Abstract
Objective
Activation of the type 1 interferon (IFN1) pathway is a prominent feature of dermatomyositis (DM) muscle and may play a role in the pathogenesis of this disease. However, the relevance of the IFN1 pathway in patients with other types of myositis such as the antisynthetase syndrome (AS), immune-mediated necrotizing myopathy (IMNM), and inclusion body myositis (IBM) is largely unknown. Moreover, the activation of the type 2 interferon (IFN2) pathway has not been comprehensively explored in myositis. In this cross-sectional study, our objective was to determine whether IFN1 and IFN2 pathways are differentially activated in different types of myositis by performing RNA sequencing on muscle biopsy samples from 119 patients with DM, IMNM, AS, or IBM and on 20 normal muscle biopsies.
Methods
The expression of IFN1- and IFN2-inducible genes was compared between the different groups.
Results
The expression of IFN1-inducible genes was high in DM, moderate in AS, and low in IMNM and IBM. In contrast, the expression of IFN2-inducible genes was high in DM, IBM, and AS but low in IMNM. The expression of IFN-inducible genes correlated with the expression of genes associated with inflammation and muscle regeneration. Of note, ISG15 expression levels alone performed as well as composite scores relying on multiple genes to monitor activation of the IFN1 pathway in myositis muscle biopsies.
Conclusions
IFN1 and IFN2 pathways are differentially activated in different forms of myositis. This observation may have therapeutic implications because immunosuppressive medications may preferentially target each of these pathways.
Myositis is a heterogeneous family of systemic autoimmune diseases that includes the following groups: dermatomyositis (DM), immune-mediated necrotizing myopathy (IMNM), the antisynthetase syndrome (AS), and sporadic inclusion body myositis (IBM).1,2 Myositis-specific autoantibodies (MSAs) help define additional myositis subgroups with unique clinical phenotypes.1 For example, anti–transcriptional intermediary factor (TIF) 1γ and anti-melanoma differentiation-associated protein (MDA) 5 autoantibodies are each found in patients with DM who have myositis and rash. However, whereas anti-TIF1γ –positive patients have a high risk of cancer and a low risk of lung involvement, anti-MDA5–positive patients have a relatively low risk of cancer and a high risk of lung involvement. Additional MSAs associated with distinct clinical phenotypes include those found in patients with DM (anti-Mi2 and anti–nuclear matrix protein [NXP] 2), IMNM (anti–signal recognition particle [SRP] and anti–3-hydroxy-3-methylglutaryl-CoA reductase [HMGCR]), and AS (anti-Jo1, anti-PL7, and anti-PL12).
The pathogenic mechanisms underlying the different types and subtypes of myositis are incompletely understood. However, the type 1 interferon (IFN) pathway has emerged as potentially relevant to DM pathogenesis.3 Specifically, a marked overexpression of IFN1-inducible genes has been demonstrated in the muscle,3 peripheral blood,4,5 and skin6 of patients with DM. Moreover, the expression levels of IFN1-inducible genes correlate with indicators of DM disease activity.4,5
Three different families of ligands may activate the IFN pathway by binding to cell surface receptors: type 1 IFNs (IFN1; including IFN-α and IFN-β), type 2 IFNs (IFN2; i.e., IFN-γ), and type 3 IFNs (IFN3; i.e., IFN-λ).7 These proteins bind to their corresponding surface receptors, which, via the Janus kinase (JAK)/signal transducer and activator of transcription (STAT) signaling pathway, stimulate the expression of IFN-inducible genes.8 Although there is considerable overlap between the sets of genes induced by the different types of IFN,9,10 a handful of genes are specifically stimulated by either IFN1 (e.g., ISG15,11,12 IFI6,13 and MX114) or IFN2 (e.g., GBP1, GBP2,10,15 and PSMB816).
Prior studies have established the preferential activation of the IFN1 pathway in DM muscle.3 However, activation of the IFN1 pathway has not been compared between patients with DM with different DM subtypes defined by the presence of different DM autoantibodies. Furthermore, the IFN1 pathway activation was found to be relatively low in IBM but has not been systematically explored in AS or IMNM.3,17,18 Similarly, although IFN2 pathway activation has been implicated in IBM muscle,19,20 activation of IFN2 pathways in muscle biopsies from patients with IMNM, AS, and IBM has not been systematically analyzed. In this study, we assessed activation of both IFN1 and IFN2 pathways by analyzing gene expression data from RNA sequencing performed on a large number of muscle biopsies from patients with DM, IMNM, AS, and IBM, as well as normal comparator tissue.
Methods
Patients, samples, and autoantibody testing
All the available muscle biopsies from patients enrolled in investigational review board–approved longitudinal cohorts of the NIH (Bethesda, MD), the Johns Hopkins Myositis Center (Baltimore, MD), the Clinic Hospital (Barcelona), and the Vall d'Hebron Hospital (Barcelona) were included in the study if the patients fulfilled IBM criteria according to Lloyd et al.21 or had one of the following MSAs: anti-NXP2, -Mi2, -TIF1γ, -MDA5, -HMGCR, -SRP, or -Jo1. Autoantibody testing was performed as previously described for anti-HMGCR22 and by line blot for the others (EUROLINE Myositis Profile 4). Patients were classified as having AS if they had autoantibodies against Jo-1; as being in the DM group if they had autoantibodies recognizing Mi2, NXP2, TIF1γ, or MDA5; and as being in the IMNM group if they tested positive for anti-SRP or anti-HMGCR autoantibodies. Creatine kinase (CK) levels and strength assessments obtained closest to the time of muscle biopsy were used to assess the clinical activity of the disease. Muscle strength was evaluated by the examining physician using the Medical Research Council scale. This scale was transformed to the Kendall 0-to-10 scale; the right- and left-side measurements for arm abduction and hip flexion strength were combined, and the average was used for calculations (possible range 0–10) as previously described.23 Normal muscle biopsies were obtained from the Johns Hopkins Neuromuscular Pathology Laboratory (n = 10) and the Skeletal Muscle Biobank of the University of Kentucky (n = 10).
