SUMMARY
Ionizing radiation (IR) is a mutagen that promotes tumorigenesis in multiple exposure contexts. One severe consequence of IR is the development of second malignant neoplasms (SMNs), a radiotherapy-associated complication in survivors of cancers, particularly pediatric cancers. SMN genomes are poorly characterized and the influence of genetic background on genotoxin-induced mutations has not been examined. Using our mouse models of SMNs, we performed whole exome sequencing of neoplasms induced by fractionated IR in wildtype and Nf1 mutant mice. Using non-negative matrix factorization, we identified mutational signatures that did not segregate by genetic background or histology. Copy number analysis revealed recurrent chromosomal alterations and differences in copy number that were background dependent. Pathway analysis identified enrichment of non-synonymous variants in genes responsible for cell assembly and organization, cell morphology and cell function and maintenance. In this model system, ionizing radiation and Nf1 heterozygosity each exerted distinct influences on the mutational landscape.
INTRODUCTION
Therapy-induced malignancies, or second malignant neoplasms (SMNs) are severe late complications developing in survivors of childhood cancers (Crump and Hodgson, 2009). SMNs develop after prior exposure to mutagenic agents such as radiotherapy and some chemotherapies, and are a major cause of morbidity and mortality in childhood cancer survivors (Armstrong et al., 2009a; Armstrong et al., 2009b; Bhatia and Sklar, 2002; Meadows et al., 2009). Most SMNs are associated with radiotherapy, which typically delivers fractionated, focal treatment using ionizing radiation (IR) (Bhatia and Sklar, 2002). While genotoxin exposure clearly drives SMN development, the genetic basis for this late complication is not understood.
IR is also a well-recognized and relevant mutagen in numerous aspects of modern human life, and diverse exposures include clinical radiotherapy, occupational exposure to nuclear-powered devices, nuclear fallout/waste, and diagnostic medical imaging. It is thus important to characterize the genetic consequences of IR exposure in the context of malignancy induction, a complication with significant implications for both the affected individual and society.
Next generation sequencing has permitted the characterization of genomes and mutational processes in diverse cancers, generating insights into the genomic events in tumor formation and identifying genetic mechanisms directly responsible for many human cancers. Known mutagens, such as ultraviolet radiation (UV) and tobacco, have been shown to produce characteristic mutational signatures (Alexandrov et al., 2013a). Mutagen dose, length of exposure, timing in relation to an individual’s lifespan, and genetic background are important variables influencing mutagenicity, however studies of mutagen-associated human cancers generally cannot precisely define these variables or attribute mechanisms to a specific variable. In human SMNs, IR localization, dosing and timing are clinically defined and differ greatly between patients. This heterogeneity can complicate characterizations of IR-induced tumorigenesis.
The genome-wide consequences of exposure to IR, and whether malignancies induced by IR-exposure share large or small-scale mutational motifs, are not known. IR produces multiple types of DNA injury, including double-strand breaks (Yong et al., 2014), which differ from UV-induced genetic lesions, and thus IR-induced malignancies might possess a mutational landscape that is distinguishable from those of UV or other mutagens.
In prior studies we developed experimental mouse models of SMNs by delivering focal fractionated radiation similar to that used in clinical radiotherapy to both Nf1 mutant and wildtype mice. Irradiated mice of both genotypes developed diverse solid and hematologic malignancies consistent with radiotherapy-induced SMNs that arise in irradiated cancer survivors (Choi et al., 2012; Nakamura et al., 2011). These neoplasms are unique biospecimen because in contrast to clinical SMNs samples, both the genetic background of the mouse model and the mutagen (IR) exposure were well-defined. To characterize the genomes of these malignancies we performed whole exome sequencing, comparing the exomes of malignancies arising in wildtype and Nf1 mutant backgrounds to determine whether germline tumor suppressor loss, present in the Nf1 heterozygous background, influences the mutational spectrum. These data represent in-depth genomic characterization of malignancies initiated by fractionated focal IR recapitulating set-up and dosimetry found in radiotherapy. Distinct, reproducible mutational signatures characterize malignancies arising after IR. The genetic background influenced copy number alterations in IR-induced malignancies, with the genomes of tumors arising in wildtype mice having significantly more copy number gains compared to tumors arising in Nf1+/− mice. These data suggest distinct contributions of IR and background in the mutational landscape of IR-induced malignancies.
RESULTS
Mutation frequency in tumors generated by fractionated IR
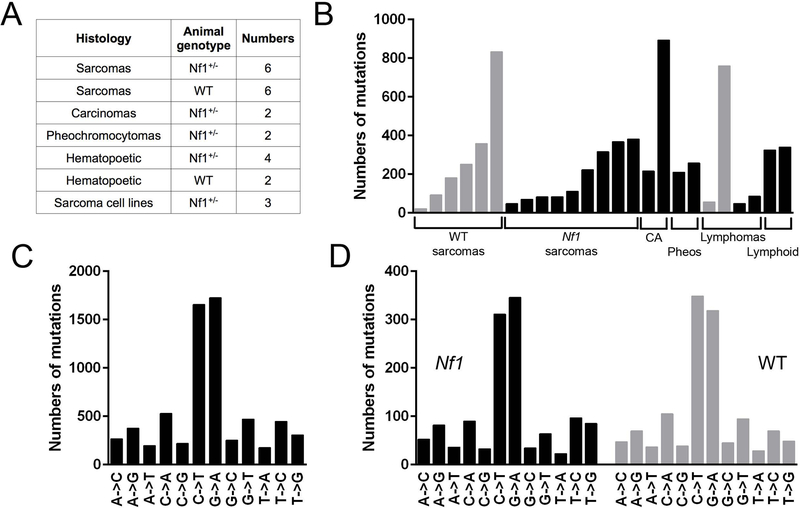
We sequenced 25 malignancies arising from our mouse models of IR-induced SMNs as diverse histologies, including: soft tissue sarcomas, squamous cell carcinoma, mammary carcinomas, pheochromocytomas and hematopoietic malignancies (Figure 1A). All malignancies were induced by focal, fractionated IR as previously described (Choi et al., 2012; Nakamura et al., 2011). Briefly, mice were irradiated at 5–8 weeks of age, targeted to the abdominal wall, specifically the mammary glands. In addition to anatomic targeting replicating that received by patients, the mice received identical dosing schema, which consisted of 3 Gy daily fractions (5 days a week) to total doses of 30 Gy. We sequenced malignancies from 19 mice, with 15 malignancies arising in F1 Nf1 mutant mice and 7 malignancies arising in wildtype mice. Three tumor cell lines established from three primary tumors (matched primary tumor – cell line pairs) were also sequenced. 158-fold mean exome coverage was obtained. A total of 6,623 single nucleotide variants (SNVs) were identified, of which 4,633 were non-synonymous for an average of 265 total SNVs and 184 non-synonymous SNVs/sample (Table S1). Histologies varied with regard to the numbers of somatic non-synonymous SNVs detected (range, 22 – 594), with all classes of malignant histologies demonstrating similarly variable numbers (Figure 1B). IR-induced sarcomas from wildtype mice had on average more SNVs (mean, 290; range, 22 – 834) than sarcomas from Nf1+/− mice (mean, 128; range, 48 – 368) (p-value < 0.00001 by z-test for rate ratios). Comparable averages for all cancer types were 320 and 239 (p-value < 0.00001). The genetic backgrounds did not differ in relative frequency of synonymous and non-synonymous SNVs (Table S2). Most nucleotide substitutions were C→T or G→A transition mutations (Figure 1C). These substitutions predominated in both synonymous and non-synonymous variants (Table S3).
