An emergent property of wing morphology bears signature of adaptation to flight ecology more strongly than morphology itself.
Abstract
Avian wing shape is highly variable across species but only coarsely associated with flight behavior, performance, and body mass. An underexplored but potentially explanatory feature is the ability of birds to actively change wing shape to meet aerodynamic and behavioral demands. Across 61 species, we found strong associations with flight behavior and mass for range of motion traits but not wing shape and strikingly different associations for different aspects of motion capability. Further, static morphology exhibits high phylogenetic signal, whereas range of motion shows greater evolutionary lability. These results suggest a new framework for understanding the evolution of avian flight: Rather than wing morphology, it is range of motion, an emergent property of morphology, that is predominantly reshaped as flight strategy and body size evolve.
INTRODUCTION
An enduring mystery in the field of animal flight is that diversity in wing shape across species is not well explained by flight style or by body mass. Some simple metrics of wing shape, particularly aspect ratio, correspond with aerodynamic performance, albeit coarsely (1, 2). Other more refined measurements of wing geometry show even weaker relationships between shape and flight style or body size across clades (3).
A potential explanation for the weak association between flight style and wing shape is that static metrics are inherently insufficient to capture the dynamic capabilities of mobile limbs. Birds can dynamically change wing shape, both actively and passively during routine flight, which enhances control over aerodynamic forces and moments acting on the wing (4, 5). Because the range of motion in the avian wing may fundamentally determine flight capabilities, examination of its evolution should inform how natural selection has reshaped wing design. To address this hypothesis, we asked how well wing morphing capability is explained by flight behavior and body mass diversity in comparison to static measures of wing shape. Across a diverse sampling of avian taxa, we first quantified wing shape and a variety of static traits of the wing. We then measured the capability of the wing to extend and flex across both the elbow and wrist joints, as well as the ability to bend or twist at the wrist. Through a phylogenetic comparative framework, we determined how well flight behavior, body mass, and/or phylogenetic history explained variation in wing shape and range of motion traits. By recording wing kinematics from two species, we then determined how in vivo flight relates to osteological range of motion. We conclude by measuring phylogenetic signal among measures of static and dynamic morphology to determine how shared evolutionary history underlies trait variation itself.
RESULTS
To measure static and dynamic morphological variation, we obtained wings from adult cadavers of 61 species (125 individuals) of birds spanning 20 avian orders (data S1). Although recent studies have produced well-supported hypotheses of relationships among avian clades (6, 7), our sampling of taxa was not well represented among published phylogenies, and our ability to account for shared history (and topological uncertainty) in our comparative analyses was limited. We therefore applied a Bayesian framework (8) to infer time-calibrated phylogenies from genetic sequences (nuclear and mitochondrial) and fossil calibrations for a 220-taxon set of species (data S2 and S3 and table S1) including representatives from every avian order and the focal 61 taxa (Fig. 1A and data S4).
Fig. 1. Flight behavior and body mass influence the range of motion in wing extension and flexion but not wing shape.
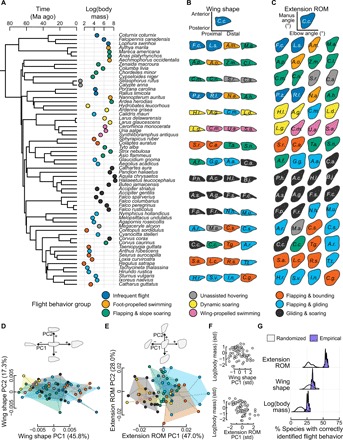
(A) Time-calibrated phylogeny of 61 focal species, pruned from our 220-taxon maximum clade credibility tree. Circles to the right of the tree show natural log–transformed body mass (measured in grams) colored by flight behavior. Ma, million years. (B) Wing shape [two-dimensional (2D) outline shape at maximum extension] and (C) extension range of motion (ROM) of each species (outer boundary of possible elbow versus manus extension or flexion) after resizing shapes via Procrustes superimposition. Initials correspond to genus and species. (D) Phylogenetic principal components analysis (pPCA) of avian wing shape; birds of varying flight behavior show extensive overlap in morphospace. (E) pPCA of extension ROM shows higher differentiation among groups. Top diagrams each depict shape changes along major axes. (F) Mass varies little with wing shape (top) but more strongly with extension ROM shape (bottom). Each variable was standardized (z-transformed). Std, standardized. (G) Phylogenetic flexible discriminant analyses found that extension ROM had superior performance to predicting flight behavior group compared to wing shape or body mass. Purple kernel densities show jackknifed empirical prediction accuracies; gray kernel densities show prediction accuracies of 61 randomized permutations.
Each of the 61 species was assigned to one of nine flight behavior groups following a literature survey (data S5 and fig. S1). To account for the potential ability of each species to exhibit multiple flight modes, we recorded whether each species has been documented to perform each of a larger set of 12 potential flight behaviors (9). We then implemented hierarchical clustering on the dissimilarity matrix of flight modes, used the gap statistic (10), and determined that nine was the optimal number of distinct groups, which we named using the behaviors that best distinguished each group.
We next asked how well flight behavior or body mass associate with either wing shape or the capacity to extend or flex the wing. We captured static wing shape [outline shape in two dimensions (2D) at full extension of the wing] and recorded the capability of extension and flexion at both the elbow and manus (wrist) joints in 3D (fig. S2 and movies S1 to S3). Range of motion was determined by manually applying force at a joint until resistance was met, ensuring that hyperextension of joints did not occur (11). Applying further force would have risked dislocating or breaking bone, ligament, or other tissue. The extension range of motion (ROM) was then defined as the space occupied by the outer boundary of possible elbow angles against possible manus angles for this type of motion. Via elliptical Fourier methods (12), we captured outline shape diversity of each of these two datasets (Fig. 1, B and C) and then used phylogenetic principal components analyses (pPCAs) (13) to determine major axes of shape diversity across species (Fig. 1, D and E). We then determined how variance in species wing or extension range of motion (ROM) shapes could be explained by flight behavior and/or body mass using Bayesian generalized linear mixed models (14). This allowed us to determine the extent to which fixed effects (flight behavior group, natural log–transformed body mass, and/or their interaction) explained variance among principal components of wing shape or extension ROM (tables S2 and S3). Phylogenetic history was used to inform the covariance matrix of the random effects and subsequently calculate phylogenetic signal in models (15).
Flight behavior and body mass explained little variation among species’ static wing shapes but substantial variation in extension ROM shapes (Fig. 1, D to F). For wing shape, a “null” but phylogenetically informed intercept-only model provided the best fit [Pagel’s λ mean, 0.64; 95% highest posterior density (HPD), 0.48 to 0.78]. Despite differences in species examined and in geometric morphometric methods, our results reinforce a finding that wing shapes bear little correspondence to flight behaviors or body size; rather, diversity lies largely along phylogenetic lines (3). In contrast, extension ROM shape was best described by including both body mass and flight behavior group in concert with phylogenetic effects (λ, 0.63; 95% HPD, 0.39 to 0.87) (Fig. 1, E and F). A key difference in extension ROM shape lies between species that use gliding, soaring, or other motion-restricted usage of the wing and those that use higher-excursion motions, often with higher-frequency flapping. This pattern was driven by differences among three flight behavior groups. Two groups (flapping and bounding; flapping and gliding), which showed lower body masses, had enhancements to range of motion in extension and flexion at extreme wing postures. The third group, flapping and slope soaring, tended to have higher body mass and showed limited range of motion throughout the full range of wing unfolding.
The strength of association between flight style and range of motion motivated us to ask how well flight behavior itself could be predicted from morphological features. We used phylogenetic flexible discriminant analyses (16) to test the predictive performance of extension ROM, wing shape, and body mass (separately), coupled with sensitivity analyses and assessments against random chance (17). Extension ROM shape considerably outperformed both wing shape and body mass when used to predict species’ flight behavior groups (Fig. 1G), with correct predictions in over half of cases.
The capacity to couple extension (or flexion) across the elbow and manus via musculoskeletal linkage is a well-documented feature of the avian wing, albeit only investigated in cadavers of pigeons (Columba livia) (18). We asked whether this coupling motion also shows concordance with flight behavior or body mass. By manually extending and flexing the wing at the elbow and tracking corresponding changes to manus extension/flexion, we determined that all 61 species have the ability to couple motion across these joints, the resultant motion path that we here term the “linkage trajectory” (Fig. 2A).
Fig. 2. Coupling of avian elbow and wrist motion is concordant with flight behavior but not body mass.
