5+ DLBCL in our Korean cases accounted for 7.2%, which is similar to previous series from Japan and other Western countries [1, 8, 9, 13–15]. Most of our series were de novo cases, because the occurrence of CLL/SLL and mantle cell lymphoma is extremely low in the Korean population, occupying only 2.1% and 1.3% of malignant lymphoma, respectively [19]. Many previous studies identified CD5 expression as an independent prognostic factor regardless of rituximab use [1, 7, 9, 10, 13, 20, 21]. Others also reported that more intensive chemotherapy other than R-CHOP or stem cell transplantation did not overcome its dismal outcome [12, 15, 22]. In the present study, CD5+ DLBCL showed female predominance, higher IPI and Ann Arbor stages, and other aggressive clinical features, which are in line with previous reports [1, 5, 7–9, 13, 14, 20, 21, 23]. These clinical characteristics are also commonly noted in double hit lymphoma (DHL) or DE cases [17, 24, 25]; both have been known as high-risk lymphomas although the clinical behaviors of DE are not as much aggressive as DHLs [26]. In particular, there is considerable pathologic overlap between DE lesions and CD5+ DLBCL regarding cell of origin and complicated mutational events [1, 9, 27–29]. Therefore, we paid special attention to the relationship of CD5 with BCL2 or MYC expression due to the aforementioned resemblance between CD5+ DLBCL and DE lymphomas. We found a strong association with CD5 positivity and BCL2 expression, which was reported in previous series [9, 13, 15, 23], and a correlation of CD5 with MYC, which has not been widely investigated before. However, only 2 (6.9%) cases of MYC rearrangement were detected and no MYC amplification or BCL2 rearrangement was identified in contrast to high levels of protein expression. Although there have been few reports, most revealed scarce mutations in the MYC gene in CD5+ DLBCL [9, 12]. Alteration of STAT3 and nuclear factor-kappaB (NF-kB) pathway was suggested for a possible explanation for overexpression of BCL2 in CD5+ DLBCL [9]. Similar genetic alterations have also been demonstrated in recent studies suggesting that the pathology of DE lymphomas reflect cumulative mutations involving B-cell receptor signaling and NF-kB [28, 30, 31]. In DE tumors, alternative transcriptional mechanisms, posttranscriptional, or posttranslational pathways involving miRNA, histone binding proteins, and ubiquitination were proposed as the basis of MYC activation [32–35]. Although direct evidence of CD5 expression influencing BCL2 or MYC activation cannot be presented in the present study, the aggressive biologic behavior of CD5+ DLBCL might be related to the higher frequency of DE in this group.
CD5 is a glycoprotein mainly expressed on the membrane of mature T cells and is only dimly expressed on a subset of late stage hematogones/normal B-lineage precursors [36–38]. Most CD5+ DLBCLs may originate from early B-cells prior to the germinal center stage with some exceptions [39]. Recently published paper by a Japanese group revealed a significantly lower prevalence of MYD88 and CD79B gene mutations in CD5+ DLBCL in contrast to other extranodal DLBCLs [40]. These findings support the idea that the cellular origin of CD5+ DLBCL might be different from that of ordinary DLBCL. Although the cellular origin is unclear, expression of CD5 is known to induce interleukin-10 production and has anti-apoptotic function maintaining B-cell survival [41], which is attributed to the aggressive biology of CD5+ DLBCL. Currently, there is no specific guideline on management of CD5+ DLBCL, although many cases are controlled under high risk stratification. Based on recent studies suggesting that MYC upregulates immune checkpoint pathways such as programmed death-ligand 1 (PD-L1) [42], immunotherapy can be added to CD5+ DLBCL patients overexpressing MYC.
In summary, CD5+DLBCL is a distinct subgroup of DLBCL, characterized by aggressive clinical courses that can easily be missed in routine clinical practice. To date, this is the largest study of CD5+ DLBDLs in Korea, and we confirmed that CD5 is an easily available, cost-effective prognostic biomarker for DLBCL. Although the exact mechanism of CD5 expression and its connection to BCL2 or MYC and DE requires further investigation at the molecular level, our results will provide a basis for new therapeutic regimens including small molecule inhibitors that block MYC or BCL2 and even immunotherapy.