Figure 1. Numbers and types of substitutions in sequenced samples.
Summary of tumors sequenced and the frequencies of mutations seen. A. Types and numbers of primary radiation-induced malignancies from wildtype and Nf1 mutant mice that were analyzed by whole exome sequencing. B. Synonymous and non-synonymous SNVs in each sample, malignancies from Nf1 mutant mice in black, wildtype in grey (CA – carcinomas, Pheos – Pheochromocytomas, Lymphoid – Lymphoid malignancies). C. Frequencies of specific types of base substitutions in SNVs pooled from all samples. D. Composition of SNVs in Nf1 mutant and wildtype sarcomas, corrected for total number of mutations. See also Tables S1–4.
The cohort composition precluded a robust comparison of SNV numbers between each of the different specific histologies. However, because sarcomas arose frequently in our mouse models (Choi et al., 2012; Nakamura et al., 2011) and are well-recognized SMNs after radiotherapy (Henderson et al., 2007; Tucker et al., 1987), we were able to carry out more in-depth analyses for this histology (Figure 1A). Both sarcomas derived from Nf1 mutant and wildtype mice demonstrated similar types and frequencies of base substitutions (Figure 1D), suggesting that the predominant types of base substitutions were not Nf1-dependent. Dinucleotide substitutions, common in UV-associated malignancies (Alexandrov et al., 2013a), were relatively uncommon in our samples (Table S4). Non-synonymous SNVs can have diverse consequences, and we used SNPEff software (Cingolani et al., 2012) to summarize the predicted impact of non-synonymous somatic variants arising in our cohort of malignancies (Table 1). This analysis revealed that most variants were missense variants of medium impact, followed by stop gains classified as high impact.
Table 1. Predicted impact of variants.
SNPEff software was used to score the somatic substitutions for predicted biologic impact based on the resulting codon change. Predicted impact is organized into High, Medium and Low impact.
| High impact | Medium impact | Low impact | |||
|---|---|---|---|---|---|
| Stop gained | 245 | Missense variant | 4326 | Mature miRNA variant | 2 |
| Stop lost | 6 | Initiator codon variant | 9 | ||
| Splice acceptor variant | 4 | Stop retained variant | 2 | ||
| Splice donor variant | 1 | Coding sequence variant | 1 | ||
| Total | 256 | 4338 | 2 | ||
Mutational signature analysis of IR-induced malignancies
Mutational signature analyses have not previously been performed for IR-induced tumors or NF1-associated tumors. Patterns of nucleotide substitutions in tumor genomes may reflect specific mutational mechanisms (Alexandrov et al., 2013a; Alexandrov et al., 2013b; Wei et al., 2011). In addition, mutational asymmetries between transcribed and untranscribed strands can result from intrinsic biases in mutation and repair mechanisms (Green et al., 2003) and have been shown to be associated with specific tumor types (Rubin and Green, 2009; Sjoblom et al., 2006; Stephens et al., 2005), although the precise molecular mechanisms responsible for the enrichment of specific mutational patterns are not well-understood. IR-induced tumors might be hypothesized to display unique mutational signatures on the basis of the distinct type of DNA damage associated with IR. We applied non-negative matrix factorization (NMF), as developed at the Wellcome Trust Sanger Institute (WTSI) (Alexandrov et al., 2013a; Alexandrov et al., 2013b) to whole exome sequencing data from 25 malignancies. Six possible substitutions are considered, based on the pyrimidine in the reference position and including the proximal sequence context (one nucleotide 5’ and 3’). We also performed separate analyses of non-synonymous substitutions only versus combined non-synonymous and synonymous substitutions. Substitutions were also analyzed by whether the altered pyrimidine was on the transcribed or untranscribed strand.
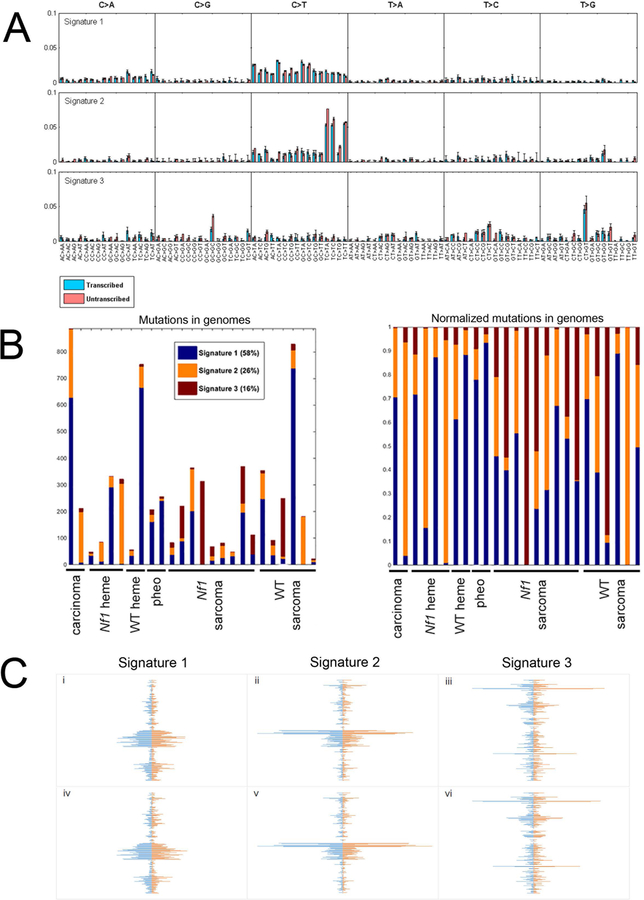
This analysis extracted three stable mutational signatures, as assessed by plotting Signature Stability and the Average Frobenius Reconstruction Error, measures introduced by WTSI to assess quality features of NMF (Alexandrov et al., 2013b)(Figure S1). Figure 2A shows the distribution of the 6 possible mutation types in the three signatures. Each sub-graph represents one substitution (e.g., A→C when A in the reference genome is mutated to C in the sample). The bars within each sub-graph include the nucleotides in the reference genome on either side of the mutation location and strand (transcribed versus untranscribed). This analysis reveals that the incidence of specific substitutions varies in relation to the flanking nucleotides, or 5’ and 3’ neighbors. Signature 1 harbored the most variants of the three signatures and is characterized by C→T substitutions distinguishable from Signatures 2 and 3 because neither the flanking 5’ nor 3’ base significantly influence the substitution frequency (Figure 2A). Signature 2 is enriched for C→T substitutions when the flanking 5’ base is thymine (Figure 2A). Signature 3 is notable for a significantly increased frequency of discrete G(C→G)C and C(T→G)T substitutions.
Figure 2. Mutation signature analysis.
