Abstract
Whitefly-transmitted begomoviruses cause serious damage to many economically important food, feed, and fiber crops. Numerous vegetable crops are severely affected and chilli leaf curl virus (ChiLCV) is the most dominant and widely distributed begomovirus in chilli (Capsicum annuum) throughout the Indian subcontinent. Recently, CRISPR-Cas9 technology was used as a means to reduce geminivirus replication in infected plants. However, this approach was shown to have certain limitations such as the evolution of escape mutants. In this study, we used a novel, multiplexed guide RNA (gRNA) based CRISPR-Cas9 approach that targets the viral genome at two or more sites simultaneously. This tactic was effective in eliminating the ChiLCV genome without recurrence of functional escape mutants. Six individual gRNA spacer sequences were designed from the ChiLCV genome and in vitro assays confirmed the cleavage behaviour of these spacer sequences. Multiplexed gRNA expression clones, based on combinations of the above-mentioned spacer sequences, were developed. A total of nine-duplex and two-triplex CRISPR-Cas9 constructs were made. The efficacy of these constructs was tested for inhibition of ChiLCV infection in Nicotiana benthamiana. Results indicated that all the constructs caused a significant reduction in viral DNA accumulation. In particular, three constructs (gRNA5+4, gRNA5+2 and gRNA1+2) were most effective in reducing the viral titer and symptoms. T7E1 assay and sequencing of the targeted viral genome did not detect any escape mutants. The multiplexed genome-editing technique could be an effective way to trigger a high level of resistance against begemoviruses. To our knowledge, this is the first report of demonstrating the effectiveness of a multiplexed gRNA-based plant virus genome editing to minimize and eliminate escape mutant formation.
Introduction
Begomoviruses (family Geminiviridae) are circular ssDNA viruses with genomes of 2.7 (monopartite) or 5.4 kb (bipartite). Begomoviruses are transmitted by whiteflies (Bemisia tabaci) and cause serious damage to numerous economically important pulse, vegetable and fiber crops in tropical and subtropical regions of the world [1–3]. Among the different vegetable crops, chilli (Capsicum annuum) suffers from a leaf curl disease caused by different begomoviruses [4]. Chilli leaf curl virus (ChiLCV) is the most predominant virus in the Indian subcontinent causing leaf curl in chilli and often accounts for 100% yield loss [5].
Begomoviruses have one or two genomic components (DNA-A and DNA-B). Majority of begomoviruses in the Old World have monopartite genomes containing one DNA component, similar to the DNA-A of bipartite begomoviruses. Often, monopartite begomoviruses are associated with a betasatellite that enhances virus virulence [6]. The genome of monopartite begomoviruses has six open reading frames (ORFs), two (V1 and V2) in viral sense and four (C1, C2, C3 and C4) in complementary sense. All proteins encoded by monopartite begomoviruses are multifunctional. V1, codes for a coat protein (CP), is involved in encapsidation of the genome into geminate particles, insect transmission and long distant movement. V2 is believed to play a role in cell-to-cell movement and suppression of RNA silencing of the host. C1, codes for a replication initiator protein (Rep), is involved in viral DNA replication. C2, codes for a transcription activator protein (TrAP), activates viral DNA transcription and is also a host RNA silencing suppressor. C3, codes for a replication enhancer protein (REn), plays a role in enhancing viral DNA replication, and the protein coded by C4 is known to be a silencing suppressor [7–9]. Besides these ORFs, the begomovirus genome has a non-coding intergenic region (IR), which possesses a bidirectional promoter and a conserved nona-nucleotide sequence (TAATATTAC), recognized by the C1 protein, acting as the origin of replication. The nona-nucleotide sequence is recognized by the AC1/C1 protein, and it serves as the origin of replication.
Management of viral diseases, especially those caused by begomoviruses, is challenging and expensive. Some of the approaches include control of the whitefly vector through insecticides, and resistant cultivars [10–13]. In tomato, host plant resistance to begomoviruses mainly was based on the use of Ty genes [14]. Another approach is the use of RNAi-based transgenic technology against bean golden mosaic virus (BGMV) in common bean in Brazil [15]. These approaches are specific to a begomovirus or its close relatives. However, in a field situation, multiple begomovirus species are known to cause the same disease. Hence, it is necessary to develop efficient and durable tactics for conferring broad spectrum resistance against begomoviruses.
CRISPR-Cas9 based genome editing shows great promise to reduce geminiviral infection [16]. The CRISPR-Cas9 system is an antiviral defence mechanism in bacteria that protects against invading DNA viruses and/or plasmids [17]. Recently, it was used for target-specific mutagenesis by inducing double strand breaks (DSBs) in DNA at a specific location in different organisms [18]. Such DSBs are either repaired by the inaccurate non-homologous end-joining (NHEJ) repair machinery or by precise homology-directed repair (HDR) [19,20]. The unrepaired DSBs are finally degraded. In most cases, mutated virus variants generated through NHEJ cannot replicate and move systemically due to the induced frameshift in the ORFs leading to a non-functional translated product. The wide ranging application of genome editing in plant virus management was discussed in recent reviews [21,22].
Over the last few years, the use of CRISPR-Cas9 editing for the management of ssDNA and dsDNA viruses was reported [23–28]. Examples include bean yellow dwarf virus (BeYDV, genus Mastrevirus) in Nicotiana benthamiana [25], beet severe curly top virus (BSCTV, genus Curtovirus) in Arabidopsis thaliana and in N. benthamiana [26] and tomato yellow leaf curl virus (TYLCV, genus Begomovirus) in Nicotiana benthamiana [23]. The gRNA-Cas9 system was also successful in simultaneously targeting three geminiviruses: beet curly top virus (BCTV) (genus Curtovirus), merremia mosaic virus (MeMV) and TYLCV (genus Begomovirus) when gRNAs specific for the IR sequence of each virus were used [23]. Ali et al. [24] demonstrated the efficiency of genome editing in cotton leaf curl Kokhran virus (CLCuKoV) and MeMV and evaluated the efficiency of the CRISPR-Cas9 machinery for targeting different coding and non-coding sequences of geminivirus genomes. However, they indicated that if gRNA is designed from coding regions, then such system generates geminivirus mutants that may evade CRISPR-Cas9 system. Such concern of development of functional mutant virus (escape mutant) after NHEJ got more emphasis when recently, Mehta et al. [28] applied the CRISPR–Cas approach in cassava. Unexpectedly, they found that the CRISPR-Cas9 system was insufficient to confer resistance to cassava against African cassava mosaic virus (ACMV) due to development of escape mutants, which may give rise to expansion of new virus variants. In this context, Rybicky [29] critically analysed their concern about such “CRISPR-induced virus evolution” and indicated that the study by Mehta et al. [28] missed an opportunity to fully investigate the potential of Cas9 for engineering resistance to plant viruses by focusing too narrowly on just one sgRNA, one Cas9, and transgenic cassava. He suggested that multiplexing of gRNA targeting different viral genomic region and proper expression of Cas9 could be the alternatives to achieving the goal. Targeting multiple regions of the viral genome simultaneously to restrict the formation of escape mutants has also been suggested earlier [30]. To investigate such multiplexing approach, we used a multiplexed gRNA-based strategy for simultaneously targeting different genomic regions of the ChiLCV genome and show that certain combinations of multiplexed-gRNAs provided a high degree of resistance to ChiLCV and restricted the generation of escape mutants. This approach could be broadly applicable to other geminiviruses.