Supporting information
(XLSX)
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
This study was supported by the Grant of National Research Foundation of Korea (2017R1A2B4005052). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Yamaguchi M, Seto M, Okamoto M, Ichinohasama R, Nakamura N, Yoshino T, et al. De novo CD5+ diffuse large B-cell lymphoma: a clinicopathologic study of 109 patients. Blood. 2002;99(3):815–21. Epub 2002/01/25. 10.1182/blood.v99.3.815 . [DOI] [PubMed] [Google Scholar]
- 2.Matolcsy A, Chadburn A, Knowles DM. De novo CD5-positive and Richter's syndrome-associated diffuse large B cell lymphomas are genotypically distinct. Am J Pathol. 1995;147(1):207–16. Epub 1995/07/01. [PMC free article] [PubMed] [Google Scholar]
- 3.Maeshima AM, Taniguchi H, Nomoto J, Maruyama D, Kim SW, Watanabe T, et al. Secondary CD5+ diffuse large B-cell lymphoma not associated with transformation of chronic lymphocytic leukemia/small lymphocytic lymphoma (Richter syndrome). Am J Clin Pathol. 2009;131(3):339–46. Epub 2009/02/21. 10.1309/AJCP58FETFGLCKKW . [DOI] [PubMed] [Google Scholar]
- 4.Swerdlow SH, Campo E, Harris NL, Jaffe ES, Pileri SA, Stein H, et al. The Classification of Tumors of Haematopoietic and Lymphoid Tissues France: Lyon, IARC; 2017. [Google Scholar]
- 5.Jain P, Fayad LE, Rosenwald A, Young KH, O'Brien S. Recent advances in de novo CD5+ diffuse large B cell lymphoma. Am J Hematol. 2013;88(9):798–802. Epub 2013/05/23. 10.1002/ajh.23467 . [DOI] [PubMed] [Google Scholar]
- 6.Swerdlow SH, Campo E, Harris NL, Jaffe ES, Pileri SA, Stein H, et al. WHO Classification of Tumours of Haematopoietic and Lymphoid Tissues France: IARC Press, Lyon; 2008. [Google Scholar]
- 7.Hyo R, Tomita N, Takeuchi K, Aoshima T, Fujita A, Kuwabara H, et al. The therapeutic effect of rituximab on CD5-positive and CD5-negative diffuse large B-cell lymphoma. Hematol Oncol. 2010;28(1):27–32. Epub 2009/04/10. 10.1002/hon.896 . [DOI] [PubMed] [Google Scholar]
- 8.Yamaguchi M, Ohno T, Oka K, Taniguchi M, Ito M, Kita K, et al. De novo CD5-positive diffuse large B-cell lymphoma: clinical characteristics and therapeutic outcome. Br J Haematol. 1999;105(4):1133–9. Epub 1999/11/11. 10.1046/j.1365-2141.1999.01513.x . [DOI] [PubMed] [Google Scholar]
- 9.Xu-Monette ZY, Tu MF, Jabbar KJ, Cao X, Tzankov A, Visco C, et al. Clinical and biological significance of de novo CD5+ diffuse large B-cell lymphoma in Western countries (vol 8, pg 5615, 2015). Oncotarget. 2015;6(16):14720–. 10.18632/oncotarget.4464 WOS:000359010000070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Miyazaki K, Yamaguchi M, Suzuki R, Kobayashi Y, Maeshima AM, Niitsu N, et al. CD5-positive diffuse large B-cell lymphoma: a retrospective study in 337 patients treated by chemotherapy with or without rituximab. Annals of Oncology. 2011;22(7):1601–7. 10.1093/annonc/mdq627 WOS:000292048500017. [DOI] [PubMed] [Google Scholar]
- 11.Kroft SH, Howard MS, Picker LJ, Ansari MQ, Aquino DB, McKenna RW. De Novo CD5+diffuse large B-Cell lymphomas—A heterogeneous group containing an unusual form of splenic lymphoma. Am J Clin Pathol. 2000;114(4):523–33. 10.1309/RM1Q-1T0B-WKQB-AF5A WOS:000089533700005. [DOI] [PubMed] [Google Scholar]