Mutation signature analysis was performed on non-synonymous and synonymous substitutions in 25 samples. A. Three discrete mutational signatures were identified. The plots show the distribution of the 6 mutation types defined by the pyrimidine base in each signature, as inferred from the NMF procedure. Each sub-graph within a signature represents one substitution (e.g., A→C when A in the reference genome is mutated to C in the sample). The bars within each sub-graph include the nucleotides in the reference genome on either side of the mutation location. All trinucleotide combinations are subdivided as to whether the pyrimidine is on the transcribed (blue) or untranscribed (pink) strand. B. The distribution of each of the three signatures in each of the 25 radiation-induced tumors is shown. The left panel plots the numbers of substitutions comprising each signature. The right panel displays a normalized plot. C. Spindle plots depict the similarity of signatures derived from different subsets of the data. Horizontal lines indicate the coefficients for 192 mutation types (including substitution based on pyrimidine reference, strand and flanking nucleotides) of the three signatures, sorted from bottom to top in the same order as panel A is sorted left to right. All 3 panels have the same 3 figures on the left, 3 signatures extracted from 25 samples using both synonymous and non-synonymous mutations. i-iii compare the three signatures extracted using both synonymous and non-synonymous SNVs (left, blue) versus signatures extracted using only non-synonymous SNVs (right, red). iv-vi compare the three signatures for non-synonymous and synonymous SNVs using 25 samples (left, blue) versus the three signatures for non-synonymous and synonymous SNVs in 22 samples (excluding the three most mutated samples)(right, red). See also Figures S1 and S2, and Tables S5–7.
The mutational signature exposures, or proportion of each neoplasm’s mutations that are represented in one of the extracted mutational signatures, is shown in Figure 2B. All three signatures were represented in most of the neoplasms, although to variable extents. There was no enrichment of a single mutational signature by either histologic type or genetic background. The mutational signature exposures failed to differentiate by the number of SNVs present in a sample.
To determine whether the three mutational signatures were driven disproportionately by those samples harboring the greatest numbers of substitutions, NMF analysis was performed after excluding the three most mutated samples, leaving 22 total (Figure S2). This yielded the same three mutation signatures, indicating that these signatures were not driven by hypermutated samples.
Restricting the analysis to non-synonymous substitutions (4,633 variants in 25 samples) also yielded the same three signatures, and restricting to synonymous substitutions (1,990 variants in 25 samples) was unable to reliably extract the third, but robustly produced the first and second signatures, suggesting that the mutational signatures did not discriminate between non-synonymous and synonymous variants. Correlation coefficients were calculated for signatures generated from non-synonymous SNVs only and both non-synonymous and synonymous SNVs, demonstrating that for each mutational signature, the correlation coefficients are greater than 0.9 between these analyses of non-synonymous only and both non-synonymous and synonymous SNVs (Table S5). Spindle graphs (Figure 2C) display the similarity among the coefficients with non-synonymous only (blue) and both non-synonymous and synonymous (orange) analyses.
In order to compare the 3 signatures derived from our mouse tumors with signatures previously described by WTSI for human cancers (Alexandrov et al., 2013a), the 3 signatures were normalized by the trinucleotide frequencies in the mouse genome (Frenkel et al., 2011). The coefficients of the 22 signatures reported by WTSI were correlated with the coefficients of our 3 signatures (Table S6). Our signatures 1 and 2 were not highly correlated with any of the WTSI (maximum correlations of 0.685 and 0.593). Signature 3, on the other hand, correlated 0.89 with WTSI signature 17. WTSI describe this signature as found in colorectal, liver, lymphoma and stomach cancers (Alexandrov et al., 2013a). While 0.89 is a fairly strong correlation, it should be noted that the signatures considered as separate in the WTSI analysis have correlations as high as 0.90 (Table S6).
These signatures also demonstrated a transcriptional strand bias for specific substitution contexts (Table S7). Fisher’s Exact Test returned a p-value of <0.000001 for transcribed/untranscribed strand by mutation type. Specific patterns of substitutions (G(T→C)T, T(C→A)G, T(C→A)T, T(T→G)C, T(T→G)G (p< 0.01)) were highly significant for preferentially involving the transcribed strand.
Copy number analysis
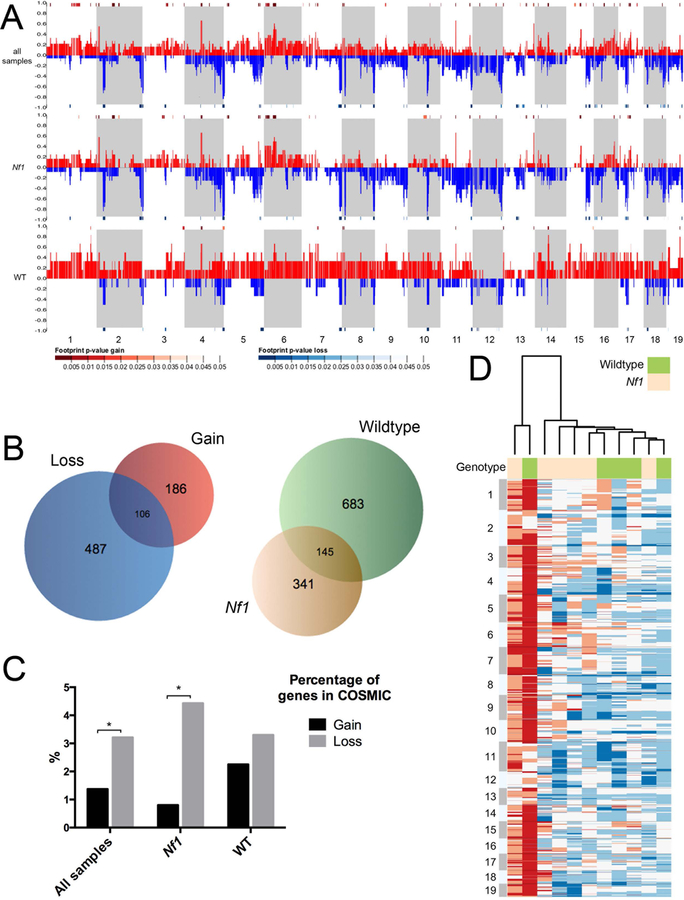
Copy number variations (CNV) were detected employing Control-FREEC (Boeva et al., 2012), using standard parameters and the germline sequencing as a control. We observed both large-scale and focal copy number changes, as well as tumors with relatively normal ploidy, across all histologies (Figure 2A, Table S8). To compare patterns of copy number alterations between sarcomas developing in Nf1 mutant or wildtype mice, we generated a dendrogram showing sample relatedness when clustering the data by Ward’s method (squared Euclidean distance, variables normalized using z-scores) using the WGCNA R software package (Figure 2B) (Langfelder and Horvath, 2008). Apart from one wildtype sarcoma showing almost genome-wide copy number gain, sarcomas from the wildtype background cluster (Figure 2D). Sarcomas arising in Nf1 mutant mice also cluster, suggesting an influence of Nf1 heterozygous background on CNV in IR-induced sarcomas.
We also used Control-FREEC to analyze CNVs in the group as a whole, in pooled Nf1-mutant derived tumors only and pooled wildtype tumors only (Figure 2A). Overall, Nf1 mutant-derived samples showed far more copy number losses than gains compared to wildtype-derived samples (29% versus 11%), while wildtype derived tumors showed the opposite pattern, harboring more gains than losses (33% versus 8%) (Table S9). This observation holds true when considering sarcomas only (Table S9). One notable area of copy number loss in Nf1 mutant-derived samples is found in chromosome 11 (Figure 2B, Table S8), whose loss spans the Nf1 and Trp53 genes. In earlier work we found this area to be involved by loss of heterozygosity involving the wildtype Nf1 allele (Choi et al., 2012).