Materials and methods
Plant materials and infectious virus construct
N. benthamiana, was used to assess the efficacy of multiplexed gRNA-based CRISPR-Cas9 modules to inhibit ChiLCV. An infectious clone of ChiLCV isolate (Accession number MK882926), generated by cloning a partial tandem repeat (PTR) of the viral genome. Briefly, first a complete viral genome was amplified by rolling circle amplification (RCA) using standard methodology followed by digestion of the RCA product using BamHI [31]. The full length viral genome (ChiLCV-BamHI) was cloned in pUC18 vector and sequenced. A BamHI -EcoRI digested partial genomic fragment (1.1 kb) containing the origin of replication was cloned into pCAMBIA2300 vector. Further the BamHI digested full length genome was ligated in tandem orientation into the BamHI -EcoRI digested partial fragment cloned in pCAMBIA2300 and thus the PTR clone was obtained. The PTR clone, when co-agroinfiltrated with a croton yellow vein mosaic betasatellite to different chilli genotypes, produced severe leaf curl symptoms (unpublished). In N. benthamiana, the infectious clone alone caused typical leaf curl and stunting symptoms, and the symptom severity increased many fold in the presence of the betasatellite (unpublished). This infectious clone was used in the study.
Selection of targeted regions and design of spacer sequences for editing the ChiLCV genome
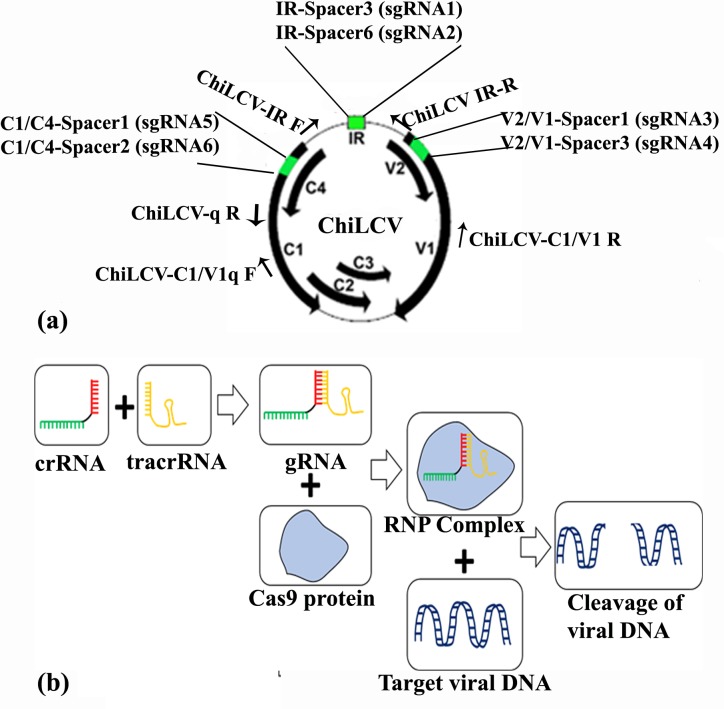
Three regions in the ChiLCV genome were selected for designing gRNAs. The three regions targeted are as follows: i. Intergenic region (IR) including the origin of replication, ii. Overlapping regions between V2 and V1 genes (referred to as V2/V1), and iii. Overlapping regions between C1 and C4 genes (C1/C4) (Fig 1A). The 20-nt spacer sequences were chosen based on the efficacy and specificity to the targeted region and the presence of an adjacent N-G-G sequence (PAM motif) (Table 1). All spacer sequences were analysed for possible off-targets in the N. benthamiana genome through a BLAST search. Spacer sequences with relatively higher GC content and no detectable off-targets were chosen.
Fig 1. Position of different primers and spacer sequence in the chilli leaf curl virus (ChiLCV) genome and plan for in vitro cleavage assay.
a) Three genomic regions (green color) of the ChiLCV were selected to identify the spacer, including i. Intergenic region (IR) with the origin of replication, ii. Overlapping regions between V2 and V1 genes (V2/V1) and iii. Overlapping regions between C1 and C4 genes (C1/C4); b) schematic representation of in vitro cleavage assay.
Table 1. Details of spacer sequences selected from chilli leaf curl virus (ChiLCV) genome and their final selection based on off-target analysis and GC%.
| Genomic region | Nucleotide coordinate and spanning length | Spacer name | Seq | PAM | Off Target analysis (10 top hit) | GC% | |||
|---|---|---|---|---|---|---|---|---|---|
| Position in the genome | Target match in NB genome | Seed seq match | PAM match | ||||||
| IR region with origin of replication | 1–145 nt, span: 145 nt and 2611–2762, span: 152 nt | IR-Spacer1 | CAATCGGTACTCAACAAACT | TGG | 2622–2641 | no | 9–20 match | No | 40 |
| IR-Spacer2 | AACAAACTTGGCTATGTAAT | CGG | 2634–2653 | no | 11–21 match | No | 30 | ||
| IR-Spacer3 | *GCCATCCGCACTAATATTAC | CGG | 2745–2762, 1–2 | no | 9–20 match | No | 45 | ||
| IR-Spacer4 | TGGCCGCGATTTTTTTACCG | TGG | 7–26 nt | no | 12–23 match | Yes | 50 | ||
| IR-Spacer5 | CTCATAGCTTAATTATTTCA | TGG | 81–100 | no | 12–23 match | No | 25 | ||
| IR-Spacer6 | *CATGGTCCCCCCTATAAACT | TGG | 99–118 | no | 12–21 match | No | 50 | ||
| Overlapping betwwen V2 and V1 ORFs (V2/V1) | 306 nt– 502 nt, span: 197 nt | V2/V1-Spacer1 | *CATTTCCACGCCCGCCTCGA | AGG | 332–351 | no | 12–20 match | No | 65 |
| V2/V1-Spacer2 | CCCCACTGTCCGCGTCACAA | AGG | 404–423 | no | 14–23 match | Yes | 65 | ||
| V2/V1-Spacer3 | *AGGCCAGAGCATGGGTGAAC | AGG | 424–443 | no | 12–21 match | No | 60 | ||
| V2/V1-Spacer4 | ACAGGAAGCCCAGGATGTAC | AGG | 454–473 | no | 10–20 match | No | 55 | ||
| Overlapping between C1 and C4 ORFs (C1/C4) | 2160–2459 nt, span: 300 nt | C1/C4-Spacer1 | *GGACCCTGAATTGATTGCCT | CGG | 2185–2204 | no | 10–21 match | No | 50 |
| C1/C4-Spacer2 | *TAGCTGATCTTCCATCGACT | TGG | 2241–2260 | no | 10–20 match | No | 45 | ||
| C1/C4-Spacer3 | CTTTTAGCTCCCTGAATGTT | CGG | 2324–2343 | no | 9–20 match | No | 40 | ||
| C1/C4-Spacer4 | GGTCGAAGAATCTGTTGTTT | TGG | 2379–2398 | no | 11–23 match | Yes | 40 | ||
| C1/C4-Spacer5 | ATTTACCTTCGAATTGTATG | AGG | 2409–2428 | no | 12–23 match | Yes | 30 | ||
| C1/C4-Spacer6 | TGTATGAGGACGTGGAGATG | AGG | 2423–2442 | no | 13–23 match | Yes | 50 | ||
*Spacer sequences finally selected
In vitro cleavage assay for determining the efficacy of gRNAs
To verify the efficacy of the gRNAs designed, the genomic region of ChiLCV containing all three target sites was amplified using primers ChiLCV-C1/V1q F and ChiLCV-C1/V1 R (Table 2). The locations of different primers in the ChiLCV genome are shown in Fig 1A. An in vitro cleavage assay of the viral target amplicon was performed using Alt-R® CRISPR-Cas9 system (Integrated DNA Technologies, Coralville, IA, USA) following the manufacturer’s protocol. Briefly, Alt-R CRISPR-Cas9 crRNAs were synthesized against all the selected spacer sequences designed from the ChiLCV genome. Equimolar amounts of the individual Alt-R CRISPR-Cas9 crRNA and universal Alt-R CRISPR-Cas9 tracrRNA were incubated for 5 min. at 95°C in a nuclease-free duplex buffer and the solution was allowed to cool down to room temperature to form the gRNA (Alt-R CRISPR-Cas9 crRNA:tracrRNA). The gRNA complex was then combined with Alt-R S/p Cas9 Nuclease 3NLS to form a ribonucleoprotein (RNP) complex (Fig 1B), which was then incubated with the viral target amplicon for 1 hr at 37°C. Post-cleavage fragments were analysed by agarose gel electrophoresis. The assay was performed with individual RNP complexes or with different duplex and triplex combinations.