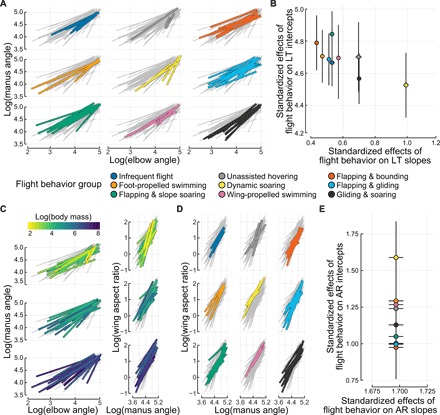
(A) Facet plots of each flight behavior group’s linkage trajectories against those of all other taxa (gray). Each line represents a motion path for one species; color corresponds to flight behavior. Elbow and manus angles are natural log–transformed for linearization. (B) Standardized effects (estimated marginal means) of flight behavior on linkage trajectory (LT) intercepts and slopes. Black lines indicate SEs of estimates. (C) Facet plots of linkage trajectory (left) and the correspondence between wing aspect ratio and linkage trajectory (right). Body mass (color bar) explains little variation. To enhance visual clarity, data are faceted by tercile. (D) Aspect ratio versus linkage trajectory, colored by flight behavior. Although this visualization suggests distinct patterns among groups, there is extensive overlap of estimated effects. (E) Estimated marginal means of flight behavior on the transfer function between linkage trajectory and wing aspect ratio (AR). Phylogenetic mixed models indicated that neither flight behavior nor body mass appears to affect these relationships; SEs of all estimates overlap extensively.
Linkage trajectory was strongly associated with flight behavior group but, unlike with extension ROM, was not explained by body mass (Fig. 2, B and C, and tables S2 and S3). Flight behavior group had strong effects on both the slopes and intercepts of the mixed model, but phylogenetic history also showed strong influence (λ, 0.85; 95% HPD, 0.80 to 0.91). Dynamic soaring species showed particularly high slopes, indicating that they have a more extreme capability to tune wing shape via the linkage system to potentially negotiate trade-offs in aerodynamic efficiency and static pitch stability (5).
The strong association between flight behavior and linkage trajectory suggests that variation in these motion paths should have functional consequences. Given that aspect ratio influences aerodynamic power and efficiency (2, 19), we next asked whether the relationship between aspect ratio and linkage trajectory could be explained by flight behavior or body mass (Fig. 2, C and D, and tables S2 and S3). Neither predictor, however, substantially improved variance explained, and instead, our null, intercept-only model with phylogenetic random effects provided the best fit (λ, 0.80; 95% HPD, 0.74 to 0.86). Although Fig. 2D suggests distinct patterns among flight behavior groups, estimated effects overlapped extensively in models informed by flight group (Fig. 2E). Thus, extension or flexion via the avian wing’s linkage system causes similarly proportional changes to aspect ratio across species of varying size and flight behaviors.
In addition to the largely planar motion of extension and flexion, we asked whether the wing’s out-of-plane capabilities also show associations with flight behavior or body mass. We examined the capacity to bend (elevate or depress) or twist (pronate or supinate) simultaneously by manually manipulating wings over the full range of extension and flexion of both the elbow and manus (fig. S3). Because of the increased complexity of data capture, out-of-plane ranges of motion were determined only for the hand-wing (i.e., at the manus joint) and for 30 of the 61 species.
Although both bending and twisting capabilities showed complex topographies across extension ROM, all species showed restrictions to both bending and twisting as the wing was extended fully (Fig. 3). This led us to ask whether such restrictions, the osteological mechanisms of which have been generally described (20), vary according to body mass or flight behavior. We found that heavier species showed greater overall restrictions to both bending and twisting ROM (analyzed separately; Fig. 3, A and C, and tables S2 and S3) and that these relationships were little affected by phylogeny (λ for bending, 0.03 and 95% HPD, 0 to 0.14; λ for twisting, 0.05 and 95% HPD, 0 to 0.13). Incorporation of flight behavior in models hinted at some separation in standardized effects between species that glide and soar against those that primarily use flapping (Fig. 3, B and D, and tables S2 and S3), but these effects were weak and not statistically distinguishable.
Fig. 3. The bending and twisting capacity of the wing is position dependent and is more strongly associated with body mass than with flight behavior.
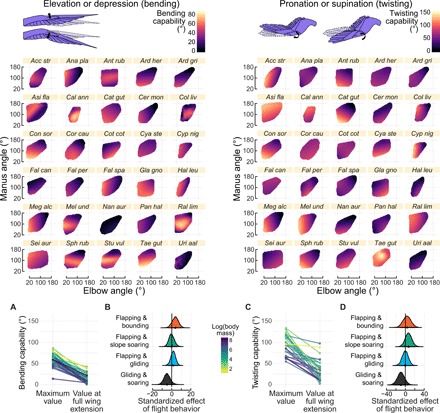
Elevation/depression capability (left) and pronation/supination capability (right) at the manus joint were assessed over the extension ROM of each of 30 species. The combined capacity to elevate or depress at this joint was defined as bending capability; the combined capacity to pronate or supinate was defined as twisting capability. Species are labeled according to the first three letters of each of their genus and species names (as Gen spe); see Fig. 1 for full binomial nomenclature. In all 30 species, both (A) bending and (C) twisting capacity are reduced when the wing is at full extension. Heavier species (darker colored) tend to show greater overall restrictions to these types of motion. “Maximum value” indicates the highest capacity for a given motion irrespective of wing extension. Within each plot, lines connect data for each species. Estimated marginal means of flight behavior on the reduction in (B) bending and (D) twisting capacity after accounting for effects of natural log–transformed body mass. Given our data, flight behavior does not appear to substantially affect bending or twisting capacity.
Because flight behavior associated with some range of motion traits but not others, we asked how in vivo wing kinematics relate to osteological range of motion. We recorded 3D wing motion during in vivo flight from two species: the pigeon (C. livia, n = 1) and the zebra finch (Taeniopygia guttata, n = 4). On each bird, we marked a single wing in a similar fashion to that used in our cadaveric study (see Materials and Methods). Five cameras recorded wing motion at 240 frames/s. The pigeon was observed alone, whereas all four zebra finches were released into the flight cage together, but data were collected only on one individual at a time. To attain as comprehensive a sampling of wing usage as possible, we allowed birds to fly freely and did not limit recordings to singular flight behaviors. Rather, we collected data on an indiscriminate mix of behaviors including ascending flight, level flapping, transient hovering, turning, and descending flight (although we note that most recordings were of forward flight).
Not only did we find that in vivo wing configurations fell within osteological range of motion but also each species showed distinct patterns of wing use (Fig. 4). Zebra finches used a broad array of wing configurations, many of which overlapped the cadaveric linkage trajectory (Fig. 4A). The pigeon, however, showed a narrower range of both elbow and manus extension, with a tendency toward low-elbow, high-manus configurations (Fig. 4B). It is conceivable that high-elbow, high-manus configurations are used during gliding, but we did not observe this behavior (likely due to the limits of our flight arena). Data from the pigeon also showed strong departure from the linkage trajectory, indicating that wing morphing is not merely restricted to the path achieved by the coupling of elbow and wrist motion. Out-of-plane motion in both species also fell within the limits defined by our cadaveric study (Fig. 4, D to F), although we note that because of difficulties in marker visualization, we were unable to track wing twist in the zebra finches. Neither species showed bending or twisting as extreme as the underlying osteological capability, which aligns with the lack of correspondence we found between flight behavior and out-of-plane range of motion.
Fig. 4. In vivo wing kinematics reveal that natural motions fall within ROM limits.
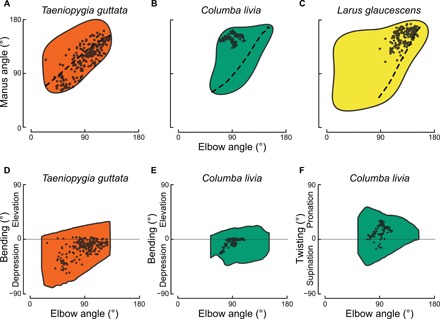
Joint angles were tracked during in vivo flight in zebra finches (n = 4) and a pigeon (n = 1). Cadaveric extension ROM is shown as a colored shape for (A) the zebra finch and (B) the pigeon. Each point shows elbow extension against manus extension from a single frame of recorded flight; data from multiple individuals and flight behaviors are pooled. Dashed lines indicate linkage trajectories from cadavers. (C) Similar data from in vivo observations of gliding (points) in the glaucous-winged gull obtained from (5), shown against cadaveric extension ROM (colored shape) and linkage trajectory (dashed line) determined in this study. (D to F) Observations of in vivo bending or twisting (points) shown against ROM envelopes from cadavers (colored shapes).
Although other studies of avian wingbeat kinematics do not typically track joint angles within the wing, a study of the gliding behaviors of glaucous-winged gulls (Larus glaucescens) inferred elbow and wrist extension through matching in vivo wing shapes to those shown by manipulating cadavers through their range of motion (5). Inferred in vivo data from this study similarly fall within the extension ROM of our cadavers for this species, tending toward fuller wing extension and overlapping somewhat with linkage trajectory configurations (Fig. 4C).
Despite observations from only a few species, comparing in vivo wing usage against osteological range of motion suggests three hypotheses. First, the shape of the wing when at full extension poorly describes the airfoil being used during most of the behaviors we recorded. Full extension was rarely, if ever, shown during flight. Second, different regions within the range of motion seem to be used by species of differing flight behaviors and body mass. These patterns reveal exciting potential to use range of motion as a substrate upon which to compare in vivo wing usage both within and across taxa. Third, in vivo studies of wing kinematics typically cannot guarantee to elicit the full range of behaviors that a species shows and instead present a more limited view of wing usage and morphing capability.