We assessed whether genes most frequently affected by copy number change were similarly altered between sarcomas arising in different genotypes (Figure 2C). Only a small fraction of genes involved both gain and loss in the pooled samples (Figure 2C), suggesting that these genes are unlikely to function as cancer drivers. Interestingly, sarcomas arising in Nf1-mutant or wildtype backgrounds shared a significant fraction of genes with copy number alterations. To investigate whether known cancer-causing mutations are in regions with known CNVs, we calculated the percentage of genes in the COSMIC database that involve either copy number gain or loss in all sarcomas, Nf1 mutant sarcomas, or wildtype sarcomas (Figure 2D). We saw a significantly higher percentage of COSMIC annotated genes that were affected by copy number loss than gain, suggesting that loss may drive tumorigenesis more so than gain.
To determine whether the mutational rate changed significantly across the exome, and to determine whether mutational hotspots and copy number variation correlate in general, we segmented the exome into windows of 15,000 base increments and plotted number of mutations with copy number variations as estimated by Control-FREEC software. The two were not related overall and failed to demonstrate a correlation between areas of copy number alterations and SNVs. Correlations calculated over windows within each chromosome were < 0.2 for all chromosomes and < 0.1 for all but two, indicating that somatic SNVs did not co-localize with areas of copy number alterations.
Pathway analysis
To determine whether sarcomas from mutant and wildtype backgrounds utilized common or distinct pathways for tumorigenesis we used Ingenuity Variant Analysis software (www.Ingenuity.com/variants) to identify recurrently mutated pathways (Table 2). We also analyzed the tumor exomes for recurrently mutated genes (Table 3). Sarcomas from both genetic backgrounds shared mutational involvement of pathways influencing cellular assembly and organization, and cellular function and maintenance (Table 2). Interestingly, cell cycle and cell signaling pathways were among the most significantly mutated pathways in sarcomas from wildtype mutant mice, suggesting a specific role for these pathways in promoting tumorigenesis in the widltype background.
Table 2. Pathway analysis.
Ingenuity Variant Analysis software (www.Ingenuity.com/variants) was used to identify recurrently mutated pathways for wildtype and Nf1 sarcomas. Shown are the number of molecules from each pathway where there is a non-synonymous SNV in the gene in one or more of our sarcomas. A right-tailed Fisher’s exact test was used to calculate p-values by considering (1) the number of focus genes that participate in that process and (2) the total number of genes that are known to be associated with that process in the selected reference set.
| Molecular and Cellular Functions | # molecules | p-value | |
|---|---|---|---|
| WT sarcomas | Cellular Assembly and Organization | 155 | 7.07 × 10−07 - 6.33 × 10−03 |
| Cellular Function and Maintenance | 163 | 7.07 × 10−07 - 5.52 × 10−03 | |
| Cell Morphology | 198 | 8.97 × 10−07 - 5.88 × 10−03 | |
| Cell Cycle | 28 | 2.32 × 10−06 - 5.50 × 10−03 | |
| Cell Death and Survival | 269 | 2.66 × 10−06 - 5.50 × 10−03 | |
| Nf1+/− Sarcomas | Cellular Assembly and Organization | 139 | 7.02 × 10−05 - 2.04 × 10−02 |
| Cellular Function and Maintenance | 166 | 7.02 × 10−05 - 2.18 × 10−02 | |
| Cellular Compromise | 24 | 1.17 × 10−04 - 2.18 × 10−02 | |
| DNA Replication, Recombination, and Repair | 25 | 1.17 × 10−04 - 1.99 × 10−02 | |
| Cell Death and Survival | 68 | 1.53 × 10−04 - 2.09 × 10−02 | |
Table 3. Recurrently mutated genes.
The most frequently mutated genes are listed in the table, with the total number of variants identified across all 25 samples and number of non-synonymous only. Variants in genes known to be pseudo-genes, in repetitive regions, or seen at high frequency across all samples and thus likely to be errors were removed.
| Chr | Gene Name | Variant count all | Variant count non-syn | Protein name/function |
|---|---|---|---|---|
| 1 | Mroh2a | 38 | 27 | Maestro heat-like repeat family member 2A |
| 1 | Hjurp | 35 | 29 | Holliday junction recognition protein |
| 1 | Ugt1a1 | 30 | 22 | UDP glucuronosyltransferase 1 family, polypeptide A1 |
| 9 | Mtmr2 | 25 | 21 | Myotubularin related protein 2 |
| 6 | Tmcc1 | 24 | 16 | Transmembrane and coiled coil domains 1 |
| 1 | Itln1 | 22 | 8 | Intelectin 1 |
| 16 | Muc4 | 21 | 17 | Mucin 4 |
| 11 | Obscn | 20 | 10 | Obscurin, cytoskeletal calmodulin and titin-interacting RhoGEF |
| 1 | Cd244 | 16 | 13 | CD244 natural killer cell receptor 2B4 |
| 8 | Kars | 16 | 10 | Lysyl-tRNA synthetase |
| 5 | Nxpe5 | 14 | 4 | Neurexophilin and PC-esterase domain family, member 5 |
| 17 | Foxn2 | 14 | 12 | Forkhead box N2 |
| 4 | Skint6 | 13 | 12 | Selection and upkeep of intraepithelial T cells 6 |
| 6 | Klra1 | 13 | 11 | Killer cell lectin-like receptor, subfamily A, member 1 |
| 7 | Psg18 | 12 | 7 | Pregnancy specific glycoprotein 18 |
| 9 | Cbl | 12 | 12 | Casitas B-lineage lymphoma |
| 6 | Klra10 | 11 | 10 | Killer cell lectin-like receptor subfamily A, member 10 |
| 6 | Klra4 | 11 | 11 | Killer cell lectin-like receptor, subfamily A, member 4 |
| 7 | Mrgpra9 | 11 | 8 | MAS-related GPR, member A9 |
| 17 | Plin4 | 11 | 2 | Perilipin 4 |
| X | Tro | 11 | 11 | Trophinin |
| 4 | Thrap3 | 10 | 6 | Thyroid hormone receptor associated protein 3 |
| 13 | Mplkip | 10 | 9 | M-phase specific PLK1 intereacting protein |
| 13 | Taf9 | 10 | 5 | TAF9 RNA polymerase II, TATA box binding protein (TBP)-associated factor |
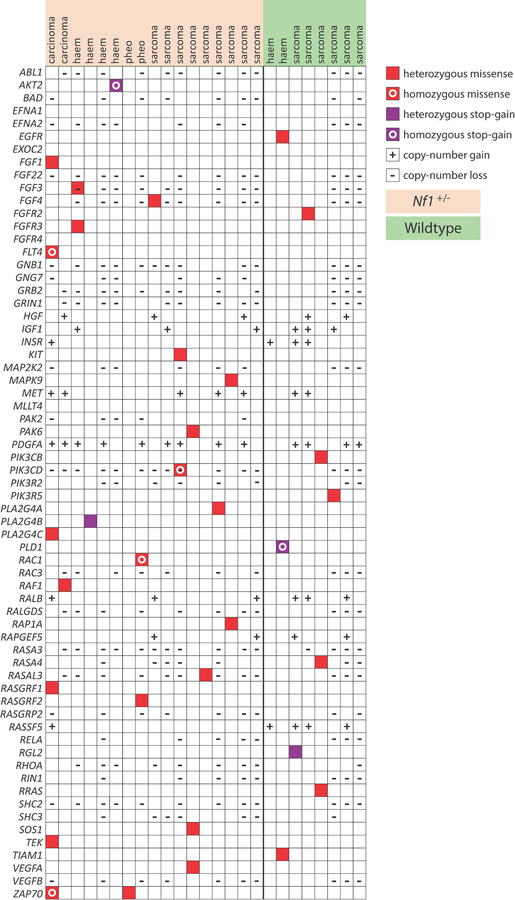
Mutations involving the Ras pathway are common in human cancers (Stephen et al., 2014). Analysis of Ras pathway genes demonstrating significant copy-number changes (either loss or gain), missense mutations, and/or stop-gain mutations in IR-induced neoplasms indicate Ras pathway mutations as common in neoplasms arising from either the wildtype or Nf1 mutant backgrounds (Figure 3). Interestingly, H-ras, K-ras and N-ras mutations are absent from neoplasms arising in Nf1 mutant mice, consistent with Nf1-driven tumorigenesis being exclusive of somatic ras mutations.