Table 2. Details of primers used in the study.
| Purpose | Primer combinations and sequence (5’-3’) | Amplification condition* | Expected amplicon size (kb) | |
|---|---|---|---|---|
| Annealing temperature (0C) | Extension time (Sec) at 72°C | |||
| Amplification of all the target site for in vitro cleavage assay | ChiLCV-C1/V1q F (GGGCTAAGGTCGAGATGTCC)ChiLCV-C1/V1 R (CATCCATCCATATCTTCCCTAATAC) | 59 | 45 | 1.5 |
| Generating amplicon for T7E1 assay and sequencing | ChiLCV-C1/V1q F (GGGCTAAGGTCGAGATGTCC)ChiLCV IR-R (GTCGCTTGGACATAATTCTTAGC) | 61.2 | 37 |
1.2 |
|
ChiLCV-IR F (ATTGTATGAGGACGTGGAGATGAG) ChiLCV-C1/V1 R (CATCCATCCATATCTTCCCTAATAC) |
60.7 | 30 | 1.0 | |
| qPCR of virus quantification | ChiLCV-C1/V1q F (GGGCTAAGGTCGAGATGTCC)ChiLCV-q R (CCGGAGGAACTTGAAGAATGG) | 62 |
30 | 0.2 |
| qPCR of gRNA expression | ||||
| i. gRNA-1 | IR-spacer3-Top (GGTCAGCCATCCGCACTAATATTAC)gR-qR (GCACCGACTCGGTGCCAC) | 64 | 30 | 0.096 |
| ii. gRNA-2 | IR-spacer6-Top (GATTGCATGGTCCCCCCTATAAACT)gR-qR: (GCACCGACTCGGTGCCAC) | 64 | 30 | 0.096 |
| iii. gRNA-3 | V2/V1-Spacer1-Top (GGTCACATTTCCACGCCCGCCTCGA)gR-qR: (GCACCGACTCGGTGCCAC) | 64 | 30 | 0.096 |
| iv. gRNA-4 | V2/V1-Spacer3-Top (GATTGAGGCCAGAGCATGGGTGAAC)gR-qR: (GCACCGACTCGGTGCCAC) | 64 | 30 | 0.096 |
| v. gRNA-5 | C1/C4-spacer1-Top (GGTCAGGACCCTGAATTGATTGCCT)gR-qR: (GCACCGACTCGGTGCCAC) | 64 | 30 | 0.25 |
| vi. gRNA-6 | C1/C4-spacer2-Top (GATTGTAGCTGATCTTCCATCGACT)gR-qR: (GCACCGACTCGGTGCCAC) | 64 | 30 | 0.50 |
| qPCR for Cas9 | Cas9 qF (GGACCACTTGCTAGAGGAAACTCTC) Cas9 qR (GGAAGGTTCTTATCGAAGTTGGTCATTC) | 64 | 30 | 0.15 |
| PP2A gene primer of N. benthamiana(reference gene control on qPCR assay for gRNA and Cas9) | PP2A F (GACCCTGATGTTGATGTTCGCT)PP2A R (GAGGGATTTGAAGAGAGATTTC) | 59 | 30 | 0.2 |
| Actin gene primer of N. benthamiana(reference gene control on qPCR assay for ChiLCV) |
Actin-DNA-qF(AATGATCGGAATGGAAGCTG) Actin-DNA-qR: (TGGTACCACCACTGAGGACA) |
61.2 | 30 | 0.116 |
*other parameters of PCR are followed from standard methods. Phusion Taq polymerase (Thermo) was used.
Development of plants expressing multiplexed gRNA-Cas9 modules by using a Golden Gate and Gateway compatible vector toolkit
Plants expressing multiplexed gRNA-Cas9 constructs (duplex and triplex) were generated following the methodology and toolkit established by Lowder et al.[32]. The gRNAs and their complementary sequences were synthesized with the addition of linker sequences at the 5’ or 3’ ends (Table 3). Double-stranded spacer sequences with flanking linkers were produced by annealing the complementary strands of each spacer sequences. The process of developing multiplexed gRNA-Cas9 constructs is as follows: individual double-stranded spacer sequences with flanking linkers were cloned between either the U3 or U6 promoter and the gRNA scaffold sequence of one of the Golden Gate entry plasmids (pYPQ131B, pYPQ132A or pYPQ133B) to generate an individual gRNA clone. One gRNA under the U3 promoter (pYPQ131B derived) and another gRNA under the U6 promoter (pYPQ132A derived) were cloned in tandem into a Gateway entry vector (pYPQ142) via Golden Gate cloning to generate duplexed gRNA clones. Similarly, three Golden Gate entry clones were constructed by cloning the three gRNAs into their respective plasmids (pYPQ131B, pYPQ132A and pYPQ133B). Three gRNA cassettes were subsequently cloned in tandem into pYPQ143 to form a triplexed gRNA Gateway entry clone. A Gateway recombination reaction was performed using a gRNA cassette entry clone (pYPQ142 or pYPQ143), a plant codon-optimized Cas9 (under 35S promoter) entry clone (pYPQ150), and a plant expression Gateway destination vector (pEarleyGate100) to generate duplexed or triplexed gRNA-Cas9 modules. After every step, the resultant clones containing either gRNAs or gRNA-Cas9 expression cassettes were confirmed by sequencing.
Table 3. Sequence of double stranded spacer oligos along with linker and the corresponding Golden Gate entry vectors where they were cloned to generate gRNAs.
| Selected Spacers | Two strands of spacer oligo sequence with linker (5'-3')* | Entry vector (promoter) | Name of gRNA Constructs |
|---|---|---|---|
| IR-Spacer3 | IR-spacer3-Top:ggtcaGCCATCCGCACTAATATTAC | pYPQ131B (U3) | gRNA1 |
| IR-spacer3-Bot:aaacGTAATATTAGTGCGGATGGCt | |||
| IR-Spacer6 | IR-spacer6-Top: gattgCATGGTCCCCCCTATAAACT | pYPQ132A (U6) | gRNA2 |
| IR-spacer6-Bot: aaacAGTTTATAGGGGGGACCATGc | |||
| V2/V1-Spacer1 | V2/V1-Spacer1-Top: ggtcaCATTTCCACGCCCGCCTCGA | pYPQ131B (U3) | gRNA3 |
| V2/V1-Spacer1-Bot: aaacTCGAGGCGGGCGTGGAAATGt | |||
| V2/V1-Spacer3 | V2/V1-Spacer3-Top: gattgAGGCCAGAGCATGGGTGAAC | pYPQ132A (U6) | gRNA4 |
| V2/V1-Spacer3-Bot: aaacGTTCACCCATGCTCTGGCCTc | |||
| C1/C4-Spacer1 | C1/C4-spacer1-Top: ggtcaGGACCCTGAATTGATTGCCT | pYPQ131B (U3) and pYPQ133B (U3) | gRNA5 gRNA5-1 |
| C1/C4-spacer1-Bot: aaacAGGCAATCAATTCAGGGTCCt | |||
| C1/C4-Spacer2 | C1/C4-spacer2-Top: gattgTAGCTGATCTTCCATCGACT | pYPQ132A (U6) | gRNA6 |
| C1/C4-spacer2-Bot: aaacAGTCGATGGAAGATCAGCTAc |
*Spacer sequences (Top) and its complimentary sequences (Bot) were synthesized with addition of linker (underlined) as required for their cloning into GoldenGate entry vectors. The Top and Bot sequences were annealed to each other to develop double stranded spacer molecule for cloning. The vector has the promoter and gRNA scaffold. After cloning the spacer sequence along with scaffold form the functional gRNAs.