Collectively, we find that different features of the wing and its range of motion associate in notably different ways with body mass and flight behavior. Computing effect sizes (21) highlights the differences in relative importance of these variables in explaining static and dynamic morphological variation (Fig. 5A and table S4). The inclusion of flight behavior grouping and body mass did not explain variation in static wing shape. In contrast, flight behavior was key to explaining wing extension capabilities, particularly in the linkage trajectory achieved via coupling elbow and wrist motion. The limits to out-of-plane motion were explained by variation in body mass but not by flight behavior or even phylogeny.
Fig. 5. Wing range of motion is more strongly associated with flight behavior and body mass and is more evolutionarily labile than wing shape.
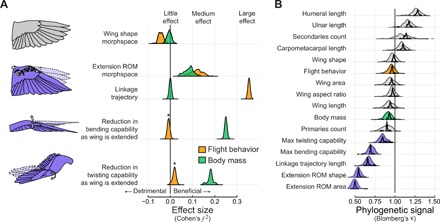
(A) Effect sizes (Cohen’s f2) for each of the fixed effects considered. Increasingly positive effect sizes indicate that the addition of that variable substantially improved variance explained; increasingly negative indicates the opposite. Flight behavior or body mass did little to explain wing shape. Flight behavior has a pronounced effect in explaining extension/flexion patterns, whereas body mass substantively explains trends in bending and twisting capability of the hand-wing. Density plots show distributions of f2 as phylogeny is varied. Asterisks indicate that analyses were restricted to four well-represented flight behavior groups. (B) Range of motion traits (purple) have lower phylogenetic signal (Blomberg’s κ) than those related to static morphology or flight behavior. A κ value of 1 indicates strong phylogenetic signal that ostensibly follows Brownian motion. Traits with κ values increasingly greater than 1 are more phylogenetically conserved; κ values increasingly lower than 1 indicate greater lability. We performed two sensitivity analyses: one in which phylogeny was varied (dashed distributions) and one in which data were jackknifed (solid).
The fluctuating importance of phylogenetic effects across our models led us to ask how shared history among species underlies trait variation itself. We uncovered a stark difference in phylogenetic signal for static and dynamic traits (Fig. 5B and table S5). Static morphological traits and flight behavior group showed Blomberg’s κ values (22, 23) that were close to or overlapping 1.0, the value expected under a Brownian motion model of evolution. Different evolutionary processes can produce similar values of phylogenetic signal; therefore, κ values close to 1.0 should not necessarily be ascribed to Brownian motion (24). Nevertheless, we determined that static morphology in the avian wing shows greater evolutionary conservatism, whereas dynamic range of motion traits show more dynamic histories.
DISCUSSION
To examine whether variation in flight style and body mass is more strongly associated with static or dynamic traits of the wing, we measured the range of motion in extension, bending, and twisting for wings from birds of diverse sizes and flight styles. We found that variation in range of motion in extension was explained by both flight style and body mass, but extension coupling corresponded only with flight style. Both bending and twisting were influenced by body mass but not flight style. In contrast, static wing shape did not correspond with either factor and showed stronger evolutionary conservatism than all measurements of wing range of motion.
Collectively, these results provide new context for not only understanding the avian wing specifically but also, more generally, for testing hypotheses of morphological evolution. Biomechanical systems comprise both underlying morphology and their emergent functional properties, the latter of which has stronger ties to performance of behavior (25). In other vertebrate clades, static traits show similar extents of phylogenetic signal to that seen in the avian wing (22, 23, 26), suggesting that morphology may generally be expected to show evolutionary conservatism. Range of motion, an emergent feature of morphology, has been little examined in a macroevolutionary context. Its evolutionary lability and concordance with ecological factors point to its efficacy in determining how selection for performance affects morphological evolution in the avian wing. In birds, specialization for flight and size does not impose strong limitations on wing shape (3) but instead reshapes an emergent feature: the wing’s range of motion.
Although we considered multiple axes of wing motion capability, birds can move other wing elements that we did not measure here. Some feathers, e.g., the alula and the last primary, can be further controlled by motion of the major and minor digits of the hand-wing (27), and the forearm, much like that of other vertebrates, can also pronate and supinate (9). We chose to focus on the largest motions in the distal half of the wing because the center of pressure is located in this region during flapping flight. Pressure location has been measured on revolving wing specimens, which confirmed that larger differential pressure occurs where velocity along the wing is greatest (28). Thus, changes to the distal half of the wing will strongly influence aerodynamic force magnitude and orientation.
Within aircraft design, there is increasing interest to explore the potential for morphing wings to increase aerodynamic performance and response to perturbations relative to static wings (29–32). The results of our comparative study suggest specific design targets for drone aircraft depending on flight mode and mass. Across species, we find that increases in mass correspond to increasing restrictions within all axes of range of motion. Motion capabilities are also limited in species that dynamically soar or glide, indicating that these flight styles may demand a more rigid maintenance of posture akin to the wings found on engineered gliders and sailplanes. Although flight behavior did not generally explain variation in out-of-plane range of motion, we note that such capabilities are particularly limited among species that use their wings to swim underwater and face a relatively viscous environment. In contrast, taxa that use high-excursion flapping and bounding behaviors show relatively fewer restrictions to any of the axes of variation we measured, a correspondence that appears in multiple avian clades. Among engineered vehicles, particularly those that rely on flapping, a primary challenge in moving from static to morphing wing design is the increased complexity of dealing with unsteady flows (29, 32). By considering the limitations that natural selective forces have placed on birds of a particular size and flight style, we find that passive control capability within the avian wing provides convenient reference toward engineering of next-generation morphing wings on unmanned aerial vehicles.
MATERIALS AND METHODS
Specimens
For 58 of the 61 species examined, frozen cadavers of adult specimens were each acquired from one of five sources: (i) University of British Columbian Beaty Biodiversity Museum (Cowan Tetrapod Collection), Vancouver, BC, Canada; (ii) Royal British Columbia Museum, Victoria, BC, Canada; (iii) Wildlife Rescue Association, Burnaby, BC, Canada; (iv) Wild Animal Rehabilitation Centre, Victoria, BC, Canada; or (v) BC Ministry of Forests, Lands, Natural Resource Operations and Rural Development, Nanaimo, BC, Canada.
For the remaining three species, fresh cadavers of adult specimens (Anna’s hummingbird, Calypte anna; pigeon, C. livia; and zebra finch, T. guttata) were sourced from ongoing studies in the Altshuler Laboratory. All procedures with these species were approved by the Animal Care Committee of the University of British Columbia.
Before data collection, each specimen was carefully assessed to determine that its wings were fully intact (including all feathers) and did not exhibit effects of desiccation or decomposition. Adult male specimens were sampled for species in which appreciable sexual dimorphism (by coloration or mass) occurs. Specimens that had been frozen were fully thawed, and all subsequent data collection occurred within 10 min. Masses were measured on intact carcasses before any dissection. Wings were then removed at the shoulder joint, taking special care to ensure that each wing’s skin, propatagial elements, and feathers remained intact. Although in many cases, both wings of a cadaver were removed, data collection occurred only on a single wing per specimen. Details of sample sizes, body masses, and other static morphological measurements (see the “Data collection: Static morphology” section) of specimens are provided in data S1.
Tree inference
We reconstructed phylogenetic relationships using a molecular dataset that comprised six mitochondrial (12S, 16S, COI, CytB, ND1, and ND2) and six nuclear gene regions (B-Fib, MUSK, ODC, RAG1, TGFB2, and ZENK), with 18,246 total base pairs. We obtained sequences for 220 birds and a two-taxon out-group (crocodiles). All 61 species for which we examined morphology and ROM were included in this sampling. The additional 159 species of birds (220 to 61) were included to aid in the estimation of divergence times among avian clades. All sequences were downloaded from GenBank (see data S2 for accession numbers and information on genetic sampling).
We aligned each gene sequence separately using the built-in algorithm in Geneious v9.1.6 (33). Flanking regions that contained sequences from less than 60% of taxa were excluded. To identify the best-fitting model of nucleotide substitution for each gene, we used PartitionFinder v2.0 (34). In each case, we found the best fit (assessed via Akaike’s information criteria and Bayesian information criteria scores) to be a GTR + I + Γ model or a close variant thereof.
Using SequenceMatrix v1.7.8 (35), we concatenated the aligned sequences into a supermatrix. We partitioned this supermatrix by individual molecular markers and performed a maximum-likelihood analysis in RAxML (36), using a bootstrap analysis under a GTR + Γ model with 1000 pseudoreplicates. The phylogenetic tree with the best likelihood score was retained to guide further analyses (data S3).