Standard protocol approvals and patient consents
This study was approved by the Institutional Review Boards at participating institutions, and written informed consent was obtained from each participant.
RNA sequencing
RNA sequencing was performed as previously described.24 Briefly, RNA was prepared with TRIzol (Thermo Fisher Scientific). Libraries were prepared with the NeoPrep system according to the TruSeqM Stranded mRNA Library Prep protocol (Illumina, San Diego, CA) and sequenced with the Illumina HiSeq 2500 or 3000. Reads were aligned with STAR version 2.525; the abundance of each gene was quantified with StringTie version 1.3.326; and the differential gene expression was performed with DESeq2 version 1.20.0.27 The Benjamini-Hochberg correction was used to adjust for multiple comparisons, and a corrected value of p (q value) ≤0.05 was considered statistically significant.
IFN genes and pathways
IFN pathway genes were collected from the Reactome biorepository (reactome.org/). General IFN-related genes and genes from the IFN1 and IFN2 pathways were merged into a single list. The 13 genes included in the previously proposed IFN score in myositis were also added to the list.3 The expression of the genes of this list was analyzed in the different autoantibody and clinical myositis subsets.
Data analysis
Gene expression (fragments per kilobase of transcript per million mapped reads [FPKM]) values were log-transformed (logFPKM: log2[FPKM + 1]) for visualization purposes with the Python programming language and the Numpy, Pandas, and Seaborn packages. Correlation among continuous variables was measured with the Spearman ρ.
Data availability
Any anonymized data not published within the article will be shared by request from any qualified investigator.
Results
Ranking IFN-inducible gene expression in myositis muscle biopsies
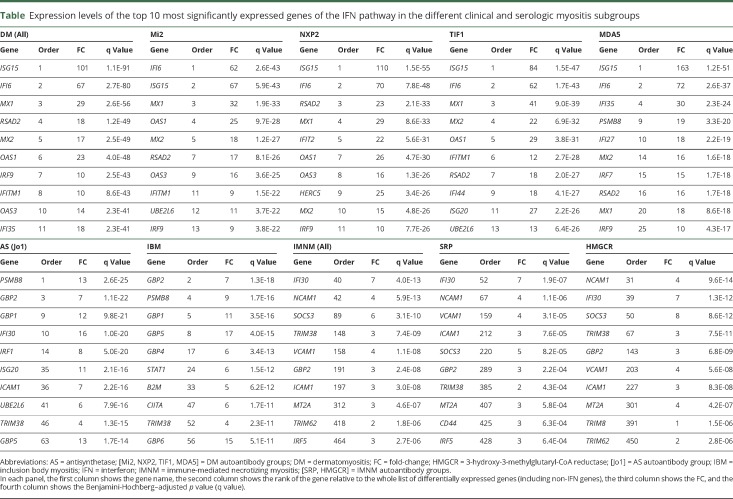
Muscle biopsy specimens were available from 119 myositis patients, including 39 with DM (11 anti-Mi2–, 12 anti-NXP2–, 11 anti-TIF1γ–, and 5 anti-MDA5–positive), 49 with IMNM (9 anti-SRP– and 40 anti-HMGCR–positive), 18 with anti-Jo1–positive AS, and 13 with IBM. Twenty normal muscle biopsy specimens were used as comparators. Expression levels of all genes were determined by RNA sequencing. The expression level of each gene from each major type of myositis (i.e., DM, IMNM, AS, and IBM) and each autoantibody group (i.e., anti-Mi2, -NXP2, -TIF1γ, -MDA5, -SRP, and -HMGCR) was compared to the expression level of the same gene in the comparator group. Differentially expressed genes were rank ordered by the degree of significance according to the adjusted p value. From among the complete list of differentially expressed genes, IFN-inducible genes were identified; the top 10 upregulated IFN-inducible genes for each group are listed in the table.
Table.
Expression levels of the top 10 most significantly expressed genes of the IFN pathway in the different clinical and serologic myositis subgroups
Expression levels of IFN1-inducible genes
The most significantly upregulated IFN-inducible genes in DM muscle biopsies were ISG15, IFI6, MX1, RSAD2, MX2, OAS1, IRF9, IFITM1, OAS3, and IFI35 (table), all of which are preferentially induced by IFN1 (IFN-α/β signaling of reactome.org/).11–14 Among all differentially expressed genes in DM (not just IFN-induced genes), these 10 IFN1-inducible genes were also among the most significantly upregulated (with all of them in the top 12 overall differentially expressed genes) (table).