Plant inoculation
Multiplexed gRNA-Cas9 modules in the pEarleyGate100 vector and the infectious ChiLCV partial tandem dimer in the pCAMBIA2300 vector were individually transformed into Agrobacterium tumefaciens strain GV3103 by electroporation. Single colonies from the transformed cells were grown overnight in a selective medium. A fresh culture was prepared from the overnight grown culture by growing it for 3–4 h. The bacterial cells were collected and resuspended in infiltration buffer (10 mM MES [pH 5.7], 10 mM MgCl2, and 150 μ M acetosyringone) to obtain OD600 ~0.5. The culture was incubated at room temperature in the dark for 2–4 h and then used to infiltrate the leaves of 4–5 week-old N. benthamiana plants using a 1.0 mL needleless syringe. Agro-infiltration was performed on three plants for each treatment. Three leaves of each plant were co-inoculated simultaneously with one multiplex gRNA-Cas9 construct and ChiLCV infectious construct. However, to assure that the multiplexed gRNA-Cas9 construct cuts the replicating virus only and not the plasmid backbone of the ChiLCV construct, the inoculation was spatially separated in a way that the multiplexed gRNA-Cas9 module was inoculated onto the portion of lamina adjacent to the petiole, while the ChiLCV infectious construct was inoculated towards the apex of the lamina. It was assumed that after releasing from the plasmid backbone, when the ChiLCV starts replication and moves to the bottom portion of the leaf, it encounters the gRNA-Cas9. Inoculation of ChiLCV infectious construct with an empty gRNA-Cas9 vector backbone served as a positive control, while only the vector backbone inoculated plants served as mock negative controls. At 4 days post-inoculation (dpi), leaf tissues from the gRNA-Cas9 inoculated portion (lamina adjacent to petiole) was harvested and further molecular analysis were performed to determine the virus accumulation and the presence of targeted modifications of the viral sequences. Additionally, symptom development in the newly emerging leaves was recorded. Symptoms were categorized based on two parameters: curling of leaves and stunting of plants. Curling of leaf is graded into the following scale: no visible symptom: 0; flecking, vein thickening and vein twisting: 1; mild puckering and folding of leaves from margin: 2; cupping, rolling and size reduction of leaves: 3. Stunting of plant is compared to that of healthy plant and graded into following scale: no stunting: 0; stunting less than 20%: 1, stunting 20–50%: 2, stunting more than 50%: 3. Only virus inoculated CRISPR-Cas9 non-treated diseased plant will have the highest scale for both categories.
qPCR assay to detect viral accumulation
Genomic DNA was extracted with cetyl trimethyl ammonium bromide (CTAB) buffer from the gRNA-Cas9 inoculated portion of the N. benthamiana leaves at 4 dpi. To estimate the effect of genome editing on virus accumulation, quantitative PCR (qPCR) was performed using the SsoAdvanced Universal SYBR Green Supermix and the CFX96 Touch™ Real-Time PCR detection system (Bio-Rad, Hercules, CA, USA). The N. benthamiana Actin gene was used as a reference control. To quantify the ChiLCV from the systemic leaf of promising multiplexed-gRNA inoculated plants, qPCR was also conducted at 20 dpi. The primers used for the qPCR study are listed in Table 2. qPCR assay was performed in three replicates for all the samples.
Quantitative reverse transcriptase PCR (qRT-PCR) to analyse expression of gRNAs and Cas9
Total RNA was extracted from the gRNA-Cas9 inoculated portion of the N. benthamiana leaves at 4 dpi using Trizol reagent (Thermo Fisher Scientific). To detect the expression of gRNAs and Cas9 into the inoculated area, qRT-PCR was carried out (Table 1). Briefly, first strand cDNA was synthesized using the iScript™ Reverse Transcription Supermix (Bio-Rad). Quantitative RT-PCR was performed using the SsoAdvanced Universal SYBR Green Supermix (Bio-Rad). qRT-PCR was carried out with the spacer-specific primers and the gRNA scaffold primers (Table 2). The N. benthamiana PP2A gene was used as a reference control [33]. Relative gene expression was calculated using the ΔCT method. qRT-PCR for analyzing the expression of both gRNA and Cas9 was performed in three replicates for all the samples.
Rolling circle amplification
Virus accumulation was determined in the systemic leaves of promising multiplexed gRNA-Cas9 treated and ChiLCV inoculated plants. Total DNA was isolated 20 dpi and rolling circle amplification (RCA) was carried out using a TempliPhi 100 RCA Kit (GE Healthcare, USA) following the manufacturer’s protocol. DNA from the systemic, symptomatic leaves of mock vector treated, ChiLCV-inoculated plants served as positive control. Each RCA product was separately digested with two enzymes (BamHI and XbaI) with unique restriction sites, which were expected to generate full-length virus genome-sized fragment. Digestion products were resolved on 1% agarose gel.
T7EI assay for detecting mutations
To detect the mutation resulting from DSB repair through the NHEJ pathway, a T7E1 assay was performed as described previously [23]. T7EI assay detect on-target genome editing and estimate editing efficiency using T7 endonuclease I (T7EI), which can cleave a heteroduplex DNA having mutation in any strand. In the T7EI assay, CRISPR-induced mutant gene is amplified by PCR. The PCR products are denatured and reannealed to allow heteroduplex formation between wild-type DNA and CRISPR–mutated DNA. Mutations are then detected using T7EI, which recognizes and cleaves mismatched DNA heteroduplexes. T7EI assay results are analyzed by visualizing cleavage products and full-length amplicons by gel electrophoresis. In this study genomic DNA was isolated from ChiLCV-challenged plants treated either with gRNA-Cas9 or with mock vector (control) samples collected at 4 dpi and was used as the template for PCR using primers ChiLCV-C1/V1q F and ChiLCV-IR R (Table 1). Amplicons of the ChiLCV genomic fragment obtained from those samples were denatured, renatured, and treated with T7EI using the Alt-R™ Genome Editing Detection Kit (Integrated DNA Technologies) following the manufacturer’s protocol. Results were evaluated using agarose gel electrophoresis. A control heteroduplex DNA sample provided in the kit served as positive control for the assay.
TIDE analysis
The ChiLCV genomic portion was amplified from the harvested tissue using two sets of primers ChiLCV-C1/V1q F and ChiLCV IR-R and ChiLCV-IR F and ChiLCV-C1/V1 R using Phusion Taq polymerase (Thermo Fisher Scientific). Three amplicons for each samples were sequenced and the sequence was analysed through Tracking of Indels by Decomposition (TIDE) programme (https://tide.nki.nl/) as described previously [34]. TIDE analysis provides rapid and reliable assessment of genome editing efficiencies. It quantifies the rates of NHEJ-mediated repair in an edited sample by decomposing the sequence trace data. It identifies the predominant types of insertions and deletions (indels) in the DNA. Briefly, the virus sequences from gRNA-Cas9 treated and untreated plants were compared for detecting mutation in the region flaking the gRNA target site. All analyses were performed with a default setting.
Results
In vitro cleavage of ChiLCV DNA by the gRNA-Cas9 RNP complex
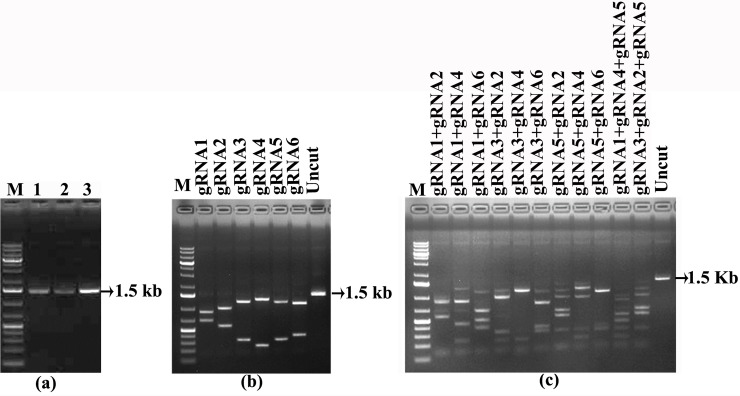
Initially six, four and six spacer sequences were identified based on the PAM sequence from IR, V2/V1 and C1/C4 regions of ChiLCV, respectively (Table 1). After off-target analysis and %GC calculation, a total of six spacer sequences, two each from the above three regions in the ChiLCV genome were selected for further analysis (Table 1). To ensure that these spacer sequences could cleave the ChiLCV genome, a 1.5 kb viral genomic fragment containing all six target spacer sequences was amplified from the N. benthamiana plants inoculated with an infectious clone of ChiLCV (Fig 2A). In vitro cleavage assay of the 1.5 kb template using the individual gRNA-Cas9 RNP complex showed efficient cleavage of the template, yielding bands of expected size (Fig 2B). As the main aim was to apply a multiplexed gRNA to cleave the viral genome, we used two or three RNP complexes at a time to see the cleavage products under in vitro conditions. The result showed that the multiple RNP complex could cleave the 1.5 kb viral genomic portion into many fragments (Fig 2C). This confirmed that all the spacer sequences that were used to create the RNP complexes could effectively disintegrate the viral genome either individually or in multiplexed modes.