We then used BEAST v2.4.7 (8) to simultaneously estimate topology, branch lengths, and divergence times via Bayesian Markov chain Monte Carlo (MCMC). Using a relaxed log normal clock model approach, we partitioned the supermatrix by sequence and fit a separate model for each partition based on our results from PartitionFinder. To estimate divergence times, we placed informative parametric priors on nodes of the tree to reflect the available paleontological history of the group (table S1). Descendant members of each specified node were based on the topology of the maximum likelihood tree from RAxML.
To ensure that each BEAST MCMC sampling converged on the target distribution, we ran nine separate chains, each from a different random starting tree. Each MCMC chain ran for 200 million generations, sampling every 20,000 generations. We assessed convergence by plotting likelihood versus generation and estimating the effective sample size (ESS) of each parameter. A similar analysis in which the supermatrix was not partitioned resulted in most MCMC chains not attaining stationarity.
Once we discarded the burn-in from each chain (the first ~20%), we combined chains via LogCombiner v2.4.7 (8). Within each chain, the ESSs of all parameters were generally >200, with the lowest ESS still >100. After we combined all results, most of the parameters had ESS of >2000. The combined set included >500 million trees, which we used to assemble the maximum clade credibility tree in TreeAnnotator v2.4.7 (8). This resulting maximum clade credibility tree (data S4) was used for all phylogenetically informed analyses of comparative data (and used to generate main text figures). In addition, 1000 trees were sampled from the posterior distribution to aid in the understanding of phylogenetic sensitivity in all comparative analyses (see Fig. 5).
Determination of flight behavior grouping
We determined 12 potential flight behaviors using descriptions from (9). These behaviors were the following: unassisted hovering, ability to hover without being reliant on wind conditions; dynamic hovering, reliance on wind speed gradients to hover in place (often transiently using a head wind); bounding, short bursts of flapping flight alternating with (short) intervals where the wings are folded against the body; flap gliding, short bursts of flapping flight alternating with (short) intervals where the wings are held an extended position; gliding, wings are held in an extended position (no flapping), often resulting in losing altitude; thermal soaring, bird propelled upward by moving air on rising thermals; slope soaring, bird propelled upward by rising air along a slope; dynamic soaring, use of wind speed gradients and bird’s own momentum to maintain or increase altitude; stooping, controlled, rapid, head-first dives; foot-propelled swimming, bird uses feet but not wings to propel itself underwater; wing-propelled swimming, bird uses wings to propel and steer itself underwater; (relatively) infrequent flight, behavior meets any of the following: (i) completely incapable of flight, (ii) ground-dwelling, (iii) incapable of flight for several weeks (often due to molt), (iv) aversion to flight, and/or (v) previous classification of “poor flight.”
To account for the ability of each species to exhibit multiple flight behaviors, we scored each species’ ability to exhibit a given flight behavior on a binary scale. For a given behavior, species were scored a “1” if that behavior was regularly or repeatedly found in natural observations, descriptions, and/or previous categorizations of that species (9, 37–43). A 61 × 12 flight behavior matrix (data S5) was thereafter created with each species occupying a row and each flight behavior occupying a column.
We then categorized taxa into flight behavior groups by first generating a dissimilarity matrix based on the flight behavior matrix using the Gower distance between rows (44). We then performed hierarchical clustering based on Ward’s clustering criterion (45) on the dissimilarity matrix. To determine the optimal number of flight behavior groups, we computed the gap statistic, which compares the change in within-cluster dispersion with that expected under a null distribution (10), using 500 bootstrap replicates. Because the gap statistic identified nine groups as the best performing scheme, we then cut our hierarchical cluster tree at nine groups (fig. S1A). To aid in the identification of behaviors that best described each group, we then performed pPCA (13) on the flight behavior matrix and used the loadings of the pPCA to determine how specific flight behaviors corresponded to each group (fig. S1B). Groups were thereafter named using the flight behaviors that best corresponded to that group’s ordination in pPCA.
Data collection: Range of motion
To determine their range of motion, wings were mounted to a support frame. The head of the humerus was attached to the frame, and several motion types were actuated by hand (movies S1 to S3). To capture resulting changes in wing shape, five points were marked on the dorsal surface of each wing (fig. S2): (i) the head of the humerus, (ii) the center of motion of the elbow joint, (iii) the center of motion of the manus joint, (iv) the distal tip of the carpometacarpus, and (v) point on the dorsal wing surface that lay in line with the edge of the carpometacarpus (point 4). Point 5 was located along the leading edge of the wing. The locations of point 5 was such that a line segment along the dorsal surface of the wing connecting points 4 and 5 would lie perpendicular to the length of the carpometacarpus (i.e., the segment along the dorsal surface connecting points 3 and 4). Select covert feathers were removed to allow for a clearer visualization of the specified markers. Wings were marked either with permanent marker applied directly to the skin or on pieces of tape that were attached lightly to the wing to not restrict freedom of movement across joints. Marking occurred within 1 min, which allowed for subsequent filming of all range of motion within 10 min of thawing the wing.
Wing motion was captured in 3D at 24 frames/s at 1920 × 1080 pixel resolution using three Miro M120 video cameras (Vision Research, NJ, USA), and Phantom Camera Control (v3.3.781.0) software was used to synchronize video recording across cameras. Calibrations for 3D digitization were performed by waving a 7 cm by 7 cm checkerboard grid within the filming area before each recording session to obtain direct linear transformation coefficients via easyWand v6 (46). Axes were defined using a stationary object. Throughout all videos, the antebrachium of the wing was held in parallel with the X-Y plane. The directions of X and Y were not standardized, but the Z axis was strictly defined to align with the dorsoventral axis of the wing (+Z, dorsal). One camera (24-mm lens) was oriented to give a top-down view that aligned with the X-Y plane of wing motion. The other two cameras (both 28-mm lenses) were positioned such that all markers could be visualized throughout the range of motion actuation. We then used DLTdv v6 (47) in MATLAB (MATLAB and Statistics Toolbox Release 2016b) to digitize range of motion data in 3D. During digitization of videos, a maximum reprojection error of 1.0 pixel was strictly enforced.
For wings from all 61 species, the range of flexion and extension of both the elbow and manus joints was recorded. All measurements were made by V.B.B. Range of motion was determined by applying force manually at two locations on the wing: (i) midway along the length of the carpometacarpus to manipulate the manus and (ii) midway along the ulna to manipulate the elbow. Force was applied at a joint until resistance was met (11). In all species, the range of extension was limited by the skeletal system rather than by soft tissue such as ligaments or tendons. Thus, any differences in soft tissue compliance were not registered with this technique. Special care was taken to ensure that hyperextension of the joints did not occur; applying further force would have risked dislocating or breaking bone.
The ability to couple elbow and manus motion via a musculoskeletal linkage system in the wing (18) was additionally assessed across all specimens. The wing was initially held in a fully flexed position; the elbow was then extended until maximum elbow extension was achieved, held briefly at maximum position, and then reversed to full flexion of the elbow to complete one “cycle.” Six full cycles of movement were recorded for each wing, with each cycle actuated at approximately 0.5 Hz.
Because of increased difficulty in out-of-plane digitization, the range of motion in bending or twisting the wing was determined only for the hand-wing (i.e., at the manus joint) and for 30 of the 61 species. Out-of-plane wing motion was manipulated by applying force manually midway along the length of the carpometacarpus (see fig. S2). As for extension, the out-of-plane motion was determined by applying force until resistance was met within the wrist joint and was limited by bone rather than by soft tissue. Bending and twisting of the hand-wing were simultaneously determined over the full range of flexion and extension of both the elbow and manus.
In all videos, elbow angles were calculated as angular changes involving points 1, 2 (vertex), and 3, whereas manus angles were calculated as angular changes involving points 2, 3 (vertex), and 4. For bending motion, elevation (positive) and depression (negative) of the hand-wing were assessed as the vertical axis (Z) angular change of point 4 relative to point 3. For twisting, pronation (negative) and supination (positive) were calculated as the angular deflection of point 5 (or 6) against a horizontal drawn from point 4. This angle was calculated in degrees using the points {X5,Y5,Z5}, {X4,Y4,Z4} (vertex), and {X5,Y5,Z4} (see fig. S2).
Data collection: Static morphology
For each wing, the following linear measurements or discrete counts of static morphology were recorded (definitions in parentheses): humeral, ulnar, and carpometacarpal length (linear distance in millimeters between points 1 and 2, 2 and 3, and 3 and 4, respectively); static wing length (linear distance in millimeters between point 1 and the distalmost tip of the wing when held at full extension, measured perpendicular to the root chord of the wing); and primary and secondary feather counts (numerical counts; primary feathers, remiges located on the hand-wing; and secondary feathers, remiges located on the arm-wing).