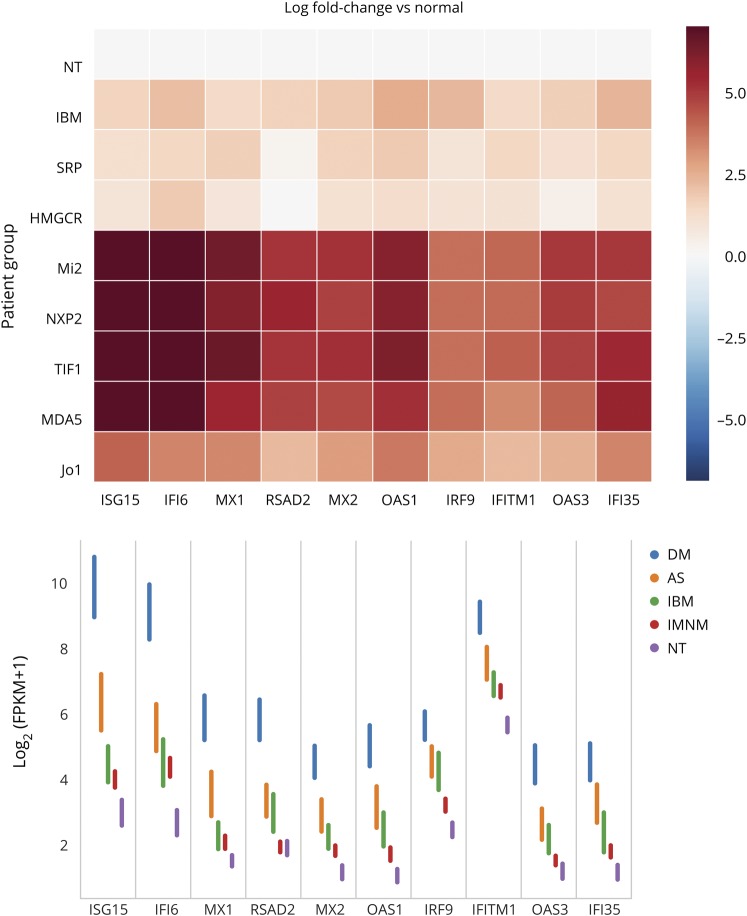
The overexpression of IFN1-inducible genes was not restricted to DM muscle biopsies (figure 1). However, the magnitude of this increase was markedly different among the different myositis types. Specifically, IFN1-inducible genes were expressed at markedly elevated levels in DM, at moderately increased levels in AS, and at minimally increased levels in IBM and IMNM (figure 1). Using ISG15 expression as an illustrative example, we found a 101-fold increase in DM (q value 1.1 × 10−91), an 8.7-fold increase in AS (q value 1.8 × 10−13), a 2.4-fold increase in IBM (q value 0.01), and a 1.8-fold increase in IMNM (q value 0.05) compared to comparator muscle biopsies (figure 1). In DM, ISG15 expression was 11 times higher than in AS (q value 5.3 × 10−27), 42 times higher than in IBM (q value 1.6 × 10−48), and 56 times higher than in IMNM (q value 9.8 × 10−109). Likewise, ISG15 expression in AS was higher than in IBM and IMNM by 4 and 4.8 times, respectively (q values 0.001 and 3.8 × 10−11).
Figure 1. Expression of IFN1-inducible genes in myositis muscle biopsies.
(A) Relative and (B) raw (95% confidence interval) expression levels of the type 1 interferon (IFN1)–inducible genes among the different clinical and serologic groups. AS = antisynthetase syndrome; DM = dermatomyositis; FPKM = fragments per kilobase of transcript per million mapped reads; IBM = inclusion body myositis; IMNM = immune-mediated necrotizing myositis; [Jo1] = AS autoantibody group; [Mi2, NXP2, TIF1, MDA5] = DM autoantibody groups; NT = normal biopsies; [SRP, HMGCR] = IMNM autoantibody groups.
We next analyzed the expression levels of IFN1-inducible genes among autoantibody subgroups. Interestingly, ISG15 and IFI6 were the most significantly upregulated genes in all DM autoantibody groups (i.e., anti-Mi2, anti-NXP2, anti-TIF1γ, and anti-MDA5) (table). In each DM autoantibody subgroup, these 2 genes were upregulated by at least 60-fold compared to healthy comparators (all q values <1 × 10−44) with no significant differences between the DM subgroups. Within IMNM, the expression of IFN1-inducible genes in those with anti-SRP autoantibodies was not significantly different compared to those with anti-HMGCR autoantibodies.
Expression levels of IFN2-inducible genes
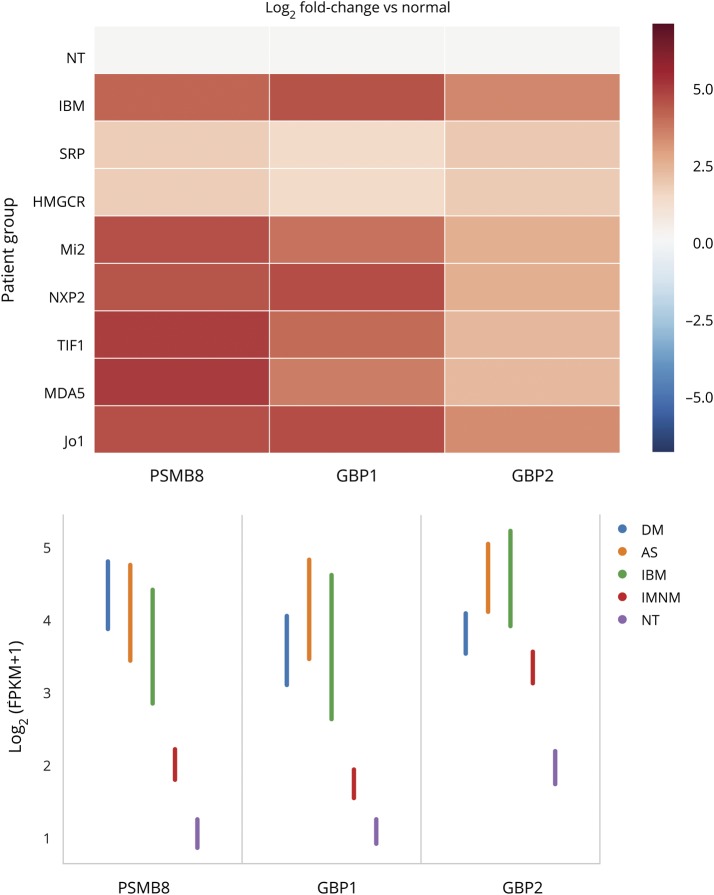
The IFN2-specific genes GBP1, GBP2, and PSMB8 were the 3 most significantly upregulated IFN-inducible genes in both AS and IBM. In addition, in muscle biopsies from both patients with AS and those with IBM, these 3 IFN2-inducible genes were within the top 10 most upregulated genes overall (table).