- 12.Alinari L, Gru A, Quinion C, Huang Y, Lozanski A, Lozanski G, et al. De novo CD5+ diffuse large B-cell lymphoma: Adverse outcomes with and without stem cell transplantation in a large, multicenter, rituximab treated cohort. Am J Hematol. 2016;91(4):395–9. Epub 2016/01/23. 10.1002/ajh.24299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yamaguchi M, Nakamura N, Suzuki R, Kagami Y, Okamoto M, Ichinohasama R, et al. De novo CD5+ diffuse large B-cell lymphoma: results of a detailed clinicopathological review in 120 patients. Haematologica. 2008;93(8):1195–202. Epub 2008/06/17. 10.3324/haematol.12810 . [DOI] [PubMed] [Google Scholar]
- 14.Westin J, McLaughlin P. De novo CD5+ diffuse large B-cell lymphoma: a distinct subset with adverse features, poor failure-free survival and outcome with conventional therapy. Leuk Lymphoma. 2010;51(1):161–3. Epub 2009/10/30. 10.3109/10428190903324244 . [DOI] [PubMed] [Google Scholar]
- 15.Thakral B, Medeiros LJ, Desai P, Lin P, Yin CC, Tang G, et al. Prognostic impact of CD5 expression in diffuse large B-cell lymphoma in patients treated with rituximab-EPOCH. Eur J Haematol. 2017;98(4):415–21. Epub 2017/01/01. 10.1111/ejh.12847 . [DOI] [PubMed] [Google Scholar]
- 16.Hans CP, Weisenburger DD, Greiner TC, Gascoyne RD, Delabie J, Ott G, et al. Confirmation of the molecular classification of diffuse large B-cell lymphoma by immunohistochemistry using a tissue microarray. Blood. 2004;103(1):275–82. Epub 2003/09/25. 10.1182/blood-2003-05-1545 . [DOI] [PubMed] [Google Scholar]
- 17.Johnson NA, Slack GW, Savage KJ, Connors JM, Ben-Neriah S, Rogic S, et al. Concurrent expression of MYC and BCL2 in diffuse large B-cell lymphoma treated with rituximab plus cyclophosphamide, doxorubicin, vincristine, and prednisone. J Clin Oncol. 2012;30(28):3452–9. Epub 2012/08/02. 10.1200/JCO.2011.41.0985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Quesada AE, Medeiros LJ, Desai PA, Lin P, Westin JR, Hawsawi HM, et al. Increased MYC copy number is an independent prognostic factor in patients with diffuse large B-cell lymphoma. Mod Pathol. 2017;30(12):1688–97. Epub 2017/08/05. 10.1038/modpathol.2017.93 . [DOI] [PubMed] [Google Scholar]
- 19.Jung H-R, Huh J, Ko Y-H, Jeon YK, Yoon SO, Kim SH, et al. Classification of malignant lymphoma subtypes in Korean patients: a report of the 4th nationwide study. Journal of Hematopathology. 2019. 10.1007/s12308-019-00354-y [DOI] [Google Scholar]
- 20.Chuang WY, Chang H, Shih LY, Wang PN, Chang YS, Lin TL, et al. CD5 positivity is an independent adverse prognostic factor in elderly patients with diffuse large B cell lymphoma. Virchows Arch. 2015;467(5):571–82. Epub 2015/09/16. 10.1007/s00428-015-1845-1 . [DOI] [PubMed] [Google Scholar]
- 21.Ennishi D, Takeuchi K, Yokoyama M, Asai H, Mishima Y, Terui Y, et al. CD5 expression is potentially predictive of poor outcome among biomarkers in patients with diffuse large B-cell lymphoma receiving rituximab plus CHOP therapy. Ann Oncol. 2008;19(11):1921–6. Epub 2008/06/25. 10.1093/annonc/mdn392 . [DOI] [PubMed] [Google Scholar]