Measures of wing shape, wing area, and wing aspect ratio (data S1) were each computed in 2D from video recordings from the “top-down view” camera only. “Static” wing shape (the shape of the wing at maximum extension of the elbow and manus) was determined by first finding the video frame in which both elbow and manus angle were maximized and then using the Freehand Selection tool in ImageJ v1.5 (48) to create a selection that included all components of the wing. This selection was first used to compute static wing area (in square millimeters). Each of these selections was then imported into R and converted into a closed outline (Coe) object using the Momocs package (12) for further analyses of wing shape (see the next section). Measures of wing area and wing length were also captured for every frame of the videos in which coupling of the elbow and manus was assessed. Again, using the Freehand Selection tool, a selection that included all components of the wing was first created and then used to compute wing area in square millimeters. The length of the wing was calculated as the longest possible line from the distalmost portion of the wing perpendicular to the wing root. Aspect ratio was then calculated as
where b was twice the length of the wing, and S was the 2D wing area.
Quantification of range of motion and wing shape
We quantified range of motion in the avian wing using three metrics: (i) extension ROM, (ii) bending capability, and (iii) twisting capability. Extension ROM was defined as the space occupied by the outer boundary of possible elbow angles against possible manus angles. To determine extension ROM for each species, the set of all possible elbow angles (X axis; ranging from 0° to 180°) was plotted against all possible manus angles (Y axis; ranging from 0° to 180°). Because we found intraspecific variation to be minimal, we pooled data from all specimens of a species. We then generated a convex hull that encapsulated all possible elbow and manus angles. To facilitate comparison of these shapes across species, we converted all convex hulls into Coe objects (12) and then used elliptical Fourier analysis (49) to quantify outline shapes without the use of homologous landmarks. Because we sought to test hypotheses of extension ROM shape, we first standardized all outlines to the same size and location using generalized Procrustes analysis (50). Elliptical Fourier coefficients that described >95% of shape variance across species were retained for further analyses involving extension ROM. The area of each extension ROM shape was also computed by taking the total area of occupation in the elbow angle × manus angle plots.
Bending capability was defined as the combined capability to elevate or depress the hand-wing at the manus joint, evaluated at each point of extension ROM. For each species, elevation (positive) and depression (negative) (both on Z axis) were simultaneously plotted in 3D against elbow angle (X axis) and manus angle (Y axis). To encapsulate the ROM, an α-hull [3D generalization of a convex hull (51)] was fit to the data (fig. S3). To describe, visualize, and compare bending capabilities across species, the vertices of each α-hull were extracted and then separated according to whether they corresponded to elevation or depression. We fit regularized neural networks (52, 53) to each set of vertices to resolve the relationship between elevation (or depression) capability and wing extension (elbow angle and manus angle, jointly). This method not only guards against potential outliers resulting from digitization error (amounting to a low-pass filter) but also allows for the interpolation of data missing at any point within the extension ROM. Each regularized neural network was trained and then cross-validated within its dataset following a machine-learning framework using the Caret package (54). In cases where datasets were too sparse to use regularized neural networks, locally estimated scatterplot smoothing (LOESS), or linear models were used. Last, bending capability was determined by summing the predictions of elevation and depression from trained models for every point of extension ROM. Two key metrics were collected: (i) bending capacity at full extension (the combined ability to elevate or depress the wing when it is fully extended) and (ii) maximum value of bending capacity (the highest value of bending capacity achieved regardless of wing extension).
Twisting capability follows a similar definition to that of bending capability. The combined capability to pronate or supinate the hand-wing was evaluated at each point of extension ROM. All methods of α-hull and model fitting follow the above. Twisting capability across extension ROM, including its value at full extension and maximum value, was computed in a similar fashion.
Wing shape was quantified via a method akin to that used to quantify extension ROM. We first standardized all wing shape Coe objects to the same size and location using generalized Procrustes analysis (50). To standardize wing orientation, the distalmost tip of the wing was set as the “principal point” (49). Elliptical Fourier coefficients that described >95% of shape variance across species were retained for further analyses involving wing shape.
Multivariate analyses of shape
We used pPCA (13) to ordinate and facilitate comparison among species’ extension ROMs and wing shapes. Elliptical Fourier coefficients from each dataset were used as the raw data for each pPCA. In each case, we allowed the extent of phylogenetic signal (Pagel’s λ) to, first, be measured directly from the data (using either the maximum clade credibility tree from our BEAST analyses or one of the 1000 posterior distribution trees) and, second, to be used to inform the pPCA (13). For the extension ROM pPCAs, we found the resulting principal components to be invariant to phylogeny (all λ nearly zero; eigenvectors and values all identical); further analyses were carried out using the maximum clade credibility tree only.
Hypothesis testing of flight behavior and body mass on range of motion or wing shape via phylogenetic mixed models
To determine how extension ROM or wing shape vary according to flight behavior, body mass, or phylogenetic history, we used a phylogenetic Bayesian generalized linear mixed model approach via the MCMCglmm package in R (55). This allowed us to determine the extent to which fixed effects (flight behavior, body mass, and/or their interaction) explained variance among principal components of extension ROM or wing shape. In all cases, phylogenetic history was used to inform the covariance matrix of the random effects (identity matrix). For computational simplicity, only the maximum clade credibility tree was used. We used priors for the random effect and residual variances corresponding to an inverse-Wishart distribution (V = 1 and nu = 5 × 10−4) (56) and used the default uninformative priors for the fixed effects.
Because we had a multivariate response, we first used power analyses to determine the extent to which all principal components could be used (56). These analyses indicated that each interspecific dataset of 61 species was sufficient (power, 0.8) for analysis of three multivariate axes, given the number of fixed effects to be tested and approximate effect sizes. Hence, fixed and random effects were regressed against the first three principal components of the extension ROM or wing shape pPCAs.
For each dataset (extension ROM or wing shape), the following models were fit
where {PC1, PC2, PC3} are the first three principal component axes of either the extension ROM or wing shape pPCA, M is natural log–transformed body mass, and F is flight behavior grouping. The first listed model corresponds to a null case where neither flight behavior nor body mass explains meaningful variation in principal component scores.
Although deviance information criteria (DIC) values were computed for each model, we opted to not use this metric as our sole criterion for model selection for two reasons. First, DIC has been identified to have weak theoretical justification and reduced predictive power (57). Second, we observed in our results that, occasionally, the most complex model was selected, but effects were poorly estimated (extremely large credible intervals for multiple fixed effects).
To guard against the selection of overly complex models, we used a cross-validation approach to model selection. For each model, folds of six species’ data were each held out as a “test” set, whereas data from the other 55 species were used for “training.” Our method of cross-validation is robust to the issues related to phylogenetic data (58) because the entire 61-taxon maximum clade credibility tree was used to inform random effects of the MCMCglmm models, thereby accounting for phylogenetic structure in both training and test datasets. Point estimates (maximum a posteriori estimates of principal component scores) for both training and test datasets were then generated from each model using the predict.MCMCglmm() function, and their associated mean squared errors (MSEs) were then computed. Although the MSEs of training data were informative to assess goodness of model fit, only the MSEs of test data were used to determine the best performing model (59). The conditional R2 of each model was also calculated (60) to aid in the description of goodness of fit. Information on model fits is available in table S2, and specific information on best-fitting models is available in table S3.
For each MCMC model, 10 separate chains of 45,000 iterations were run, with a burn-in of 4500 and thinning interval of 39. Within each chain, the ESS of each parameter was ~1000. This procedure yielded autocorrelation values ≤ 0.10 between retained iterations. To assess convergence among chains, we used the Gelman-Rubin diagnostic procedure (61) and determined that the upper 95% confidence limit for the Gelman-Rubin statistic was ≤1.02 in every case.
We calculated Cohen’s f2 (21) to determine the effect sizes of fixed effects (table S4). This process was carried out iteratively using the maximum clade credibility tree or one of the 1000 sampled posterior distribution trees. The f2 of each fixed effect was calculated as
where is the f2 specifically for the effect of interest, Vnull is the residual variance of a null model with only the intercept and random effects, Vab is the residual variance of a model that includes all fixed and random effects, and Va is the residual variance of the model that includes all fixed effects except the effect of interest. In addition, the extent of phylogenetic signal (Pagel’s λ) in each model was calculated as
where Varp is the phylogenetic (random effect) variance, and Varr is the residual variance (62, 63).
To determine how linkage trajectories, as well as the relationship between linkage trajectory and wing aspect ratio, vary according to our principal factors of interest, we again implemented Bayesian generalized linear mixed models (via MCMCglmm). Model fitting was followed by cross-validation for model selection and effect size estimation. Because these models were fit to univariate response variables (linkage trajectory or wing aspect ratio), we instead used inverse-gamma distributed priors with shape and scale of 0.02 for random effect and residual variances. For each model, 10 separate chains of 15,000 iterations were run, with a burn-in of 1500 and thinning interval of 13, resulting in ESS for parameters ~1000.
For analysis of linkage trajectories, the following models were fit
where AM is natural log–transformed manus angle, AE is natural log–transformed elbow angle, M is natural log–transformed body mass, and F is flight behavior grouping. The first listed model corresponds to a null case where neither flight behavior nor body mass explains meaningful variation in linkage trajectories. Subsequent models include body mass, flight behavior, and/or each of their interactive effects with elbow extension. Information on model fits is available in table S2, and specific information on best-fitting models is available in table S3.