Fig 2. In vitro cleavage assay.
a) Amplification of the target region from the ChiLCV (lane 1–3 indicates three replicates of same sample) b) In-vitro cleavage assay using individual gRNA-Cas9 ribonucleoprotein (RNP) complex. The 1.5-kb viral genomic fragment containing all six target spacer sequences was treated by six individual gRNA-Cas9 RNP complexes. c) In-vitro cleavage assay using multiplexed gRNA-Cas9 RNP complex. The 1.5-kb viral genomic fragment containing all six target spacer sequences was treated by duplex or triplex gRNA-Cas9 RNP complexes. Uncut: 1.5-kb viral genomic fragment without the RNP treatment.
Multiplexed gRNA-Cas9 complex vectors
In the first step of construct development, six individual gRNA clones were obtained after cloning the double-stranded spacer sequences into a Golden Gate entry vector (Table 3). These gRNAs target the viral genome in the following regions: gRNA1 and gRNA2: IR, gRNA3 and gRNA4: V2/V1 and gRNA5 and gRNA6: C1/C4 regions. In the second step, a total of nine duplex and two triplex gRNA expression clones were generated and used as Gateway entry clones (Table 4). In the third step, same numbers of multiplexed gRNA-Cas9 constructs were cloned into a plant expression vector. The final clone IDs are mentioned in Table 4.
Table 4. Duplex and triplex gRNA constructs developed and their impact on disease phenotype.
| Sl No. | Multiplex level | gRNA combination | Final clone ID after joining with Cas9 | Target region in ChiLCV genome | Disease phenotype grade* |
|---|---|---|---|---|---|
| 1 | Duplex | gRNA1+ gRNA2 | gRNA1+2 | IR | Leaf curl– 1, Stunting—2 |
| 2 | Duplex | gRNA1+ gRNA4 | gRNA1+4 | IR and V2-V1 overlap | Leaf curl– 1, Stunting—2 |
| 3 | Duplex | gRNA1+ gRNA6 | gRNA1+6 | IR and C1-C4 overlap | Leaf curl– 2, Stunting—2 |
| 4 | Duplex | gRNA3+ gRNA2 | gRNA3+2 | IR and V2-V1 overlap | Leaf curl– 2, Stunting—2 |
| 5 | Duplex | gRNA3+gRNA4 | gRNA3+4 | V2-V1 overlap | Leaf curl– 2, Stunting—2 |
| 6 | Duplex | gRNA3+gRNA6 | gRNA3+6 | V2-V1 overlap and C1-C4 overlap | Leaf curl– 2, Stunting—2 |
| 7 | Duplex | gRNA5+gRNA2 | gRNA5+2 | IR and C1-C4 overlap | Leaf Curl– 0, Stunting—1 |
| 8 | Duplex | gRNA5+gRNA4 | gRNA5+4 | V2-V1 overlap and | Leaf curl– 0, Stunting– 0 |
| 9 | Duplex | gRNA5+gRNA6 | gRNA5+6 | C1-C4 overlap | Leaf curl– 1, Stunting– 2 |
| 1 | Triplex | gRNA1+gRNA4+gRNA5-1 | gRNA1+4+5 | IR, V2-V1 overlap and C1-C4 overlap | Leaf curl– 2, Stunting—2 |
| 1 | Triplex | gRNA1+gRNA4+gRNA5-1 | gRNA3+2+5 | IR, V2-V1 overlap and C1-C4 overlap | Leaf curl– 2, Stunting—2 |
*In mock vector infiltrated ChiLCV inoculated plant (positive control) both the leaf curl and stunting grades were 3, while only mock vector inoculated plant (negative control) both these grades were 0.
Efficacy of transiently expressed duplex and triplex gRNA-Cas9 complex
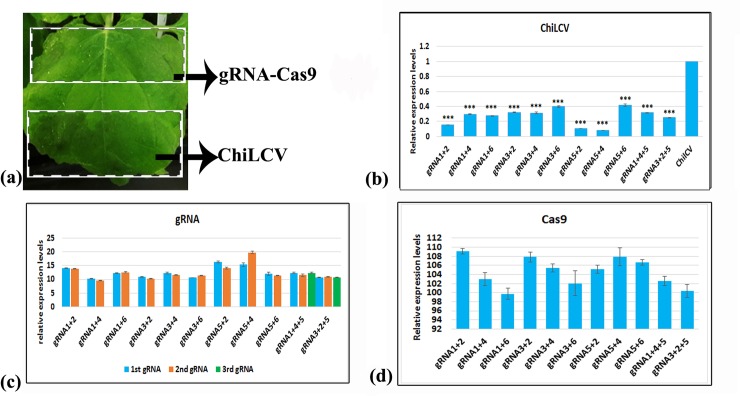
The efficacy of multiplexed gRNA-Cas9 constructs to cleave the viral DNA under in planta condition was determined by measuring the expression level of ChiLCV in the gRNA-Cas9 infiltrated portion by qPCR (Fig 3A). Compared to the mock (vector) inoculated plant, virus accumulation was reduced significantly in plants treated with different combinations of duplex/triplex gRNA-Cas9 (Fig 3B). Among the different gRNA combinations, gRNA5+4 resulted in the lowest level of ChiLCV accumulation followed by gRNA5+2 and gRNA1+2. The triplex gRNA-inoculated plants did not show any added advantage with respect to reduction of ChiLCV accumulation. All combinations had similar levels of gRNA and Cas9 expression (Fig 3C and 3D). In the systemic leaves of all the multiplexed gRNA-Cas9 inoculated plants, appearance of leaf curl symptoms was delayed by five to ten days compared to that in the mock-inoculated plants. The ratings for both leaf curl and stunting phenotypes were lower in all multiplexed gRNA-Cas9 combinations (Table 4). The most promising result with respect to symptom attenuation was observed in plants inoculated with gRNA5+4, where plants did not have any leaf curl symptom (grade 0) nor the plant growth was affected significantly (grade 0) (Fig 4A). The next promising combinations were gRNA5+2 (leaf curl grade:0; stunting grade:1) and gRNA1+2 (leaf curl grade:1; stunting grade:2) (Fig 4B and 4C).
Fig 3. In planta expression assay.
a) Inoculation positions of gRNA-Cas9 construct and ChiLCV, b) qPCR assay to understand reduction of viral replication, c) qRT-PCR assay for gRNA expression analysis, d) qRT-PCR assay for Cas9 expression analysis. For qPCR and qRT-PCR, three replicates were used for each data point. The error bar denotes SEM. Stars indicate significant difference (student’s unpaired t-test) of ChiLCV expression levels between gRNA-Cas9 treated and non-treated plants (*P< 0.05, **P< 0.01, ***P< 0.001).
Fig 4. Disease phenotype of N. benthamiana plants and viral load assay in distal leaves.