Compared to comparators, the expression of IFN2-inducible genes was increased by 7- to 14-fold in IBM, AS, and DM biopsies (all q values <1 × 10−15) (table and figure 2). There were no significant differences between AS or IBM and DM except that GBP2 had slightly higher expression levels in IBM (fold-change 1.7, q value 0.01) and AS (fold-change 1.8, q value 0.02) compared to DM. In contrast, the magnitude of IFN2-inducible gene overexpression in IMNM compared to comparators was much lower (PSMB8 fold-change 2.5, q value 7.6 × 10−5). Compared to IMNM, IFN2-inducible genes were expressed at higher levels in DM (fold-change 5.6, q value 9.5 × 10−25), AS (fold-change 5.2, q value 2.1 × 10−12), and IBM (fold-change 3.7, q value 9.2 × 10−7). There were no significant differences in the expression of IFN2-inducible genes between the different autoantibody subgroups within IMNM or DM.
Figure 2. Expression of IFN2-inducible genes in myositis muscle biopsies.
(A) Relative and (B) raw (95% confidence interval) expression levels of type 2 interferon (IFN2)–inducible genes among the different clinical and serologic groups. AS = antisynthetase syndrome; DM = dermatomyositis; FPKM = fragments per kilobase of transcript per million mapped reads; IBM = inclusion body myositis; IMNM = immune-mediated necrotizing myositis; [Jo1] = AS autoantibody group; [Mi2, NXP2, TIF1, MDA5] = DM autoantibody groups; NT = normal biopsies; [SRP, HMGCR] = IMNM autoantibody groups.
Interestingly, the IFN2-inducible gene IFI30 was 1 of the 2 most significantly upregulated IFN genes in both anti-SRP– and anti-HMGCR–positive patients with IMNM. Compared to normal biopsies, this gene showed a 7-fold increase in IMNM (q value 4 × 10−13), a 16-fold-increase in DM (q value 5.7 × 10−32), a 15.8-fold-increase in AS (q value 1 × 10−20), and a 7-fold-increase in IBM (q value 2.1 × 10−9) (table). Apart from IFI30 gene expression, the relative magnitude of IFN-related genes among all differentially expressed genes in IMNM was modest compared to other types of myositis. In fact, the first-ranked IFN-inducible gene in IMNM was ranked 40th in the list of all differentially expressed genes. In contrast, the first-ranked IFN-inducible gene was also first among all differentially expressed genes in DM and AS and the second among all differentially expressed genes in IBM (table).
Expression levels of genes associated with inflammation and muscle regeneration
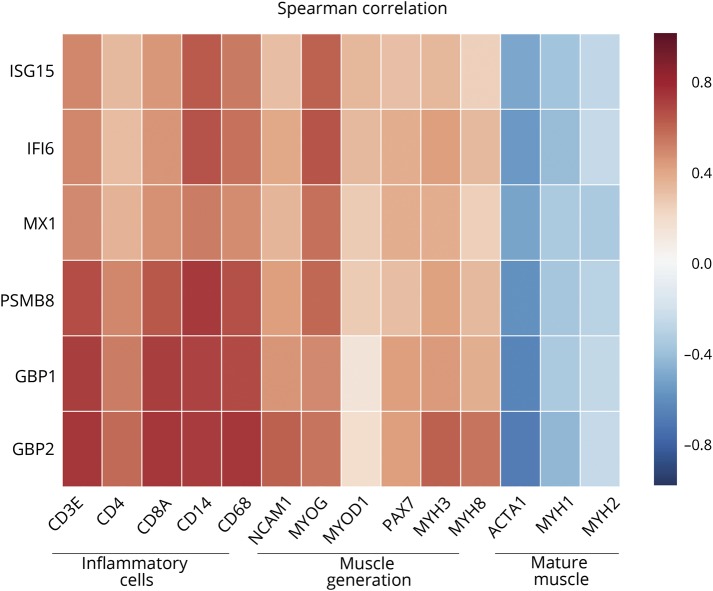
In each of the clinical and autoantibody subgroups studied, the expression of both IFN1- and IFN2-inducible genes was positively correlated with the expression of genes associated with inflammatory cells (T-cells [CD3E, CD4, CD8A] and macrophages [CD14, CD68]) and genes associated with muscle regeneration (NCAM1, MYOG, MYOD1, PAX7, MYH3, and MYH8) (all q values <0.05) (figure 3). Conversely, IFN-inducible genes were inversely correlated with mature-muscle structural proteins (ACTA1, MYH1, and MYH2) (all q values <0.05).
Figure 3. Correlation of IFN-inducible gene expression with expression of inflammatory cell and muscle regeneration genes.
Correlation of type 1 and type 2 interferon (IFN)–inducible genes with the expression of genes related to T cells (CD3E, CD4, and CD8A), macrophages (CD14 and CD68), muscle regeneration (NCAM1, MYOG, MYOD1, PAX7), and adult muscle structural proteins (ACTA1, MYH1, MYH2).