Figure 3. Copy number alteration shared between WT and Nf1 mutant backgrounds.
Control-FREEC software was used to compare read numbers between tumors and germline control in order to estimate copy number alterations in tumors. A. Genome-wide copy number alterations in all malignancies (top row), Nf1 tumors alone (middle row) and wildtype tumors alone (bottom row). Alternating stripes indicate consecutive chromosomes, beginning with chromosome 1 at the far left. The proportion of samples showing gain is displayed in red, and loss in blue. Significantly altered areas, as calculated using STAC (Diskin et al., 2006),are indicated by heatmap bars directly above the chromosomal area for gain, and below for loss (p < 0.05 shown). Overall, Nf1 mutant-derived samples showed far more copy number losses than gains (23% versus 8%), while wildtype-derived tumors showed the opposite pattern, demonstrating fewer losses than gains (9% versus 29%).
B. Unsupervised hierarchical clustering analysis was performed on copy number variation (CNV) data to organize sarcomas from wildtype and Nf1 mutant mice into groups sharing similar patterns of copy number alterations. The heatmap shows CNVs smoothed into 15 kb windows with white indicating normal copy number, blue indicating loss and red indicating gain. See also Tables S8 and S9.
C. Venn diagrams depicting: left, the numbers of genes affected by significant copy number change that are gained in all sarcomas (red), lost in all sarcomas (blue), or both (intersection); right, the numbers of genes affected by significant copy number change that are altered in Nf1 sarcomas (tan), wildtype sarcomas (green), or altered in both (intersection).
D. Bar graph displays the percentage of genes in the COSMIC database that are involved by either copy number gain or loss in all sarcomas (left), Nf1 mutant sarcomas (middle), or wildtype sarcomas (right). Student’s t-test, *p < 0.05.
DISCUSSION
These data describe 1) the mutational landscape of malignancies induced by IR, and 2) the influence of the genetic background, specifically heterozygosity for Nf1, upon the mutational process. Our experimental paradigm reproducibly delivered IR to wildtype or Nf1 mice, allowing for the comparison of mutational processes between these backgrounds.
In humans, IR-induced malignancies arise after varying amounts of radiation exposure, fractionation and anatomic localization. This variation can potentially complicate efforts to compare mutational landscapes in the diverse neoplasms associated with IR-induced mutagenesis. Thus, the controlled parallels between our mouse models and human IR-induced malignancies are important and underpin the relevance of this study. Murine studies of radiation mutagenesis traditionally have been limited in their abilities to replicate clinical parameters. For example, murine studies of radiation-induced tumors have employed low dose total body irradiation (TBI), a simple technique (Mao et al., 2005; Mao et al., 2008; Ullrich et al., 1996; Ullrich et al., 1987). Unfortunately this bears little resemblance to clinical practice, where most irradiated patients receive fractionated, focal, high dose irradiation (40–70 Gy) to a site of disease, and adjacent normal tissues at risk for mutagenesis can receive up to 100% of the prescribed dose. Replicating the dosimetry and anatomic targeting of radiotherapy is critical to modeling SMNs because multiple studies indicate an important relationship between radiation dose and SMN risk in cancer survivors (Tucker et al., 1987; Tukenova et al., 2011), with increasing doses associated with increasing risk of solid tumors. Furthermore, as discussed above, focal radiation and the anatomic space irradiated are independent factors influencing the risk of SMNs. To overcome the limitations in traditional models of radiation-induced tumorigenesis we replicated radiotherapy approaches used in patients to develop clinically relevant models of radiotherapy-induced malignancies in mice, developing the first SMN models that recapitulate anatomically appropriate SMNs in a dose-dependent fashion (Choi et al., 2012; Nakamura et al., 2011).
The Nf1 mutant background is markedly sensitized to IR-induced malignancies compared to wildtype mice, as reflected by both higher total numbers of IR-induced tumors and worse overall survival due to cancer development (Choi et al., 2012). Our analysis provides an opportunity to begin to understand the basis for this background-dependent susceptibility to IR. The apparent susceptibility of Nf1 mutant mice to IR-induced malignancies might suggest that malignancies from the Nf1 mutant background can reach detectable size (suggestive of a growth advantage) even when harboring fewer SNVs than IR-induced malignancies arising in the wildtype background. Alternatively, if Nf1 heterozygosity compromised key mechanisms of genomic stability, neoplasms arising in Nf1 mutant mice might appear consistently hypermutated. Our data supports the former prediction that an Nf1-dependent increase in the cancer-promoting efficiency of IR is associated with reduced numbers of SNVs. Notably, sarcomas arising in irradiated Nf1 mutant mice harbored significantly fewer somatic variants than sarcomas arising in irradiated wildtype mice, indicating that heterozygosity for the Nf1 tumor suppressor gene reduces the average number of mutations present in a tumor.
Prior studies have shown that different tumor types can differ substantially in the types of base substitutions that dominate their mutational signature (Sjoblom et al., 2006). For example, Sjoblom et. al. showed that C to T transitions were the dominant substitution in colorectal cancers, while these substitutions were significantly less prevalent in breast cancers. Discrete mutagens can also produce distinct signatures which reflect the molecular lesions produced by the chemical mechanism of action, as is the case of MNU-induced GG(T→A) transitions (Westcott et al., 2014). The underlying mechanisms for these differences are not well-defined, although the nature of these differences may implicate specific processes. Mutational signatures found in tumors arising after exposure to IR or other known mutagens are likely to reflect not only the DNA damage specific to the mutagen, but the influence of repair mechanisms, and selection pressures inherent in tumorigenesis. The best understood mutational signature is associated with UV-induced mutagenesis, which is associated with the production of dipyrimidine dimers that are clearly apparent in UV-associated tumors (Alexandrov et al., 2013a; Bykov et al., 1998). Although IR is utilized in modern medicine, in contrast to the aforementioned mutagens its effects are poorly characterized from a genome or exome-wide perspective.
Analyzing non-synonymous variants, we identified stable three mutational signatures suggestive of operative mutational processes in IR-induced tumorigenesis. Mutational analysis detected biases in the frequencies of particular base substitutions within discrete sequence contexts. Our data indicate that flanking bases can influence the rate of C→G and T→G substitutions, as evidenced by the prominence of these substitutions patterns in signature 3. These data suggest the importance of the local sequence conformation influencing stability and enzymatic repair. Signature analysis also identified a transcriptional strand bias for specific substitution types and contexts, which may implicate processes such as transcription-coupled repair, that operate in a strand-specific fashion.