Test plants were challenged by ChiLCV and infiltrated with three promising gRNA-Cas9 constructs, respectively. Plants treated with mock vector served as negative control while plants treated with mock vector followed by challenged by ChiLCV served as positive control. a) Similar to mock plants, ChiLCV inoculated N. benthamiana treated with gRNA5+4 did not show any visible leaf curl (grade:0) or stunting (grade:0) symptoms. b) Similar to mock plants, ChiLCV inoculated N. benthamiana treated with gRNA5+2 did not show visible leaf curl symptom (grade:0), however, a mild stunting (grade:1) phenotype was observed. c) Similar to mock plants, ChiLCV inoculated N. benthamiana treated with gRNA 1+2 did not show typical leaf curl symptom. However, a mild vein thickening (grade:1) and moderate stunting (grade:2) phenotype was observed. All experiments had three replicates, and they all gave similar results. d) Rolling circle amplification (RCA)-generated ChiLCV genome length fragment (2.7 kb) from systemic leaves of three promising multiplexed gRNA-Cas9 treated and mock vector inoculated plants. RCA product of ChiLCV digested with BamHI (Lane 1–4) and XbaI (Lane 5–8). Lane 1,5: gRNA1+2 treated, Lane 2,6: gRNA5+2 treated, Lane 3,7: gRNA5+4 treated and Lane 4,8: mock vector treated and ChiLCV inoculated plants e) qPCR assay for analysing viral load from the systemic leaf of plant inoculated with these three gRNA combinations. Three replicates were conducted for each data point. The error bar denotes SEM. Stars indicate significant difference (student’s unpaired t-test) of ChiLCV expression levels between gRNA-Cas9 treated and non-treated plants (*P< 0.05, **P< 0.01, ***P< 0.001).
To estimate the viral DNA accumulation in systemic leaves, RCA analysis was performed. The RCA product, when digested with BamHI and XbaI, two unique restriction sites present in ChiLCV genome, yielded 2.7 kb viral genome length fragments in all the samples. However, concentration of the virus was distinguishably lower in gRNA-Cas9 treated plants (Fig 4D) than that from the mock vector-treated plant. To determine the relative quantification of the viral load in the systemic leaves, qPCR assay of ChiLCV was done, which showed that, in all these three gRNA combinations, there was a significant decrease in virus accumulation (Fig 4E).
Multiplex gRNA-Cas9 complexes inhibit escape mutant formation
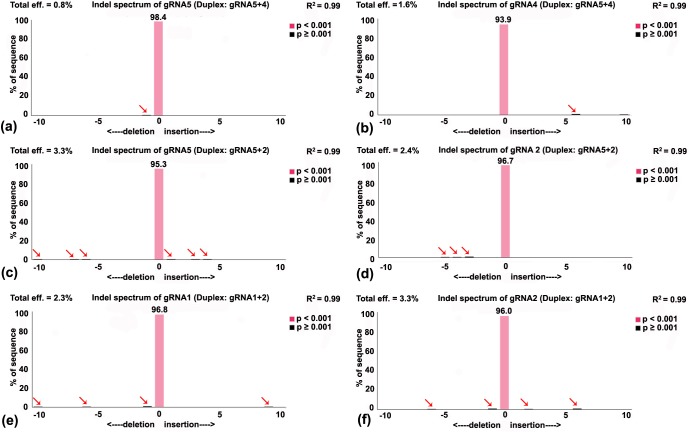
To evaluate the genome editing, the three promising gRNA-Cas9 combinations mentioned earlier (gRNA 5+4, gRNA 5+2, gRNA 1+2) were tested by T7E1 assay and sequencing. T7E1 assay generated a distinct homoduplex band (wild type virus population) from all three combinations of gRNA-Cas9 but failed to yield recognisable escape mutant fragments even after repeated attempts (Fig 5). The control heteroduplex DNA provided in the kit yielded the expected bands resulting from cleavage of heteroduplex by T7EI enzyme, indicating the effectiveness of the assay. To confirm the T7E1 result, sequencing of the PCR-amplified viral genomic fragment containing the gRNA target revealed presence of only the wild type sequences. To examine the probability of the presence of escape mutants in the PCR amplified product, sequences were analysed using TIDE, which showed that the frequency of the mutation surrounding the PAM sequence is insignificant (Fig 6), further confirming the non-existence of escape mutants in those multiplexed gRNA-Cas9 combinations.
Fig 5. Detection of genome editing by T7E1 assay.
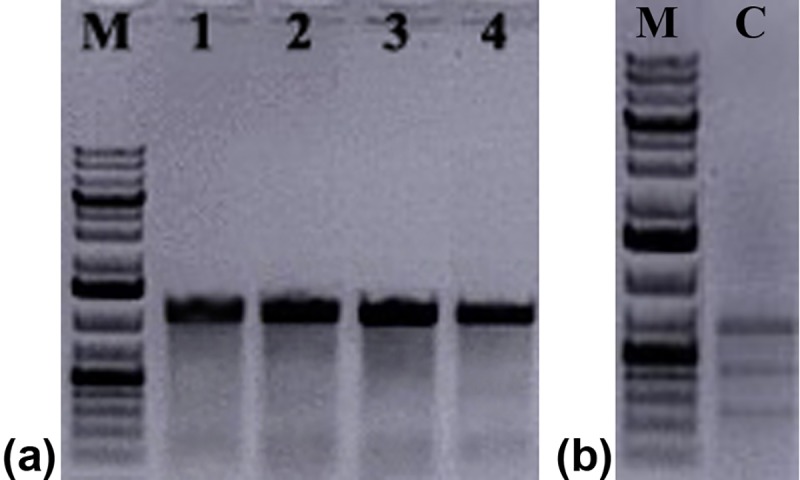
a) T7EI assay from gRNA treated ChiLCV challenged plant along with ChiLCV inoculated plant. Lane M, GeneRuler 1kb plus DNA ladder. Samples in lane 1 through 4 were all amplified using primers ChiLCV-C1/V1q F &ChiLCV-IRR and then treated with T7E1. Lane 1, amplicon from mock vector treated ChiLCV inoculated N. benthamiana. Lane 2, amplicon from gRNA5+4-Cas9 construct treated ChiLCV challenged plants. Lane 3, amplicon from gRNA5+2-Cas9 construct treated ChiLCV challenged plants. Lane 4, amplicon from gRNA1+2-Cas9 construct treated ChiLCV challenged plants. b) Control heteroduplex DNA (C) from the kit showed cleavage of the control heteroduplex by the T7EI.
Fig 6. Indel spectrum analysis through TIDE programme.
Analyses showed probability of presence of NHEJ generated mutated sequences in upstream or downstream to the PAM motifs in ChiLCV genome after treating with three promising duplex gRNA-Cas9 constructs. Three replicates of the viral genomic sequences from individual plant were identical and hence analysis was done with one sample. Indel spectra generated surrounding the cleavage site of (a) gRNA5 (duplex: gRNA5+4), (b) gRNA4 (duplex: gRNA5+4), (c) gRNA5 (duplex: gRNA5+2), (d) gRNA2 (duplex: gRNA5+2), (e) gRNA1 (duplex: gRNA1+2), (f) gRNA2 (duplex: gRNA1+2). The analysis showed in all combinations no significant escape mutants were present. Arrow indicated insignificant occurrence of mutated sequences surrounding the PAM motif of each gRNA targeting region of ChiLCV.
Discussion
CRISPR-Cas9 is the most widely adopted system for genome editing and has been used successfully in microbes, plants and animals [35]. Recent studies, using geminiviruses, showed that targeting the viral genome at a single site is not a robust strategy to limit viral replication as the escape mutants are generated during the process [24,28]. One of the most interesting features of the CRISPR-Cas9 system is its flexibility to assemble multiple gRNA modules for targeting several genes simultaneously [36,37]. However, there were no known reports of targeting geminiviruses using such multiplex approaches. Baltes et al. [25] showed that when two individual gRNA modules targeting the two regions of same viral genome were inoculated together, they reduce the viral load more effectively than the individual gRNA. In another attempt by Ali et al. [23], a polycistronic tRNA–gRNA based approach was used for simultaneous delivery two gRNAs in a NB-Cas9OE plants. Here, we developed a single cassette containing both Cas9 and combinations of two or three gRNAs and examined if such multiplexed gRNAs specifically targeting more than one gene/region would efficiently reduce the disease development and lower the viral load. It should be easier to use multiplexed gRNAs-Cas9 constructs directly to generate transgenic plants for stable editing.