- 22.Tzankov A, Leu N, Muenst S, Juskevicius D, Klingbiel D, Mamot C, et al. Multiparameter analysis of homogeneously R-CHOP-treated diffuse large B cell lymphomas identifies CD5 and FOXP1 as relevant prognostic biomarkers: report of the prospective SAKK 38/07 study. J Hematol Oncol. 2015;8:70 Epub 2015/06/14. 10.1186/s13045-015-0168-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tang H, Zhou H, Wei J, Liu H, Qian W, Chen X. Clinicopathologic significance and therapeutic implication of de novo CD5+ diffuse large B-cell lymphoma. Hematology. 2019;24(1):446–54. Epub 2019/05/11. 10.1080/16078454.2019.1614289 . [DOI] [PubMed] [Google Scholar]
- 24.Rosenthal A, Younes A. High grade B-cell lymphoma with rearrangements of MYC and BCL2 and/or BCL6: Double hit and triple hit lymphomas and double expressing lymphoma. Blood Rev. 2017;31(2):37–42. Epub 2016/10/09. 10.1016/j.blre.2016.09.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Perrot A, Monjanel H, Bouabdallah R, Quittet P, Sarkozy C, Bernard M, et al. Impact of post-brentuximab vedotin consolidation on relapsed/refractory CD30+ Hodgkin lymphomas: a large retrospective study on 240 patients enrolled in the French Named-Patient Program. Haematologica. 2016;101(4):466–73. Epub 2016/01/16. 10.3324/haematol.2015.134213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sarkozy C, Traverse-Glehen A, Coiffier B. Double-hit and double-protein-expression lymphomas: aggressive and refractory lymphomas. Lancet Oncol. 2015;16(15):e555–e67. Epub 2015/11/08. 10.1016/S1470-2045(15)00005-4 . [DOI] [PubMed] [Google Scholar]
- 27.Ye Q, Xu-Monette ZY, Tzankov A, Deng L, Wang X, Manyam GC, et al. Prognostic impact of concurrent MYC and BCL6 rearrangements and expression in de novo diffuse large B-cell lymphoma. Oncotarget. 2016;7(3):2401–16. Epub 2015/11/18. 10.18632/oncotarget.6262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Peroja P, Pedersen M, Mantere T, Norgaard P, Peltonen J, Haapasaari KM, et al. Mutation of TP53, translocation analysis and immunohistochemical expression of MYC, BCL-2 and BCL-6 in patients with DLBCL treated with R-CHOP. Sci Rep. 2018;8(1):14814 Epub 2018/10/06. 10.1038/s41598-018-33230-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Islam S, Qi W, Morales C, Cooke L, Spier C, Weterings E, et al. Disruption of Aneuploidy and Senescence Induced by Aurora Inhibition Promotes Intrinsic Apoptosis in Double Hit or Double Expressor Diffuse Large B-cell Lymphomas. Mol Cancer Ther. 2017;16(10):2083–93. Epub 2017/06/16. 10.1158/1535-7163.MCT-17-0089 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hu S, Xu-Monette ZY, Balasubramanyam A, Manyam GC, Visco C, Tzankov A, et al. CD30 expression defines a novel subgroup of diffuse large B-cell lymphoma with favorable prognosis and distinct gene expression signature: a report from the International DLBCL Rituximab-CHOP Consortium Program Study. Blood. 2013;121(14):2715–24. Epub 2013/01/25. 