Synonymous variants are commonly viewed as passenger mutations but recent literature suggests that some synonymous mutations may function as drivers of cancer development (Supek et al., 2014). To determine whether synonymous variants might harbor unique mutational signatures, we performed NMF analysis on the synonymous variants alone (1,990 SNVs). In this analysis, the original first two signatures were reproducibly extracted from the synonymous variants, but the third signature was unstable and was not reproduced. This signature instability might reflect an underlying difference between synonymous and non-synonymous mutations, a possibility we are unable to confirm or reject given the small number of variants. Overall, these findings indicate significant similarities with regard to trinucleotide signature between synonymous and non-synonymous. However, further study is clearly needed to better define possible differences between synonymous and non-synonymous variants and their biologic impact.
The three mutational signatures identified were present in malignancies from both wildtype and Nf1 mutant mice, and our analysis failed to identify an Nf1-specific trinucleotide signature. This suggests that the mutagenic IR exposure, rather than the tested genetic backgrounds, primarily influenced the development of these signatures. Our signatures are also distinct from those seen in previous analyses of mutagen-induced tumors (Alexandrov et al., 2013a).
These data contrast with other studies comparing mutational landscapes of carcinogen-based models of cancer. Westcott, et. al. compared mutation burden between mutant Kras, methyl-nitrosurea and urethane-induced lung cancer models and found that Kras mutant lung cancers harbored significantly fewer SNVs compared to carcinogen-associated tumors (Westcott et al., 2014). Our analysis of IR-induced tumors demonstrated variable numbers of SNVs and comparatively fewer SNVs than identified in MNU-induced malignancies. The relatively low rate of somatic SNVs in IR-induced malignancies was unexpected, given that mutagen-associated malignancies in mice and humans often have relatively high mutation rates (Alexandrov et al., 2013a; Westcott et al., 2014). Our data indicate that IR-associated malignancies do not necessarily harbor high numbers of somatic SNVs, and that diverse organs mutagenized by IR share similar numbers of mutations.
The SNV rate in different genetically mutant mouse backgrounds may reflect underlying fundamental differences between tumorigenesis driven by loss of a tumor suppressor gene as compared to activation of an oncogene. Nf1 loss and Kras activation both promote ras signaling, however Nf1 loss does not phenocopy oncogenic Kras expression, and is clearly a weaker driver of tumor formation (Cichowski and Jacks, 2001; Morcos et al., 1996). A potential consequence of this inherent difference in molecular function is that tumorigenesis driven by Nf1 loss, as occurs in our mouse models, may require a greater number of cooperating mutations as compared to Kras-driven tumors. The requirement for accumulation of cooperating mutational events could certainly influence tumor development and behavior by limiting the growth potential of developing tumors. The clinical course of the NF1 syndrome may reflect this possibility, as many tumors in NF1 patients are slowly progressive (Friedman, 2002). In order to understand disease and specifically cancer progression in the context of inherited tumor predisposition syndromes, additional studies are needed to characterize and explain the mutational consequences of germline mutations in NF1 and other germline mutations known to affect tumor susceptibility in humans.
The effects of genotoxin exposure also extend to cancer therapy, because IR and genotoxic chemotherapy are standard treatments in the management of many malignancies. Indeed, the mutational signatures associated with genotoxin therapy can be found in treated cancers. Johnson, et. al, (Johnson et al., 2014) examined how glioma therapies, particularly treatment with the alkylator temozolomide, influence the genomes of human gliomas. This work revealed that temozolomide treatment is associated with the development of a hypermutated genome, a finding that raises concern about how alkylator therapy itself promotes the evolution of a low grade glioma into a more aggressive and histologically high grade malignancy. Our results suggest that IR-treated cancers may similarly harbor mutational signatures distinguishable from other genotoxic cancer therapies such as alkylating chemotherapy, and further work in this area may yield important insights into tumor evolution. These issues are important considerations as cancer therapy increasingly must consider potential mechanisms of resistance to therapy. If genomic injury resulting from initial cancer therapy shapes cancer evolution and subsequent resistance, understanding this process may inform attempts to mitigate this process and improve treatment efficacy.
Previous studies have characterized the somatic copy number profiles of tumors from different histologies (Zack et al., 2013). We saw comparable levels of genomic disruption in our IR induced tumors, and similarly observed no recurrent changes that segregate with histology. However, we did observe significantly more copy number gain than loss in our wildtype tumors, and the converse in tumors from Nf1 heterozygous mice. This was particularly pronounced on chromosome 11, which shows loss in close to 50% of Nf1 tumors sequenced. We have shown in a previous LOH study (Choi et al., 2012) using our mouse model that tumors were rendered effectively Nf1 null due to preferential loss of the wildtype Nf1 allele on chromosome 11. Interestingly, this finding is consistent with the observation made in earlier work that lung tumors arising from Kras mutant mice harbor copy number alterations distinguishing them from mutagen-induced tumors (Westcott et al., 2014). Taken together, these findings suggest that copy number alterations represent an efficient route to tumorigenesis and may be selected for in tumors with discrete driver mutations.
We speculate that the bias of Nf1-derived tumors from our mouse model to harbor copy number loss and wildtype-derived tumors to favor copy number gain may reflect multiple possible mechanisms. Given that neurofibromin is not known to have a role in DNA repair, and earlier experiments we performed failed to detect a difference in dsDNA break formation or repair with Nf1 loss (Nakamura et al., 2011), it is appears unlikely that the difference in copy number alteration reflects an intrinsic difference in the basal genomic stability of Nf1+/− cells. It is possible that in neoplasms arising from Nf1 mutant backgrounds, alterations in pathways responsible for genomic stability are preferentially altered, and that this could lead, over successive divisions, to cumulative chromosomal losses. It is also possible that ionizing radiation induces different patterns of genomic injury in wildtype and Nf1 mutant cells on the basis of chromatin architecture and environments. These ideas remain speculative, as the influence of germline Nf1 mutations on genome stability and structural variation is not well defined. Malignant peripheral nerve sheath tumors (MPNSTs), which are malignancies characteristic of the NF1 syndrome (Kresse et al., 2008; Mantripragada et al., 2009; Mantripragada et al., 2008; Thomas et al., 2015), have been described to harbor both copy number gains and losses, with gains out-numbering losses (Kresse et al., 2008; Mantripragada et al., 2009). The underlying basis for this is also not understood, and whether this pattern of copy number alterations is generalizable to all NF1-mutant tumors remains unclear. These questions highlight the need for further analyses of NF1-driven tumorigenesis and the impact of germline tumor suppressor mutations in general.
In summary, this work characterizes the mutational processes in IR-induced malignancies and yields important information concerning both mutagenic exposure and the influence of a germline mutation in the Nf1 tumor suppressor gene. We identify stable mutational signatures that can serve as a basis for understanding how IR exposure and Nf1 heterozygosity can each promote the development of malignancies. This work also highlights the utility of genetically-engineered mouse models of cancer to study the mutational consequences of germline heterozygosity for tumor suppressor genes.