We first identified spacer sequences from three important regions of the viral genome with an aim to simultaneously target more than a single gene of the virus. These regions are critical for the replication, movement and RNA silencing suppression ability of the virus. Editing of these regions resulted in interfering with the establishment of the infection, reducing viral load and generation of any functional escape mutants. Initially, we demonstrated that synthesized gRNAs, either individually or in combinations, were capable of mediating targeted cleavage of the ChiLCV genome in vitro. This finding lead us to believe that similar multiple cleavages must have occurred in the viral genome under our experimental set up in planta. Since it was already shown by Ali et al. [24] and Mehta et al. [28] that individual gRNAs could result in escape mutants, and the potential implications for this phenomenon, we did not repeat such individual gRNA strategy, and we focused on expressing multiplxed gRNAs to suppress the generation of escape mutants. Using the multiplex gRNA tool-kit developed by Lowder et al. [32], duplexed and triplexed gRNA-Cas9 constructs were developed and were tested for their ability to reduce the accumulation of ChiLCV genomic DNA as well as leaf curl symptoms in N. benthamiana. qPCR and qRT-PCR analyses showed that simultaneous targeting of the different coding and non-coding sequences of the ChiLCV resulted in a significant reduction in viral titre (at least a 10-fold reduction for two gRNA combinations). This reduction accompanied an attenuation of viral symptoms in the systemic leaves. It would be interesting to examine the extent of reduction of viral load at later days post inoculation.
Earlier studies showed that the most promising gRNAs were the ones targeting the IR of geminiviruses. In this study, we also obtained severe reduction of viral load with gRNAs targeting the IR region alone (gRNA1+2) or IR region in combination with C1/C4 (gRNA1+6, gRNA5+2) and V2/V1 (gRNA1+4, gRNA3+2). However, the highest level of virus inhibition was observed when gRNAs simultaneously targeting the V2/V1 and C1/C4 were used (gRNA5+4). This construct targeted coat protein and Rep protein of ChiLCV simultaneously leading to maximal deletion of the viral genome, thereby blocking the possibility of generating any escape mutant. To detect any such escape mutants, we analyzed the result of the T7E1 assay through agarose gel electrophoresis. An alternative approach would be use of capillary electrophoresis.
TIDE analysis is a simple process to detect genome editing efficiencies directly from the sequence traces of PCR product thus avoiding tedious cloning and sequencing of all the clones. TIDE can only detect overall indel frequencies, but not nucleotide substitutions or specifically designed indels and thus it can generate a priori information on whether the desired mutation(s) which have been introduced are significant or not [38]. Using the sequence traces of three most promising gRNA combinations, TIDE analysis was carried out and it did not detect any significant mutation, indicating after joining of DSB, the mutant virus, if any, was not able to survive. It remains to be determined whether the cloned product from such mixed PCR amplicons will yield any mutant viral sequences. Thus, multiplexed gRNAs provided two benefits: (i) drastic reduction in generation of escape mutants, and (ii) reduced viral load, leading to significant attenuation of viral symptoms. In some cases, such as gRNA5+4, and gRNA5+2), plants were free of any leaf curling.
In order to trigger durable virus resistance using genome editing technology, one would require constitutive and permanent expression of the ribonucleoprotein complex in the host [28]. While this requires producing of stably transformed plants, adopting a transient means, we first set out to know if the multiplex modules could be efficiently applied to inhibit begomoviruses. Similar concern for using multiplexing in a transient assay was also highlighted by Rybicki [29]. Thus, we established the proof of concept by avoiding transgenic approach and by using transient expression to rapidly screen various combinations of gRNAs. We have demonstrated that multiplexed gRNA-Cas9 constructs efficiently inhibited the virus. It is highly likely that this approach will be even more effective, robust, and durable in transgenic plants expressing a higher amount of Cas9.
Conclusion
In conclusion, we provide here the first proof of concept of multiplexing of gRNA-Cas9 modules to inhibit a begomovirus infection through a transient assay in the model plant N. benthamiana. We showed such multiplexed CRISPR-Cas9 strategy could reduce virus accumulation significantly without escape mutant formation, which also resulted in significant decrease in disease symptoms. We also identified potential multiplexed-gRNA-Cas9 combinations that were highly effective in imparting resistance. A similar multiplexing strategy could be applicable to other ssDNA viruses of plants.
Supporting information
(PDF)
Acknowledgments
AR is grateful to Department of Biotechnology, Government of India and Indo-U.S. Science & Technology Forum (IUSSTF), New Delhi for the GETin Overseas Fellowship; and the Indian Council of Agricultural Research, New Delhi, India for the necessary permission.
Data Availability
Sequence of the virus files are available from the NCBI database (accession number MK882926). All relevant data are available at https://figshare.com/articles/PLOS_ONE_data_set/9936986.
Funding Statement
PPNS No. 0783. College of Agricultural, Human and Natural Resource Sciences, Agricultural Research Center (Hatch Project # WNPO 0545), Washington State University, Pullman, WA 99164-6430, USA. Anirban Roy was supported by Indo-U.S. Genome Engineering/Editing Technology Initiative (GETin) Overseas Fellowship, funded by Department of Biotechnology, Government of India and Indo-U.S. Science & Technology Forum (IUSSTF), New Delhi, India. The ChiLCV infectious clones were developed under ICAR-Consortium Research Platform Project on Vaccine and Diagnostics.
References
- 1.Mansoor S, Briddon RW, Zafar Y, Stanley J. Geminivirus disease complexes: an emerging threat. Trends Plant Sci. 2003; 8: 128–134. 10.1016/S1360-1385(03)00007-4 [DOI] [PubMed] [Google Scholar]
- 2.Varma A, Mandal B, Singh MK. Globale and spread of whitefly (Bemisiatabaci) transmitted Geminiviruses. In: Thompson W, Editor. The Whitefly, Bemisiatabaci (Homoptera: Aleyrodidae) Interaction with Geminivirus-Infected Host Plants; 2011. pp. 205–292. [Google Scholar]
- 3.Malathi VG, Renukadevi P, Chakraborty S, Biswas KK, Roy A, Sivalingam PN, et al. Begomoviruses and Their Satellites Occurring in India: Distribution, Diversity and Pathogenesis. In: Mandal B, Rao G, Baranwal V, Jain R, edtiors. A Century of Plant Virology in India; 2017. pp. 75–177. [Google Scholar]
- 4.Zehra SB, Ahmad A, Sharma A, Sofi S, Lateef A, Bashir Z, et al. Chilli Leaf Curl Virus an Emerging Threat to Chilli in India. Int. J. Pure App. Biosci. 2017; 5: 404–414. [Google Scholar]
- 5.Senanayake DMJB, Varma A, Mandal B. Virus–vector Relationships, Host Range, Detection and Sequence Comparison of Chilli leaf curl virus Associated with an Epidemic of Leaf Curl Disease of Chilli in Jodhpur, India. J. Phytopathol. 2012; 160: 146–155. [Google Scholar]
- 6.Zhou X. Advances in understanding begomovirus satellites. Annu. Rev. Phytopathol. 2013; 51: 357–381. 10.1146/annurev-phyto-082712-102234 [DOI] [PubMed] [Google Scholar]