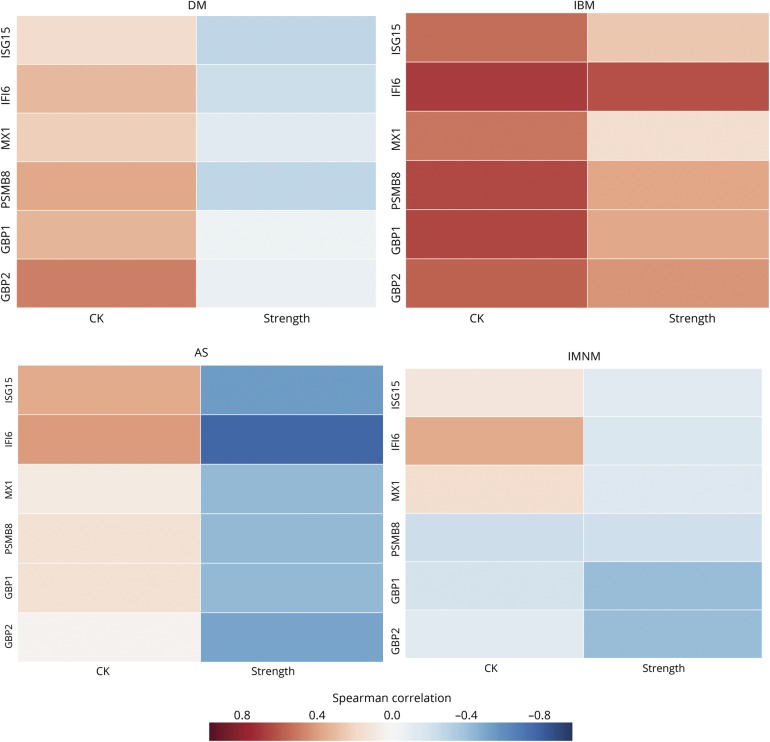
Strength measurements and CK levels obtained near the time of the muscle biopsy were available from 62 of the patients from Johns Hopkins (17 with DM, with 6 AS, 12 with IBM, and 27 with IMNM). Although there was a trend for patients with DM, AS, and IMNM with higher levels of IFN-inducible genes to have higher CK levels and decreased strength, this was not statistically significant (figure 4). However, patients with IBM with higher levels of IFN-inducible genes had significantly higher CK levels (all p ≤ 0.05) and a nonsignificant trend toward being stronger than those with lower levels of IFN-inducible genes. Because patients with IBM often have relatively preserved muscle strength early in the course of the disease, we hypothesized that IFN-inducible gene expression might also be highest early during the course of the disease. Indeed, we found that patients with IBM with a shorter interval between onset of symptoms and muscle biopsy had higher expression levels of IFN-inducible genes (data not shown).
Figure 4. Correlation of type 1 and type 2 interferon-inducible genes with the CK and strength in different types of myositis.
AS = antisynthetase syndrome; CK = creatine kinase; DM = dermatomyositis; IBM = inclusion body myositis; IMNM = immune-mediated necrotizing myositis.
ISG15 gene expression compared to composite IFN scores to quantify the IFN signature
Several gene scoring systems have been proposed to measure the activation of the IFN pathway in myositis28 and other autoimmune diseases.29 Particularly, a score combining 13 IFN1-inducible genes has been used to study the relationship between IFN1-inducible gene expression and disease activity in blood of patients with DM and polymyositis.28 We tried to test the utility of this score compared with simpler alternatives in myositis muscle biopsies.
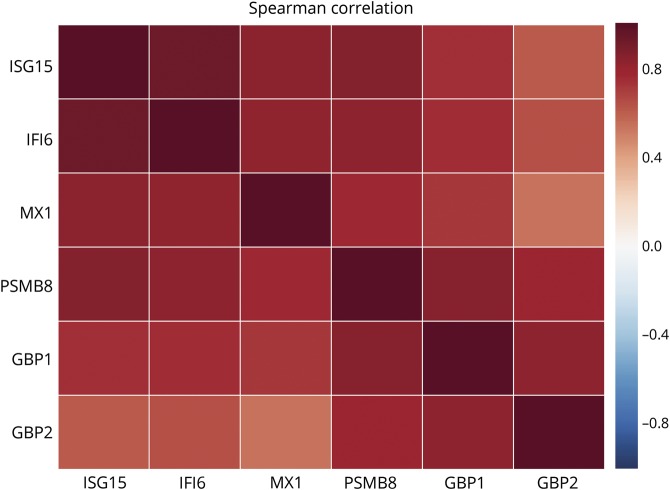
First, we analyzed the correlations between the expression levels of the different IFN-inducible genes in all of the muscle biopsies included in the study. This revealed a high correlation between expression levels of each IFN-inducible gene with all the others (figure 5). Second, because ISG15 was, overall, the most highly expressed IFN1-inducible gene, we correlated the raw expression levels of ISG15 with the previously proposed IFN1 score. This analysis revealed an almost perfect correlation between ISG15 expression levels alone and the 13-gene composite IFN1 score (Spearman ρ = 0.94, p = 1.5 × 10−64, figure 6A), suggesting that it may be unnecessary to use a more complex scoring system to measure IFN1 pathway activation levels in myositis muscle.
Figure 5. Spearman correlation of the different type 1 and type 2 interferon-inducible genes in all the biopsies included in the study.
Figure 6. ISG15 or PSMB8 expression vs composite IFN1-inducible gene scores.