EXPERIMENTAL PROCEDURES
Mouse model
Nf1+/− mice were generated as previously described. In brief, Nf1+/− mice maintained in the 129/Sv background (Jacks et al., 1994) were crossed with wild-type C57Bl/6 mice. Six to eight week old mice were given focal, fractionated irradiation delivered at a rate of five fractions per week, one fraction per day as previously described (Choi et al., 2012; Nakamura et al., 2011). Tumor samples used in this analysis were obtained after animals were euthanized, and all animal procedures were approved by the UCSF IACUC (Approved protocol numbers: AN078941 and AN080665). These practices conform to regulations defined by the Animal Welfare Act and the US Department of Agriculture.
Cell lines, cell culture conditions
All tumor cell lines were grown in Dulbecco’s modified Eagle medium (DMEM, 4500 mg/L glucose, Life Technologies, Inc.) supplemented with 10% fetal bovine serum (GIBCO-BRL, Invitrogen, Carlsbad, CA), penicillin, and streptomycin. Cells were grown in a humidified incubator containing 8% carbon dioxide at 37°C.
Whole exome sequencing
Whole exome sequencing was performed using the Agilent SureSelect mouse all exon kit. Captured material was indexed and sequenced on the Illumina GAII and HiSeq2000 platform at the Wellcome Trust Sanger Institute. Successfully sequenced reads were then aligned to mouse reference genome (GRCm38) followed by number of quality control routines (e.g. base quality score recalibration, realignment around InDels). We used Cake variants detection pipeline (Perna et al., 2015; Rashid et al., 2013) to identify somatic as well as germline variants. A stringent variant caller overlapping strategy has been adapted to minimize false positive rate. Those detected by at least four of the five algorithms (embedded within Cake: Bambino; Varscan 2; Mpileup; SomaticSniper; Mutect) were subsequently filtered for depth, known mouse variations (Keane et al., 2011), strong germline activity and functional impact to produce a cleaner list of mutations (Table S1).
Copy number analysis
Using the exome sequencing data we identified regions of copy number variation (CNV) in tumor DNAs relative to the matched normal (129/Sv-C576BL/6 heterozygote) using Control-FREEC with default parameters (Boeva et al., 2012). Analysis was restricted to the region selected by the SureSelectXT mouse exome capture kit (Agilent). CNVs were smoothed into 15kb segments. Regions with low call rate (called in less than one third of samples) and invariant regions were removed. Between sample Euclidian distances were calculated using the WGCNA R software package (Langfelder and Horvath, 2008), and samples were clustered using Ward’s method. The resulting dendogram demonstrates between sample relatedness based on genome-wide CNV patterns.
Pathway analysis
A list of Ras signalling pathway genes was obtained from KEGG pathway hsa04014 (http://www.genome.jp/dbget-bin/www_bget?pathway+hsa04014)
Statistical analysis
We performed non-negative matrix factorization (NMF) following protocols developed by WTSI (Alexandrov et al., 2013b), starting with their MATLAB scripts downloaded from http://www.mathworks.com/matlabcentral/fileexchange/38724-wtsi-mutational-signature-framework. Their scripts were modified only as necessary to accommodate our data and computer. We first replicated the signatures reported by WTSI (Alexandrov et al., 2013b; Nik-Zainal et al., 2012) to validate our procedures. We then followed these procedures on the following data sets: 1) 25 samples (22 tumors and 3 cell lines derived from 3 of the tumors) with 192 substitution types (6 substitutions using the pyrimidine as reference x 2 strands x 4 5’ neighbors x 4 3’ neighbors) for both synonymous and non-synonymous mutations; 2) the same analysis except excluding synonymous mutations; 3) 22 tissue samples, excluding 3 cell lines, using the same parameters as 1); 4) the same analysis as 1) except collapsing strands for 96 substitution types. For comparison with WTSI human coefficients (Alexandrov et al., 2013a), our coefficients were normalized to the mouse trinucleotide frequencies (Frenkel et al., 2011) as NrmSx in Table S5 and as IrrMse x in Table S6.
All but two of the frequency, contingency table and correlation analyses were performed with standard Microsoft Excel formulae and functions. One exception is Fisher’s Exact Test (or Fisher-Freeman-Halton Test) used to compare substitution frequencies by strand (Table S7), where expected frequencies were too small for a chi square statistic. The SPSS Exact Tests module completed a monte carlo estimation based on a random sample of 100,000 tables, providing a 2-sided p-value of 0.000. The other is Fisher’s Exact Test used in Table 2 for separate 2 × 2 tables done as part of the pathway analysis by IPA, the Ingenuity software.
Supplementary Material
Figure 4. Ras pathway genes mutated in IR-induced malignancies.
The table displays Ras pathway genes that demonstrate significant copy-number changes (either loss or gain), missense mutations, and/or stop-gain mutations in IR-induced neoplasms.
ACKNOWLEDGEMENTS
We thank Allan Balmain, Benjamin Braun, Kevin Shannon and their labs for helpful discussions. Kyle Halliwell provided help with bioinformatics analysis. We thank David Adams for helpful discussions and reading the manuscript. J.L.N. was supported by grants from the US National Institutes of Health, National Cancer Institute (1K08CA115476), the UCSF Breast SPORE Program, Alex’s Lemonade Stand Foundation, St. Baldrick’s Foundation Scholar Award, and Sammy and Kari Hagar and the Hagar Family Foundation.
REFERENCES
- Alexandrov LB, Nik-Zainal S, Wedge DC, Aparicio SA, Behjati S, Biankin AV, Bignell GR, Bolli N, Borg A, Borresen-Dale AL, et al. (2013a). Signatures of mutational processes in human cancer. Nature 500, 415–421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexandrov LB, Nik-Zainal S, Wedge DC, Campbell PJ, and Stratton MR. (2013b). Deciphering signatures of mutational processes operative in human cancer. Cell reports 3, 246–259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Armstrong GT, Liu Q, Yasui Y, Huang S, Ness KK, Leisenring W, Hudson MM, Donaldson SS, King AA, Stovall M, et al. (2009a). Long-term outcomes among adult survivors of childhood central nervous system malignancies in the Childhood Cancer Survivor Study. J Natl Cancer Inst 101, 946–958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Armstrong GT, Liu Q, Yasui Y, Neglia JP, Leisenring W, Robison LL, and Mertens AC. (2009b). Late mortality among 5-year survivors of childhood cancer: a summary from the Childhood Cancer Survivor Study. J Clin Oncol 27, 2328–2338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhatia S, and Sklar C. (2002). Second cancers in survivors of childhood cancer. Nat Rev Cancer 2, 124–132. [DOI] [PubMed] [Google Scholar]
- Boeva V, Popova T, Bleakley K, Chiche P, Cappo J, Schleiermacher G, Janoueix-Lerosey I, Delattre O, and Barillot E. (2012). Control-FREEC: a tool for assessing copy number and allelic content using next-generation sequencing data. Bioinformatics 28, 423–425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bykov VJ, Jansen CT, and Hemminki K. (1998). High levels of dipyrimidine dimers are induced in human skin by solar-simulating UV radiation. Cancer Epidemiol Biomarkers Prev 7, 199–202. [PubMed] [Google Scholar]