10.1182/blood-2012-10-461848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bogusz AM, Kovach AE, Le LP, Feng D, Baxter RH, Sohani AR. Diffuse large B-cell lymphoma with concurrent high MYC and BCL2 expression shows evidence of active B-cell receptor signaling by quantitative immunofluorescence. PLoS One. 2017;12(2):e0172364 Epub 2017/02/18. 10.1371/journal.pone.0172364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Toyota M, Suzuki H, Sasaki Y, Maruyama R, Imai K, Shinomura Y, et al. Epigenetic silencing of microRNA-34b/c and B-cell translocation gene 4 is associated with CpG island methylation in colorectal cancer. Cancer Res. 2008;68(11):4123–32. Epub 2008/06/04. 10.1158/0008-5472.CAN-08-0325 . [DOI] [PubMed] [Google Scholar]
- 33.Luscher B, Vervoorts J. Regulation of gene transcription by the oncoprotein MYC. Gene. 2012;494(2):145–60. Epub 2012/01/10. 10.1016/j.gene.2011.12.027 . [DOI] [PubMed] [Google Scholar]
- 34.Choe JY, Park M, Yun JY, Na HY, Go H, Kim HJ, et al. PELI1 expression is correlated with MYC and BCL6 expression and associated with poor prognosis in diffuse large B-cell lymphoma. Mod Pathol. 2016;29(11):1313–23. Epub 2016/10/28. 10.1038/modpathol.2016.128 . [DOI] [PubMed] [Google Scholar]
- 35.Choe JY, Yun JY, Na HY, Huh J, Shin SJ, Kim HJ, et al. MYC overexpression correlates with MYC amplification or translocation, and is associated with poor prognosis in mantle cell lymphoma. Histopathology. 2016;68(3):442–9. Epub 2015/06/24. 10.1111/his.12760 . [DOI] [PubMed] [Google Scholar]
- 36.Marie-Cardine A, Divay F, Dutot N, Green A, Perdrix A, Boyer O, et al. Transitional B cells in humans: Characterization and insight from B lymphocyte reconstitution after hematopoietic stem cells transplantation. Clin Immunol. 2008;127(1):14–25. 10.1016/j.clim.2007.11.013 WOS:000254501700003. [DOI] [PubMed] [Google Scholar]
- 37.Kipps TJ. The CD5 B cell. Adv Immunol. 1989;47:117–85. Epub 1989/01/01. . [DOI] [PubMed] [Google Scholar]
- 38.Antin JH, Emerson SG, Martin P, Gadol N, Ault KA. Leu-1+ (CD5+) B cells. A major lymphoid subpopulation in human fetal spleen: phenotypic and functional studies. J Immunol. 1986;136(2):505–10. Epub 1986/01/01. . [PubMed] [Google Scholar]
- 39.Catherwood MA, Venkatraman L. Follicular origin of a subset of CD5(+) diffuse large B-cell lymphomas. Am J Clin Pathol. 2006;125(6):954–5. Epub 2006/06/10. . [PubMed] [Google Scholar]
- 40.Takeuchi T, Yamaguchi M, Kobayashi K, Miyazaki K, Tawara I, Imai H, et al. MYD88, CD79B, and CARD11 gene mutations in CD5-positive diffuse large B-cell lymphoma. Cancer. 2017;123(7):1166–73. Epub 2016/12/05. 10.1002/cncr.30404 . [DOI] [PubMed] [Google Scholar]
- 41.Gary-Gouy H, Harriague J, Bismuth G, Platzer C, Schmitt C, Dalloul AH. Human CD5 promotes B-cell survival through stimulation of autocrine IL-10 production. Blood. 2002;100(13):4537–43. Epub 2002/10/24. 10.1182/blood-2002-05-1525 . [DOI] [PubMed] [Google Scholar]
- 42.Casey SC, Tong L, Li Y, Do R, Walz S, Fitzgerald KN, et al. MYC regulates the antitumor immune response through CD47 and PD-L1. Science. 2016;352(6282):227–31. Epub 2016/03/12. 10.1126/science.aac9935 [DOI] [PMC free article] [PubMed] [Google Scholar]