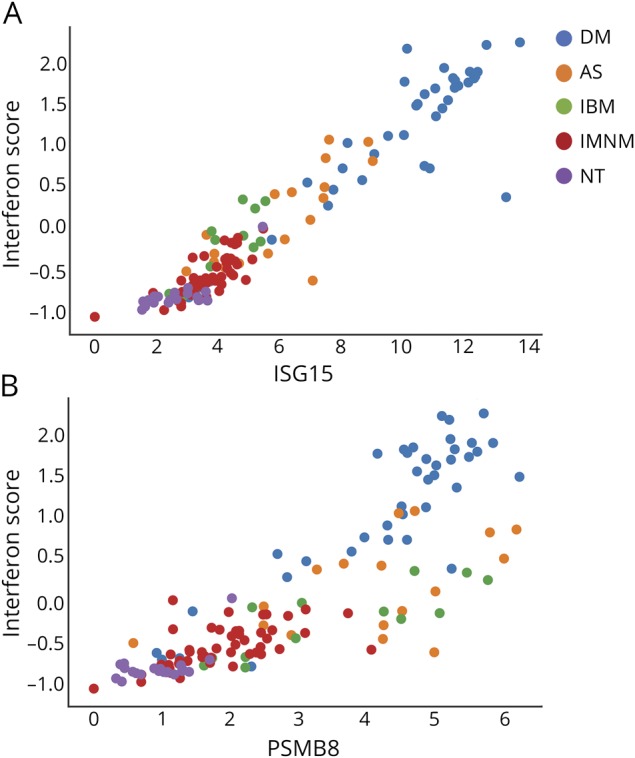
Correlation of the expression level (log2[FKPM+1]) of (A) ISG15 and (B) PSMB8 with the previously proposed 13-gene type 1 interferon (IFN1) score. AS = antisynthetase syndrome; DM = dermatomyositis; FPKM = fragments per kilobase of transcript per million mapped reads; IBM = inclusion body myositis; IMNM = immune-mediated necrotizing myositis; NT = normal biopsies.
The expression levels of the IFN2-inducible genes PSMB8, GBP1, and GBP2 were highly correlated with each other (figure 5). However, the association of these IFN2-inducible genes with the 13-gene IFN1 gene score was restricted to DM. For example, PSMB8 expression levels correlated well with the composite IFN1 gene score in patients with DM but not in patients with AS or IBM (figure 6B). This suggests that IFN1-inducible gene activation may correlate with IFN2 activation in DM but not in AS or IBM.
Discussion
In this study, using RNA sequencing data from a large number of myositis and comparator muscle biopsies, we have established that the IFN1 pathway is activated not only in patients with DM, as previously described,3–6 but also in patients with AS, IMNM, and IBM. Quantitatively, the IFN1 pathway was most upregulated in DM, with intermediate activation of the pathway in AS and lower levels of activation in IBM and IMNM. We also used RNA sequencing data to study activation of the IFN2 pathway, demonstrating robust activation in AS, IBM, and DM but not in IMNM. We were also able to show that activation of the IFN pathway was associated with increased expression of inflammatory cell and muscle regeneration genes. The correlation between this activation and muscle weakness and CK levels, however, did not reach statistical significance.
Interestingly, different collections of IFN-inducible genes were most prominently upregulated in the different groups. For example, the IFN1 genes ISG15, IFI6, and MX1 were the most upregulated IFN-inducible genes in DM. In contrast, IFI30, NCAM1, and SOCS3 were the most upregulated IFN1-inducible genes in patients with IMNM. Of note, the IFN2 genes PSMB8, GBP2, and GBP1 were the most upregulated IFN-inducible genes in both patients with AS and patients with IBM, underscoring the prominence of the IFN2 pathway in these 2 diseases.
It is well established that patients with DM with different myositis autoantibodies have unique clinical manifestations. In fact, there are differences in muscle biopsy features between patients with DM with different autoantibodies.30 For example, half of the muscle biopsies from anti-Mi2–positive patients with DM include examples of lymphocytes surrounding and invading healthy muscle fibers; this histopathologic feature was never seen in patients with DM with anti-NXP2 autoantibodies. Despite these histopathologic differences, the IFN gene signature was remarkably similar among patients with DM with different myositis autoantibodies. Indeed, ISG15 and IFI6 were the top 2 IFN-inducible genes in each of the serologically defined DM subgroups, and MX1 and MX2 were present among the top 10 IFN-inducible genes in each DM subgroup. These findings suggest that, at least with regard to activation of IFN pathways in the muscle, the different autoantibody subgroups of DM are more alike than different. Similarly, in patients with IMNM with either anti-SRP or -HMGCR autoantibodies, IFI30, NCAM1, VCAM1, ICAM1, SOC3, GBP2, and MT2A were among the top 10 IFN-inducible genes. We did not have a sufficient number of biopsies from patients with anti-PL7, anti-PL12, or other non-Jo1 antisynthetase autoantibodies to determine whether these serologic subgroups of the AS share a similar IFN gene signature pattern.
Some investigators have shown that immunostaining muscle biopsies for specific IFN-inducible proteins can be used to distinguish between different types of myositis. For example, DM but not AS muscle biopsies stain positive for MxA (MX1)31 or RIG-I (DDX58),32 both IFN1-inducible genes. Our RNA sequencing data, which show higher expression levels of these genes in DM than in AS (MX1 fold-change 4.7 and RIG-1 fold-change 3.3, both q values <5 × 10−9), are consistent with this observation. In addition, ISG15 overexpression is an established feature in muscle biopsies from patients with DM and perifascicular atrophy.17 Accordingly, we found a marked preferential overexpression of ISG15 in patients with DM (ISG15 fold-change compared to comparator biopsies 101, q value 1.1 × 10−91).
We also found that ISG15 expression levels alone can be used to reliably quantify the activation of the IFN1 pathway in myositis muscle biopsies. In fact, measuring ISG15 levels was equivalent to a composite score derived from measuring expression levels of 13 different IFN1-inducible genes, which is concordant with previous data showing the marked specificity of ISG15 muscle transcript measurements for DM with perifascicular atrophy.17 We also noted that both ISG15 expression levels and the previously proposed composite IFN1 scores were associated with activation of the IFN2 pathway in DM but not in IBM or AS.