- 7.Lazarowitz S. Geminiviruses: Genome structure andgene function. Crit. Rev. Plant Sci. 1992; 11: 327–349. [Google Scholar]
- 8.Hanley-Bowdoin L, Settlage SB, Orozco BM, Nagar S, Robertson D. Geminiviruses: models for plant DNA replication, transcription, and cell cycle regulation. Crit. Rev. Biochem. Mol. Biol. 2000; 35: 105–140. [PubMed] [Google Scholar]
- 9.Fondong VN. Geminivirus protein structure and function. Mol. Plant Pathol. 2013; 14: 635–649. 10.1111/mpp.12032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lapidot M, Legg JP, Wintermantel WM, Polston JE. Management of whitefly-transmitted viruses in open-field production systems. Adv. Virus Res. 2014; 90: 147–206. 10.1016/B978-0-12-801246-8.00003-2 [DOI] [PubMed] [Google Scholar]
- 11.Vu TV, Choudhury NR, Mukherjee SK. Transgenic tomato plants expressing artificial microRNAs for silencing pre-coat and coat proteins of a begomovirus ToLCNDV, shows tolerance to virus infection. Virus Res. 2013; 172: 35–45. 10.1016/j.virusres.2012.12.008 [DOI] [PubMed] [Google Scholar]
- 12.Singh A, Taneja J, Dasgupta I, Mukherjee SK. Development of plants resistant to tomato geminiviruses using artificial transacting small interfering RNA. Mol. Plant Pathol. 2015; 16: 724–734. 10.1111/mpp.12229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rojas MR, Macedo MA, Maliano MR, Soto-Aguilar M, Souza JO, Briddon RW, et al. World Management of Geminiviruses. Annu. Rev.Phytopathol. 2018; 56: 637–677. 10.1146/annurev-phyto-080615-100327 [DOI] [PubMed] [Google Scholar]
- 14.Gill U, Scott JW, Shekasteband R, Ogundiwin E, Schuit C, Francis DM, et al. Ty-6, a major begomovirus resistance gene on chromosome 10, is effective against Tomato yellow leaf curl virus and Tomato mottle virus. Theor. Appl. Genet. 2019; 132: 1543–1554. 10.1007/s00122-019-03298-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bonfim K, Faria JC, Nogueira EOPL, Mendes EA, Arago FJL. RNAi mediated resistance to Bean Golden Mosaic Virus in Genetically engineered common bean (Phaseolus vulguris). Mol. Plant Microbe Interact. 2007; 20: 717–726. 10.1094/MPMI-20-6-0717 [DOI] [PubMed] [Google Scholar]
- 16.Voytas DF, Gao C. Precision genome engineering and agriculture: opportunities and regulatory challenges. PLoS Biol. 2014; 12: e1001877 10.1371/journal.pbio.1001877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Barrangou R, Fremaux C, Deveau H, Richards M, Boyaval P, Moineau S, et al. CRISPR provides acquired resistance against viruses in prokaryotes. Science 2007; 315: 1709–1712. 10.1126/science.1138140 [DOI] [PubMed] [Google Scholar]
- 18.Wright AV, Nunez JK, Doudna JA. Biology and applications of CRISPR systems: harnessing nature’s toolbox for genome engineering. Cell 2016; 164: 29–44. 10.1016/j.cell.2015.12.035 [DOI] [PubMed] [Google Scholar]
- 19.Aouida M, Eid A, Ali Z, Cradick T, Lee C, Deshmukh H, et al. Efficient fdCas9 synthetic endonuclease with improved specificity for precise genome engineering. PLoS One 2015; 10: e0133373 10.1371/journal.pone.0133373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Piatek A, Mahfouz MM. Targeted genome regulation via synthetic programmable transcriptional regulators. Crit. Rev.Biotechnol. 2016; 37: 429–440. 10.3109/07388551.2016.1165180 [DOI] [PubMed] [Google Scholar]
- 21.Chaparro-Garcia A, Kamoun S, Nekrasov V. Boosting plant immunity with CRISPR/Cas. Genome Biol. 2015; 16: 254–257. 10.1186/s13059-015-0829-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hadidi A, Flores R, Candresse T, Barba M. Next-Generation Sequencing and Genome Editing in Plant Virology. Front. Microbiol. 2016; 7: 1325 10.3389/fmicb.2016.01325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ali Z, Abulfaraj A, Idris A, Ali S, Tashkandi M, Mahfouz MM. CRISPR/Cas9-mediated viral interference in plants. Genome Biol. 2015; 16: 238 10.1186/s13059-015-0799-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ali Z, Ali S, Tashkandi M, Zaidi SS, Mahfouz MM. CRISPR/Cas9-mediated immunity to geminiviruses: differential interference and evasion. Sci. Rep. 2016; 6: 26912 10.1038/srep26912 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Baltes NJ, Hummel AW, Konecna E, Cegan R, Bruns AN, Bisaro DM, et al. Conferring resistance to geminiviruses with the CRISPR–Cas prokaryotic immune system. Nature Plants 2015; 1: 15145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ji X, Zhang H, Zhang Y, Wang Y, Gao C. Establishing a CRISPR–Cas-like immune system conferring DNA virus resistance in plants. Nature Plants 2015; 1: 15144 10.1038/nplants.2015.144 [DOI] [PubMed] [Google Scholar]
- 27.Liu HJ, Soyars CL, Li JH, Fei QL, He GJ, Fei Q, et al. CRISPR/Cas9-mediated resistance to cauliflower mosaic virus. Plant Direct. 2018; 2: e00047 10.1002/pld3.47 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mehta D, Stürchler A, Anjanappa RB, Zaidi SS, Hirsch-Hoffmann M, Gruissem W, et al. Linking CRISPR-Cas9 interference in cassava to the evolution of editing-resistant geminiviruses. Genome Biol. 2019; 20: 80 10.1186/s13059-019-1678-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rybicki EP. CRISPR–Cas9 strikes out in cassava. Nature Biotechnol. 2019; 37: 727–728. [DOI] [PubMed] [Google Scholar]
- 30.Zaidi SSA, Tashkandi M, Mahfouz MM. Engineering molecular immunity against plant viruses. Prog. Mol. Biol. Transl. 2017; 149: 167–186. [DOI] [PubMed] [Google Scholar]
- 31.Shilpi S, Kumar A, Biswas S, Roy A, Mandal B. A recombinant Tobacco curly shoot virus causes leaf curl disease in tomato in a north-eastern state of India and has potentiality to trans-replicate a non-cognate betasatellite. Virus Genes 2015; 50: 87–96. 10.1007/s11262-014-1141-1 [DOI] [PubMed] [Google Scholar]
- 32.Lowder LG, Zhang D, Baltes NJ, Paul JW, Tang X, Zheng X, et al. A CRISPR/Cas9 toolbox for multiplexed plant genome editing and transcriptional regulation. Plant Physiol. 2015; 169: 971–985. 10.1104/pp.15.00636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Liu D, Shi L, Han C, Yu J, Li D, Zhang Y. Validation of reference genes for gene expression studies in virus-infected Nicotiana benthamiana using quantitative real-time PCR. PLoS One 2012; 7: e46451 10.1371/journal.pone.0046451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Brinkman EK, Chen T, Amendola M, van Steensel B. Easy quantitative assessment of genome editing by sequence trace decomposition. Nucleic Acids Rec. 2014; 42: e168 10.1093/nar/gku936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hsu PD, Lander ES, Zhang F. Development and applications of CRISPR-Cas9 for genome engineering. Cell 2014; 157: 1262–1278. 10.1016/j.cell.2014.05.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Xing HL, Dong L, Wang ZP, Zhang HY, Han CY, Liu B, et al. A CRISPR/Cas9 tool kit for multiplex genome editing in plants. BMC PlantBiol. 2014; 14: 327–339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Xie K, Minkenberg B, Yang Y. Boosting CRISPR/Cas9 multiplex editing capability with the endogenoust RNA-processing system. Proc. Natl. Acad. Sci. U.S.A. 2015; 112: 3570–3575. 10.1073/pnas.1420294112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Brinkman EK, van Steensel B. Rapid Quantitative Evaluation of CRISPR Genome Editing by TIDE and TIDER In: Luo Y. (eds) CRISPR Gene Editing. Methods in Molecular Biology, vol 1961 Humana Press, New York, NY: 2019. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(PDF)
Data Availability Statement
Sequence of the virus files are available from the NCBI database (accession number MK882926). All relevant data are available at https://figshare.com/articles/PLOS_ONE_data_set/9936986.