For analysis of wing aspect ratio, a similar suite of models was fit
where AR is natural log–transformed wing aspect ratio, AM is natural log–transformed manus angle, M is natural log–transformed body mass, and F is flight behavior group. In each of these models, the AM term represents wing extension according to the linkage trajectory; although both AM and AE describe the motion path achieved by the linkage system, we avoided using both angles in the above models because of their high degree of correlation with each other. Instead, AM was chosen simply because it showed consistently stronger relationships to AR. The first listed model corresponds to a null case where neither flight behavior nor body mass explains meaningful variation in how wing extension along linkage trajectories affects wing aspect ratio. Information on model fits is available in table S2, and specific information on best-fitting models is available in table S3.
To determine how bending or twisting capabilities vary according to our principal factors of interest, we again used Bayesian generalized linear mixed models (albeit via a simpler approach due to the reduction in species measured). We used inverse-gamma distributed priors with shape and scale of 0.02 for random (phylogenetic) effect and residual variances. For each model, 10 separate chains of 15,000 iterations were run, with a burn-in of 1500 and thinning interval of 13, resulting in ESS for parameters ~1000. We first compared models to assess how well body mass variation explained trends in the reduction of bending or twisting capability at full wing extension compared to its maximum value
where C represents bending or twisting capability, S denotes the state (a categorization of value at full extension versus maximum value), and M is natural log–transformed mass. The effect size of body mass was thereafter assessed via Cohen’s f2, iteratively across varying phylogenies.
We then fit models to assess how well flight behavior group (after controlling for body mass) explained the same trends in bending or twisting capacity. To ensure adequate statistical power (56), we restricted our analyses to flight behavior groups in which five or more species were measured (amounting to an analysis of 21 species across four flight groups)
again, where C represents bending or twisting capability, S denotes the state (a categorization of value at full extension versus maximum value), M is natural log–transformed mass, and F is flight behavior group. Information on model fits is available in table S2, and specific information on best-fitting models is available in table S3.
For all best-fitting Bayesian generalized linear mixed models, we computed standardized effects (estimated marginal means) for intercept and/or slope effects for each fixed effect (64). We computed these standardized effects not only to help summarize the effects of our principal factors of interest but also to test linear contrasts among these predictors (Fig. 3).
How accurately data predict flight behavior
To assess how well extension ROM, wing shape, or body mass could be used to predict species’ flight behavior group, we used phylogenetic flexible discriminant analyses (16). Here, the dependent variable was the set of nine flight behavior groups. A separate analysis was carried out for extension ROM, wing shape, and body mass datasets. For the analyses involving extension ROM and wing shape, scores from principal components that accounted for 95% of the parent dataset’s variance were used as predictors. For the analysis using body mass, values were natural log–transformed before being used as predictors. Across all analyses, we used two types of sensitivity analyses of prediction performance. First, the maximum clade credibility tree along with 1000 posterior distribution trees was each used to inform the discriminant analysis. Because we found the analysis invariant to phylogeny, only the maximum clade credibility tree was used thereafter. Second, species’ data were jackknifed: In each of 61 iterations, a single species was held out and discriminant axes were informed by data from the other 60 species. Prediction performance was then determined by the proportion of correct entries in each resulting confusion matrix. To determine how well each empirical dataset performed against random chance, we additionally randomized rows within each dataset and reran all discriminant analyses along with phylogenetic sensitivity analyses. Cohen’s κ, a measure of effect size in empirical versus random performance (17), was then computed for each iteration of this sensitivity analysis using the formula
where po was the prediction performance when empirical data were used, and pe was the prediction performance for when randomized data were used.
In vivo kinematic data collection and analyses
To determine how cadaveric range of motion relates to in vivo flight, we recorded wing kinematics from two species: pigeons (C. livia; n = 1; mass, 613 g) and zebra finches (T. guttata; n = 4; masses, 16 to 17 g). We obtained all birds from breeders and housed them in cages with ad libitum access to seed mix and water under 12-hour/12-hour light/dark cycle. All husbandry and data collection procedures were approved by the Animal Care Committee of the University of British Columbia.
To capture changes in wing shape during flight, up to five points were marked on the dorsal surface of one wing on each bird. We placed a 4-mm-diameter, removable, highly reflective marker at each point to allow for data capture at 240 frames/s via a five-camera tracking system (OptiTrack; NaturalPoint Inc.). Marker placement followed that in our cadaveric study (fig. S2) except the following: (i) Because of difficulties in visualizing the shoulder during flight, point 1 was placed midway along the length of the humerus instead of at the humeral head. (ii) Because of difficulties in marker visualization, point 5 was not used during zebra finch trials, and accordingly, wing twist was not assessed for this species. Each marker weighed 0.0287 g, resulting in 0.1435 g added mass for the pigeon and 0.1148 g added mass for zebra finches. These added masses amount to ~0.03 and ~0.72% of body mass, respectively.
We allowed the pigeon to fly in an arena with dimension of 118 inches by 73 inches by 55 inches, whereas zebra finches flew in a smaller cage with dimensions of 40 inches by 15.5 inches by 14 inches. During all flight trials, we provided seed ad libitum. For each bird, all data collection occurred within 15 to 30 min of wing marking and release within the flight arena or cage.
Range of motion in extension and bending (both species) and twisting (pigeon only) were then calculated for each species via similar methods to those in our cadaveric study. We obtained data only from frames in which all markers were visible. While analyzing zebra finch flights, we pooled data from all four individuals. We then determined whether any of the angular data recorded in vivo for each species fell outside the corresponding range of motion α-hulls we established with cadavers via the inashape3d() function in the alphashape3d package (51).
Phylogenetic signal
Phylogenetic signal was measured for all studied traits using Blomberg’s κ (22) and is summarized in table S5. For all univariate traits, measurements spanned at least two orders of magnitude and were therefore natural log–transformed before analyses except for maximum twisting capability and maximum bending capability. Three multivariate traits were analyzed using Adam’s generalized κ statistic (κmult), which is interpreted in the same way as Blomberg’s κ and is also robust to certain issues in evaluating phylogenetic signal in multidimensional data (23). Two of these traits (extension ROM and wing shape) were analyzed using their respective Procrustes-aligned shape sets. For the third (flight behavior), we computed κmult on the 61 × 12 flight behavior (binary) matrix.
For all measurements of signal, we ran two types of sensitivity analyses. In the first analysis, data were jackknifed: A single species’ trait value was iteratively removed, and signal was recomputed via the influ_physig() function in the R package sensiPhy (65). For ease of understanding, only the maximum clade credibility tree was used. In the second analysis, we varied phylogeny using 1000 trees from the posterior distribution of our BEAST runs to determine the sensitivity of signal to topology. To allow for κmult to be used instead of its univariate counterpart, we wrote customized versions of both the influ_physig() and tree_physig() functions; see influ_physig_kmult() and tree_physig_kmult() functions available in our Figshare repository.
Supplementary Material
Acknowledgments
We thank M. W. Pennell, C. Harvey, D. A. Skandalis, D. A. Somo, and J. Wong for helpful discussion of methods and analyses. S. Bhaidhani and S. Wanniarachchi helped digitize cadaveric ROM videos. S. Senthivasan, J. S. Theriault, E. R. Press, F. Ciocca, J. Wong, and M. Armstrong provided advice and assistance during kinematic data collection. We also thank S. Naeem and two anonymous reviewers for constructive feedback and suggestions during the review process. Funding: This work was funded by the U.S. Air Force Office of Scientific Research (under grant no. FA9550-16-1-0182, titled “Avian-inspired multifunctional morphing vehicles” monitored by B. L. Lee). Author contributions: V.B.B. and D.L.A. designed the study and wrote the manuscript. V.B.B. collected data and performed analyses. I.S. provided materials (specimens) and assisted in determination of flight behaviors. D.L.A. provided conceptual guidance. All authors edited the manuscript. Competing interests: The authors declare that they have no competing interests. Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. All other data and scripts for analyses are available on Figshare at https://doi.org/10.6084/m9.figshare.8856161. Additional data related to this paper may be requested from the authors.
SUPPLEMENTARY MATERIALS
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/5/10/eaaw6670/DC1
Fig. S1. Determination of number of flight behavior groups.
Fig. S2. Placement of markers for capture of ROM from videography.
Fig. S3. Determination of out-of-plane motion from angular data.
Table S1. Fossil and biogeographic information used for divergence time estimation in BEAST.
Table S2. Comparisons of Bayesian generalized linear mixed models.
Table S3. Descriptions of best-fitting Bayesian generalized linear mixed models.
Table S4. Distributions of effect sizes in mixed models as phylogeny varies.
Table S5. Distributions of phylogenetic signal.
Movie S1. Sample of video used to determine extension ROM in the belted kingfisher (Megaceryle alcyon) from one of the three cameras used.
Movie S2. Sample of video used to determine linkage trajectory in the spruce grouse (Falcipennis canadensis).
Movie S3. Sample of video used to determine bending and twisting capability in the mallard (Anas platyrhynchos).
Data S1. Static morphological data from specimens.
Data S2. List of genetic sequences sampled.