This study has several limitations. For example, some less common autoantibody groups (e.g., non–anti-Jo1 AS) could not be included due to an insufficient number of biopsies. In addition, we had relevant CK and strength information only for muscle biopsies obtained at Johns Hopkins, which may have limited our ability to show significant associations between IFN pathway activation and markers of clinical disease activity such as strength and CK levels.
This study demonstrates that DM muscle biopsies are characterized by high levels of both IFN1- and IFN2-inducible genes. In contrast, biopsies from patients with AS and IBM reveal gene expression patterns consistent with prominent IFN2 activation. Finally, RNA sequencing analysis reveals that IMNM biopsies show relatively low activation of the IFN pathway. These findings are consistent with recent case series suggesting the efficacy of JAK/STAT inhibitors in patients with DM.33–37 They also suggest that these agents may be effective in patients with AS or IBM. However, the relatively modest activation of IFN pathways in IMNM does not provide compelling evidence to support the use of JAK/STAT inhibitors in this patient population.
Acknowledgment
The authors acknowledge Dr. Gustavo Gutierrez-Cruz, Dr. Stefania Dell'Orso, and Faiza Naz from the National Institute of Arthritis and Musculoskeletal and Skin Disease sequencing facility for all technical collaboration in RNA sequencing library construction and sequencing. The University of Kentucky Center for Muscle Biology provided control human muscle samples.
Glossary
- AS
antisynthetase syndrome
- CK
creatine kinase
- DM
dermatomyositis
- FPKM
fragments per kilobase of transcript per million mapped reads
- HMGCR
3-hydroxy-3-methylglutaryl-CoA reductase
- IBM
inclusion body myositis
- IFN
interferon
- IMNM
immune-mediated necrotizing myopathy
- JAK
Janus kinase
- MDA
melanoma differentiation-associated protein
- MSA
myositis-specific autoantibodies
- NXP
nuclear matrix protein
- SRP
signal recognition particle
- STAT
signal transducer and activator of transcription
- TIF
transcriptional intermediary factor
Appendix. Authors
Study funding
This research was supported in part by the Intramural Research Program of the National Institute of Arthritis and Musculoskeletal and Skin Diseases of the NIH. The Johns Hopkins Myositis Center Research Database and Dr. Christopher-Stine are supported by the Huayi and Siuling Zhang Discovery Fund. Dr. Pinal-Fernandez’s research was supported by a fellowship from the Myositis Association.
Disclosure
The authors report no disclosures relevant to the manuscript. Go to Neurology.org/N for full disclosures.
References
- 1.Selva-O'Callaghan A, Pinal-Fernandez I, Trallero-Araguas E, Milisenda JC, Grau-Junyent JM, Mammen AL. Classification and management of adult inflammatory myopathies. Lancet Neurol 2018;17:816–828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mariampillai K, Granger B, Amelin D, et al. Development of a new classification system for idiopathic inflammatory myopathies based on clinical manifestations and myositis-specific autoantibodies. JAMA Neurol 2018;75:1528–1537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Greenberg SA, Pinkus JL, Pinkus GS, et al. Interferon-alpha/beta-mediated innate immune mechanisms in dermatomyositis. Ann Neurol 2005;57:664–678. [DOI] [PubMed] [Google Scholar]
- 4.Walsh RJ, Kong SW, Yao Y, et al. Type I interferon-inducible gene expression in blood is present and reflects disease activity in dermatomyositis and polymyositis. Arthritis Rheum 2007;56:3784–3792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Baechler EC, Bauer JW, Slattery CA, et al. An interferon signature in the peripheral blood of dermatomyositis patients is associated with disease activity. Mol Med 2007;13:59–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wong D, Kea B, Pesich R, et al. Interferon and biologic signatures in dermatomyositis skin: specificity and heterogeneity across diseases. PLoS One 2012;7:e29161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ivashkiv LB, Donlin LT. Regulation of type I interferon responses. Nat Rev Immunol 2014;14:36–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.O'Shea JJ, Schwartz DM, Villarino AV, Gadina M, McInnes IB, Laurence A. The JAK-STAT pathway: impact on human disease and therapeutic intervention. Annu Rev Med 2015;66:311–328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhou Z, Hamming OJ, Ank N, Paludan SR, Nielsen AL, Hartmann R. Type III interferon (IFN) induces a type I IFN-like response in a restricted subset of cells through signaling pathways involving both the Jak-STAT pathway and the mitogen-activated protein kinases. J Virol 2007;81:7749–7758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hall JC, Casciola-Rosen L, Berger AE, et al. Precise probes of type II interferon activity define the origin of interferon signatures in target tissues in rheumatic diseases. Proc Natl Acad Sci USA 2012;109:17609–17614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhang D, Zhang DE. Interferon-stimulated gene 15 and the protein ISGylation system. J Interferon Cytokine Res 2011;31:119–130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.D'Cunha J, Ramanujam S, Wagner RJ, Witt PL, Knight E Jr, Borden EC. In vitro and in vivo secretion of human ISG15, an IFN-induced immunomodulatory cytokine. J Immunol 1996;157:4100–4108. [PubMed] [Google Scholar]