Data S3. Maximum likelihood tree.
Data S4. Bayesian maximum clade credibility tree.
Data S5. Flight behavior matrix.
REFERENCES AND NOTES
- 1.J. M. V. Rayner, Form and Function in Avian Flight (Current Ornithology series, 1988), vol. 5, pp. 1–66. [Google Scholar]
- 2.Kruyt J. W., van Heijst G. F., Altshuler D. L., Lentink D., Power reduction and the radial limit of stall delay in revolving wings of different aspect ratio. J. R. Soc. Interface 12, 20150051 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wang X., Clarke J. A., The evolution of avian wing shape and previously unrecognized trends in covert feathering. Proc. Biol. Sci. 282, 20151935 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lentink D., Müller U. K., Stamhuis E. J., de Kat R., van Gestel W., Veldhuis L. L., Henningsson P., Hedenström A., Videler J. J., van Leeuwen J. L., How swifts control their glide performance with morphing wings. Nature 446, 1082–1085 (2007). [DOI] [PubMed] [Google Scholar]
- 5.Harvey C., Baliga V. B., Lavoie P., Altshuler D. L., Wing morphing allows gulls to modulate static pitch stability during gliding. J. R. Soc. Interface 16, 20180641 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Prum R. O., Berv J. S., Dornburg A., Field D. J., Townsend J. P., Lemmon E. M., Lemmon A. R., A comprehensive phylogeny of birds (Aves) using targeted next-generation DNA sequencing. Nature 526, 569–573 (2015). [DOI] [PubMed] [Google Scholar]
- 7.Jetz W., Thomas G. H., Joy J. B., Hartmann K., Mooers A. O., The global diversity of birds in space and time. Nature 491, 444–448 (2012). [DOI] [PubMed] [Google Scholar]
- 8.Bouckaert R., Heled J., Kühnert D., Vaughan T., Wu C. H., Xie D., Suchard M. A., Rambaut A., Drummond A. J., BEAST 2: A software platform for Bayesian evolutionary analysis. PLOS Comput. Biol. 10, e1003537 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.I. J. Lovette, J. W. Fitzpatrick, Handbook of Bird Biology (Wiley & Sons, ed. 3, 2016). [Google Scholar]
- 10.Tibshirani R., Walther G., Hastie T., Estimating the number of clusters in a data set via the gap statistic. J. R. Stat. Soc. Series B. Stat. Methodol. 63, 411–423 (2001). [Google Scholar]
- 11.Kambic R. E., Roberts T. J., Gatesy S. M., 3-D range of motion envelopes reveal interacting degrees of freedom in avian hind limb joints. J. Anat. 231, 906–920 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bonhomme V., Picq S., Gaucherel C., Claude J., Momocs: Outline analysis using R. J. Stat. Softw. 56, 1–24 (2014). [Google Scholar]
- 13.Revell L. J., Size-correction and principal components for interspecific comparative studies. Evolution 63, 3258–3268 (2009). [DOI] [PubMed] [Google Scholar]
- 14.Hadfield J. D., MCMC methods for multi-response generalized linear mixed models: The MCMCglmm R package. J. Stat. Softw. 33, 1–22 (2010).20808728 [Google Scholar]
- 15.Hadfield J. D., Nakagawa S., General quantitative genetic methods for comparative biology: Phylogenies, taxonomies and multi-trait models for continuous and categorical characters. J. Evol. Biol. 23, 494–508 (2010). [DOI] [PubMed] [Google Scholar]
- 16.Motani R., Schmitz L., Phylogenetic versus functional signals in the evolution of form-function relationships in terrestrial vision. Evolution 65, 2245–2257 (2011). [DOI] [PubMed] [Google Scholar]
- 17.Cohen J., A coefficient of agreement for nominal scales. Educ. Psychol. Meas. 20, 37–46 (1960). [Google Scholar]
- 18.Fisher H. I., Bony mechanism of automatic flexion and extension in the pigeon’s wing. Science 126, 446 (1957).13467223 [Google Scholar]
- 19.Kruyt J. W., Quicazán-Rubio E. M., van Heijst G. F., Altshuler D. L., Lentink D., Hummingbird wing efficacy depends on aspect ratio and compares with helicopter rotors. J. R. Soc. Interface 11, 570–581 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Vazquez R. J., Functional osteology of the avian wrist and the evolution of flapping flight. J. Morphol. 211, 259–268 (1992). [DOI] [PubMed] [Google Scholar]
- 21.J. Cohen, Statistical Power Analysis for the Behavioral Sciences (Routledge, 1988). [Google Scholar]
- 22.Blomberg S. P., Garland T. Jr., Ives A. R., Testing for phylogenetic signal in comparative data: Behavioral traits are more labile. Evolution 57, 717–745 (2003). [DOI] [PubMed] [Google Scholar]
- 23.Adams D. C., A generalized K statistic for estimating phylogenetic signal from shape and other high-dimensional multivariate data. Syst. Biol. 63, 685–697 (2014). [DOI] [PubMed] [Google Scholar]
- 24.Revell L. J., Harmon L. J., Collar D. C., Phylogenetic signal, evolutionary process, and rate. Syst. Biol. 57, 591–601 (2008). [DOI] [PubMed] [Google Scholar]
- 25.Alfaro M. E., Bolnick D. I., Wainwright P. C., Evolutionary dynamics of complex biomechanical systems: An example using the four-bar mechanism. Evolution 58, 495–503 (2004). [PubMed] [Google Scholar]
- 26.Kamilar J. M., Cooper N., Phylogenetic signal in primate behaviour, ecology and life history. Philos. Trans. R. Soc. Lond. B Biol. Sci. 368, 20120341 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Vazquez R. J., Functional anatomy of the pigeon hand (Columba livia): A muscle stimulation study. J. Morphol. 226, 33–45 (1995). [DOI] [PubMed] [Google Scholar]
- 28.Usherwood J., The aerodynamic forces and pressure distribution of a revolving pigeon wing. Exp. Fluids. 46, 991–1003 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Galantai V., Sofla A., Meguid S., Tan K., Yeo W., Bio-inspired wing morphing for unmanned aerial vehicles using intelligent materials. Int. J. Mech. Mater. Des. 8, 71–79 (2012). [Google Scholar]
- 30.J. Valasek, Morphing Aerospace Vehicles and Structures (Wiley & Sons Ltd., 2012). [Google Scholar]
- 31.Chin D. D., Matloff L. Y., Stowers A. K., Tucci E. R., Lentink D., Inspiration for wing design: How forelimb specialization enables active flight in modern vertebrates. J. R. Soc. Interface 14, 20170240 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ward T. A., Rezadad M., Fearday C. J., Viyapuri R., A review of biomimetic air vehicle research: 1984-2014. Int. J. Micro Air Veh. 7, 375–394 (2015). [Google Scholar]
- 33.Kearse M., Moir R., Wilson A., Stones-Havas S., Cheung M., Sturrock S., Buxton S., Cooper A., Markowitz S., Duran C., Thierer T., Ashton B., Meintjes P., Drummond A., Geneious Basic: An integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 28, 1647–1649 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lanfear R., Frandsen P. B., Wright A. M., Senfeld T., Calcott B., PartitionFinder 2: New methods for selecting partitioned models of evolution for molecular and morphological phylogenetic analyses. Mol. Biol. Evol. 34, 772–773 (2016). [DOI] [PubMed] [Google Scholar]
- 35.Vaidya G., Lohman D. J., Meier R., SequenceMatrix: Concatenation software for the fast assembly of multi-gene datasets with character set and codon information. Cladistics 27, 171–180 (2011). [DOI] [PubMed] [Google Scholar]
- 36.Stamatakis A., RAxML-VI-HPC: Maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics 22, 2688–2690 (2006). [DOI] [PubMed] [Google Scholar]
- 37.F. Gill, Ornithology (W. H. Freeman and company, ed. 3, 2007). [Google Scholar]
- 38.S. Podulka, R. W. Rohrbaugh Jr., R. Bonney, Handbook of Bird Biology (Princeton Univ. Press, 2004). [Google Scholar]
- 39.J. J. Videler, Avian Flight (Oxford Univ. Press, 2005). [Google Scholar]
- 40.Bruderer B., Boldt A., Flight characteristics of birds: I. Radar measurements of speeds. Ibis 143, 178–204 (2001). [Google Scholar]
- 41.Bruderer B., Peter D., Boldt A., Liechti F., Wing-beat characteristics of birds recorded with tracking radar and cine camera. Ibis 152, 272–291 (2010). [Google Scholar]
- 42.Alerstam T., Rosén M., Backman J., Ericson P. G., Hellgren O., Flight speeds among bird species: Allometric and phylogenetic effects. PLOS Biol. 5, e197 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Watanabe Y., Flight mode affects allometry of migration range in birds. Ecol. Lett. 19, 907–914 (2016). [DOI] [PubMed] [Google Scholar]
- 44.Gower J. C., A general coefficient of similarity and some of its properties. Biometrics 27, 857–871 (1971). [Google Scholar]
- 45.Murtagh F., Legendre P., Ward’s hierarchical agglomerative clustering method: Which algorithms implement Ward’s criterion? J. Classif. 31, 274–295 (2014). [Google Scholar]
- 46.Theriault D. H., Fuller N. W., Jackson B. E., Bluhm E., Evangelista D., Wu Z., Betke M., Hedrick T. L., A protocol and calibration method for accurate multi-camera field videography. J. Exp. Biol. 217, 1843–1848 (2014). [DOI] [PubMed] [Google Scholar]
- 47.Hedrick T. L., Software techniques for two- and three-dimensional kinematic measurements of biological and biomimetic systems. Bioinspir. Biomim. 3, 034001 (2008). [DOI] [PubMed] [Google Scholar]
- 48.Abramoff M. D., Magalhães P. J., Ram S. J., Image processing with Image J. Biophotonics Int. 11, 36–42 (2004). [Google Scholar]
- 49.J. Claude, Morphometrics with R (Springer, 2008). [Google Scholar]
- 50.Rohlf F. J., Slice D. E., Extensions of the Procrustes method for the optimal superimposition of landmarks. Syst. Zool. 39, 40–59 (1990). [Google Scholar]
- 51.T. Lafarge, B. Pateiro-Lopez, alphashape3d: Implementation of the 3D Alpha-Shape for the Reconstruction of 3D Sets from a Point Cloud. R package version 1.3 (2017).
- 52.Nguyen D., Widrow B., Improving the learning speed of 2-layer neural networks by choosing initial values of the adaptive weights. Proc. Int. Jt. Conf. Neural Netw. 3, 21–26 (1990). [Google Scholar]
- 53.MacKay D. J. C., Bayesian interpolation. Neural Comput. 4, 415–447 (1992). [Google Scholar]
- 54.Kuhn M., Building predictive models in R using the caret package. J. Stat. Softw. 28, 1–26 (2008).27774042 [Google Scholar]
- 55.J. Hadfield, MCMCglmm 2.24: MCMC Generalised Linear Mixed Models (2016).
- 56.Faul F., Erdfelder E., Lang A.-G., Buchner A., G*Power 3: A flexible statistical power analysis program for the social, behavioral, and biomedical sciences. Behav. Res. Meth. 39, 175–191 (2007). [DOI] [PubMed] [Google Scholar]
- 57.Spiegelhalter D. J., Best N. G., Carlin B. P., van der Linde A., The deviance information criterion: 12 years on (with discussion). J. R. Stat. Soc. Series B. Stat. Methodol. 64, 485–493 (2014). [Google Scholar]
- 58.Roberts D. R., Bahn V., Ciuti S., Boyce M. S., Elith J., Guillera-Arroita G., Hauenstein S., Lahoz-Monfort J. J., Schröder B., Thuiller W., Warton D. I., Wintle B. A., Hartig F., Dormann C. F., Cross-validation strategies for data with temporal, spatial, hierarchical, or phylogenetic structure. Ecography 40, 913–929 (2017). [Google Scholar]
- 59.T. Hastie, R. Tibshirani, J. Friedman, The Elements of Statistical Learning (Springer, 2001). [Google Scholar]
- 60.Nakagawa S., Schielzeth H., A general and simple method for obtaining R2 from generalized linear mixed-effects models. Methods Ecol. Evol. 4, 133–142 (2013). [Google Scholar]
- 61.Gelman A., Rubin D. B., Inference from iterative simulation using multiple sequences. Stat. Sci. 7, 457–472 (1992). [Google Scholar]
- 62.Housworth E. A., Martins E. P., Lynch M., The phylogenetic mixed model. Am. Nat. 163, 84–96 (2004). [DOI] [PubMed] [Google Scholar]
- 63.Hansen T. F., Orzack S. H., Assessing current adaptation and phylogenetic inertia as explanations of trait evolution: The need for controlled comparisons. Evolution 59, 2063–2072 (2005). [PubMed] [Google Scholar]
- 64.Searle S. R., Speed F. M., Milliken G. A., Population marginal means in the linear model: An alternative to least squares means. Am. Stat. 34, 216–221 (1980). [Google Scholar]
- 65.Paterno G. B., Penone C., Werner G. D. A., sensiPhy: An R-package for sensitivity analysis in phylogenetic comparative methods. Methods Ecol. Evol. 2018, 1461–1467 (2018). [Google Scholar]
- 66.You H., Lamanna M. C., Harris J. D., Chiappe L. M., O’connor J., Ji S. A., Lü J. C., Yuan C. X., Li D. Q., Zhang X., Lacovara K. J., Dodson P., Ji Q., A nearly modern amphibious bird from the Early Cretaceous of northwestern China. Science 312, 1640–1643 (2006). [DOI] [PubMed] [Google Scholar]
- 67.Micklich N., Parin N., The fish fauna of Frauenweiler (Middle Oligocene, Rupelian; Germany): First results of a review. Inst. Español. Oce. Pub. Esp. 21, 129–148 (1996). [Google Scholar]
- 68.Trunkó L., Munk W., Geologische Beobachtungen in drei tertiären Aufschlußkomplexen im Randbereich des Mittleren Rheingrabens. Carolinea 56, 9–28 (1998). [Google Scholar]
- 69.Ksepka D. T., Clarke J., Phylogenetically vetted and stratigraphically constrained fossil calibrations within Aves. Palaeontol. Electronica 18, 1–25 (2015). [Google Scholar]
- 70.C. Cavelier, Synthèse Géologique du Sud-Est de la France (Mémoires BRGM France, 1986), vol. 25. [Google Scholar]
- 71.Mayr G., Phylogeny of early tertiary swifts and hummingbirds (Aves: Apodiformes). Auk 120, 145–151 (2003). [Google Scholar]
- 72.Harrison C. J. O., A revision of the fossil swifts (Vertebrata, Aves, suborder Apodi), with descriptions of three new genera and two new species. Meded. Werkgr. Tert. Kwart. Geol. 21, 157–177 (1984). [Google Scholar]
- 73.Heilmann-Clausen C., Gersner F., Lithostratigraphy and depositional environments in the Upper Paleocene and Eocene of Denmark. Bull. Geol. Soc. Den. 33, 293–323 (1984). [Google Scholar]
- 74.Schnetler K. I., Heilmann-Clausen C., The molluscan fauna of the Eocene Lillebælt Clay, Denmark. Cainozoic Res. 8, 41–99 (2011). [Google Scholar]
- 75.Sheldon E., Gravesen P., Nøhr-Hansen H., Geology of the Femern Bælt area between Denmark and Germany. Geol. Survey of Denmark and Greenland Bull. 26, 13–16 (2012). [Google Scholar]
- 76.Mertz D. F., Renne P. R., Wuttke M., Mödden C., A numerically calibrated reference level (MP28) for the terrestrial mammal-based biozonation of the European Upper Oligocene. Int. J. Earth Sci. 96, 353–361 (2007). [Google Scholar]
- 77.Smith M. E., Caroll A. R., Singer B. S., Synoptic reconstruction of a major ancient lake system: Eocene Green River Formation, western United States. GSA Bull. 120, 54–84 (2008). [Google Scholar]
- 78.Clarke J. A., Tambussi C. P., Noriega J. I., Erickson G. M., Ketcham R. A., Definitive fossil evidence for the extant avian radiation in the Cretaceous. Nature 43, 305–308 (2005). [DOI] [PubMed] [Google Scholar]
- 79.Bertelli S., Lindow B. E. K., Dyke G. J., Chiappe L. M., A well-preserved ‘charadriiform-like’ fossil bird from the Early Eocene Fur formation of Denmark. Palaeontology 53, 507–531 (2010). [Google Scholar]
- 80.Dyke G. J., van Tuinen M., The evolutionary radiation of modern birds (Neornithes): Reconciling molecules, morphology and the fossil record. Biol. J. Linn. Soc. 141, 153–177 (2004). [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/5/10/eaaw6670/DC1
Fig. S1. Determination of number of flight behavior groups.
Fig. S2. Placement of markers for capture of ROM from videography.
Fig. S3. Determination of out-of-plane motion from angular data.
Table S1. Fossil and biogeographic information used for divergence time estimation in BEAST.
Table S2. Comparisons of Bayesian generalized linear mixed models.
Table S3. Descriptions of best-fitting Bayesian generalized linear mixed models.
Table S4. Distributions of effect sizes in mixed models as phylogeny varies.
Table S5. Distributions of phylogenetic signal.
Movie S1. Sample of video used to determine extension ROM in the belted kingfisher (Megaceryle alcyon) from one of the three cameras used.
Movie S2. Sample of video used to determine linkage trajectory in the spruce grouse (Falcipennis canadensis).
Movie S3. Sample of video used to determine bending and twisting capability in the mallard (Anas platyrhynchos).
Data S1. Static morphological data from specimens.
Data S2. List of genetic sequences sampled.
Data S3. Maximum likelihood tree.
Data S4. Bayesian maximum clade credibility tree.
Data S5. Flight behavior matrix.