- 13.Kelly JM, Porter AC, Chernajovsky Y, Gilbert CS, Stark GR, Kerr IM. Characterization of a human gene inducible by alpha- and beta-interferons and its expression in mouse cells. EMBO J 1986;5:1601–1606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Holzinger D, Jorns C, Stertz S, et al. Induction of MxA gene expression by influenza A virus requires type I or type III interferon signaling. J Virol 2007;81:7776–7785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hall JC, Baer AN, Shah AA, et al. Molecular subsetting of interferon pathways in Sjogren's syndrome. Arthritis Rheumatol 2015;67:2437–2446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hisamatsu H, Shimbara N, Saito Y, et al. Newly identified pair of proteasomal subunits regulated reciprocally by interferon gamma. J Exp Med 1996;183:1807–1816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Salajegheh M, Kong SW, Pinkus JL, et al. Interferon-stimulated gene 15 (ISG15) conjugates proteins in dermatomyositis muscle with perifascicular atrophy. Ann Neurol 2010;67:53–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Greenberg SA, Sanoudou D, Haslett JN, et al. Molecular profiles of inflammatory myopathies. Neurology 2002;59:1170–1182. [DOI] [PubMed] [Google Scholar]
- 19.Ivanidze J, Hoffmann R, Lochmuller H, Engel AG, Hohlfeld R, Dornmair K. Inclusion body myositis: laser microdissection reveals differential up-regulation of IFN-gamma signaling cascade in attacked versus nonattacked myofibers. Am J Pathol 2011;179:1347–1359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Allenbach Y, Chaara W, Rosenzwajg M, et al. Th1 response and systemic treg deficiency in inclusion body myositis. PLoS One 2014;9:e88788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lloyd TE, Mammen AL, Amato AA, Weiss MD, Needham M, Greenberg SA. Evaluation and construction of diagnostic criteria for inclusion body myositis. Neurology 2014;83:426–433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mammen AL, Chung T, Christopher-Stine L, et al. Autoantibodies against 3-hydroxy-3-methylglutaryl-coenzyme A reductase in patients with statin-associated autoimmune myopathy. Arthritis Rheum 2011;63:713–721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rider LG, Werth VP, Huber AM, et al. Measures of adult and juvenile dermatomyositis, polymyositis, and inclusion body myositis: Physician and Patient/Parent Global Activity, Manual Muscle Testing (MMT), Health Assessment Questionnaire (HAQ)/Childhood Health Assessment Questionnaire (C-HAQ), Childhood Myositis Assessment Scale (CMAS), Myositis Disease Activity Assessment Tool (MDAAT), Disease Activity Score (DAS), Short Form 36 (SF-36), Child Health Questionnaire (CHQ), Physician Global Damage, Myositis Damage Index (MDI), Quantitative Muscle Testing (QMT), Myositis Functional Index-2 (FI-2), Myositis Activities Profile (MAP), Inclusion Body Myositis Functional Rating Scale (IBMFRS), Cutaneous Dermatomyositis Disease Area and Severity Index (CDASI), Cutaneous Assessment Tool (CAT), Dermatomyositis Skin Severity Index (DSSI), Skindex, and Dermatology Life Quality Index (DLQI). Arthritis Care Res 2011;63(suppl 11):S118–S157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Amici DR, Pinal-Fernandez I, Mázala DA, et al. Calcium dysregulation, functional calpainopathy, and endoplasmic reticulum stress in sporadic inclusion body myositis. Acta Neuropathol Commun 2017;5:24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dobin A, Davis CA, Schlesinger F, et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 2013;29:15–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pertea M, Pertea GM, Antonescu CM, Chang TC, Mendell JT, Salzberg SL. StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat Biotechnol 2015;33:290–295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 2014;15:550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Greenberg SA, Higgs BW, Morehouse C, et al. Relationship between disease activity and type 1 interferon- and other cytokine-inducible gene expression in blood in dermatomyositis and polymyositis. Genes Immun 2012;13:207–213. [DOI] [PubMed] [Google Scholar]
- 29.El-Sherbiny YM, Psarras A, Md Yusof MY, et al. A novel two-score system for interferon status segregates autoimmune diseases and correlates with clinical features. Sci Rep 2018;8:5793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pinal-Fernandez I, Casciola-Rosen LA, Christopher-Stine L, Corse AM, Mammen AL. The prevalence of individual histopathologic features varies according to autoantibody status in muscle biopsies from patients with dermatomyositis. J Rheumatol 2015;42:1448–1454. [PMC free article] [PubMed] [Google Scholar]
- 31.Uruha A, Nishikawa A, Tsuburaya RS, et al. Sarcoplasmic MxA expression: a valuable marker of dermatomyositis. Neurology 2017;88:493–500. [DOI] [PubMed] [Google Scholar]
- 32.Suarez-Calvet X, Gallardo E, Pinal-Fernandez I, et al. RIG-I expression in perifascicular myofibers is a reliable biomarker of dermatomyositis. Arthritis Res Ther 2017;19:174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hornung T, Janzen V, Heidgen FJ, Wolf D, Bieber T, Wenzel J. Remission of recalcitrant dermatomyositis treated with ruxolitinib. N Engl J Med 2014;371:2537–2538. [DOI] [PubMed] [Google Scholar]
- 34.Kurasawa K, Arai S, Namiki Y, et al. Tofacitinib for refractory interstitial lung diseases in anti-melanoma differentiation-associated 5 gene antibody-positive dermatomyositis. Rheumatology (Oxford) 2018;57:2114–2119. [DOI] [PubMed] [Google Scholar]
- 35.Kurtzman DJ, Wright NA, Lin J, et al. Tofacitinib citrate for refractory cutaneous dermatomyositis: an alternative treatment. JAMA Dermatol 2016;152:944–945. [DOI] [PubMed] [Google Scholar]
- 36.Ladislau L, Suarez-Calvet X, Toquet S, et al. JAK inhibitor improves type I interferon induced damage: proof of concept in dermatomyositis. Brain 2018;141:1609–1621. [DOI] [PubMed] [Google Scholar]
- 37.Paik JJ, Christopher-Stine L. A case of refractory dermatomyositis responsive to tofacitinib. Semin Arthritis Rheum 2017;46:e19. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Any anonymized data not published within the article will be shared by request from any qualified investigator.