- Choi G, Huang B, Pinarbasi E, Braunstein SE, Horvai AE, Kogan S, Bhatia S, Faddegon B, and Nakamura JL. (2012). Genetically mediated Nf1 loss in mice promotes diverse radiation-induced tumors modeling second malignant neoplasms. Cancer Res 72, 6425–6434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cichowski K, and Jacks T. (2001). NF1 tumor suppressor gene function: narrowing the GAP. Cell 104, 593–604. [DOI] [PubMed] [Google Scholar]
- Cingolani P, Platts A, Wang le L, Coon M, Nguyen T, Wang L, Land SJ, Lu X, and Ruden DM. (2012). A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff: SNPs in the genome of Drosophila melanogaster strain w1118; iso-2; iso-3. Fly 6, 80–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crump M, and Hodgson D. (2009). Secondary breast cancer in Hodgkin’s lymphoma survivors. J Clin Oncol 27, 4229–4231. [DOI] [PubMed] [Google Scholar]
- Diskin SJ, Eck T, Greshock J, Mosse YP, Naylor T, Stoeckert CJ Jr., Weber BL, Maris JM, and Grant GR. (2006). STAC: A method for testing the significance of DNA copy number aberrations across multiple array-CGH experiments. Genome research 16, 1149–1158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frenkel ZM, Bettecken T, and Trifonov EN. (2011). Nucleosome DNA sequence structure of isochores. BMC genomics 12, 203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedman JM (2002). Neurofibromatosis 1: clinical manifestations and diagnostic criteria. J Child Neurol 17, 548–554; discussion 571–542, 646–551. [DOI] [PubMed] [Google Scholar]
- Green P, Ewing B, Miller W, Thomas PJ, Program NCS, and Green ED. (2003). Transcription-associated mutational asymmetry in mammalian evolution. Nat Genet 33, 514–517. [DOI] [PubMed] [Google Scholar]
- Henderson TO, Whitton J, Stovall M, Mertens AC, Mitby P, Friedman D, Strong LC, Hammond S, Neglia JP, Meadows AT, et al. (2007). Secondary sarcomas in childhood cancer survivors: a report from the Childhood Cancer Survivor Study. J Natl Cancer Inst 99, 300–308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacks T, Shih TS, Schmitt EM, Bronson RT, Bernards A, and Weinberg RA. (1994). Tumour predisposition in mice heterozygous for a targeted mutation in Nf1. Nat Genet 7, 353–361. [DOI] [PubMed] [Google Scholar]
- Johnson BE, Mazor T, Hong C, Barnes M, Aihara K, McLean CY, Fouse SD, Yamamoto S, Ueda H, Tatsuno K, et al. (2014). Mutational analysis reveals the origin and therapy-driven evolution of recurrent glioma. Science 343, 189–193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keane TM, Goodstadt L, Danecek P, White MA, Wong K, Yalcin B, Heger A, Agam A, Slater G, Goodson M, et al. (2011). Mouse genomic variation and its effect on phenotypes and gene regulation. Nature 477, 289–294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kresse SH, Skarn M, Ohnstad HO, Namlos HM, Bjerkehagen B, Myklebost O, and Meza-Zepeda LA. (2008). DNA copy number changes in high-grade malignant peripheral nerve sheath tumors by array CGH. Molecular cancer 7, 48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langfelder P, and Horvath S. (2008). WGCNA: an R package for weighted correlation network analysis. BMC bioinformatics 9, 559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mantripragada KK, Diaz de Stahl T, Patridge C, Menzel U, Andersson R, Chuzhanova N, Kluwe L, Guha A, Mautner V, Dumanski JP, et al. (2009). Genome-wide high-resolution analysis of DNA copy number alterations in NF1-associated malignant peripheral nerve sheath tumors using 32K BAC array. Genes Chromosomes Cancer 48, 897–907. [DOI] [PubMed] [Google Scholar]
- Mantripragada KK, Spurlock G, Kluwe L, Chuzhanova N, Ferner RE, Frayling IM, Dumanski JP, Guha A, Mautner V, and Upadhyaya M. (2008). High-resolution DNA copy number profiling of malignant peripheral nerve sheath tumors using targeted microarray-based comparative genomic hybridization. Clinical cancer research : an official journal of the American Association for Cancer Research 14, 1015–1024. [DOI] [PubMed] [Google Scholar]
- Mao JH, Li J, Jiang T, Li Q, Wu D, Perez-Losada J, DelRosario R, Peterson L, Balmain A, and Cai WW. (2005). Genomic instability in radiation-induced mouse lymphoma from p53 heterozygous mice. Oncogene 24, 7924–7934. [DOI] [PubMed] [Google Scholar]
- Mao JH, Wu D, DelRosario R, Castellanos A, Balmain A, and Perez-Losada J. (2008). Atm heterozygosity does not increase tumor susceptibility to ionizing radiation alone or in a p53 heterozygous background. Oncogene 27, 6596–6600. [DOI] [PubMed] [Google Scholar]
- Meadows AT, Friedman DL, Neglia JP, Mertens AC, Donaldson SS, Stovall M, Hammond S, Yasui Y, and Inskip PD. (2009). Second neoplasms in survivors of childhood cancer: findings from the Childhood Cancer Survivor Study cohort. J Clin Oncol 27, 2356–2362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morcos P, Thapar N, Tusneem N, Stacey D, and Tamanoi F. (1996). Identification of neurofibromin mutants that exhibit allele specificity or increased Ras affinity resulting in suppression of activated ras alleles. Mol Cell Biol 16, 2496–2503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura JL, Phong C, Pinarbasi E, Kogan SC, Vandenberg S, Horvai AE, Faddegon BA, Fiedler D, Shokat K, Houseman BT, et al. (2011). Dose-dependent effects of focal fractionated irradiation on secondary malignant neoplasms in Nf1 mutant mice. Cancer Res 71, 106–115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nik-Zainal S, Alexandrov LB, Wedge DC, Van Loo P, Greenman CD, Raine K, Jones D, Hinton J, Marshall J, Stebbings LA, et al. (2012). Mutational processes molding the genomes of 21 breast cancers. Cell 149, 979–993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perna D, Karreth FA, Rust AG, Perez-Mancera PA, Rashid M, Iorio F, Alifrangis C, Arends MJ, Bosenberg MW, Bollag G, et al. (2015). BRAF inhibitor resistance mediated by the AKT pathway in an oncogenic BRAF mouse melanoma model. Proc Natl Acad Sci U S A 112, E536–545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rashid M, Robles-Espinoza CD, Rust AG, and Adams DJ. (2013). Cake: a bioinformatics pipeline for the integrated analysis of somatic variants in cancer genomes. Bioinformatics 29, 2208–2210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubin AF, and Green P. (2009). Mutation patterns in cancer genomes. Proc Natl Acad Sci U S A 106, 21766–21770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sjoblom T, Jones S, Wood LD, Parsons DW, Lin J, Barber TD, Mandelker D, Leary RJ, Ptak J, Silliman N, et al. (2006). The consensus coding sequences of human breast and colorectal cancers. Science 314, 268–274. [DOI] [PubMed] [Google Scholar]
- Stephen AG, Esposito D, Bagni RK, and McCormick F. (2014). Dragging ras back in the ring. Cancer Cell 25, 272–281. [DOI] [PubMed] [Google Scholar]
- Stephens P, Edkins S, Davies H, Greenman C, Cox C, Hunter C, Bignell G, Teague J, Smith R, Stevens C, et al. (2005). A screen of the complete protein kinase gene family identifies diverse patterns of somatic mutations in human breast cancer. Nat Genet 37, 590–592. [DOI] [PubMed] [Google Scholar]
- Supek F, Minana B, Valcarcel J, Gabaldon T, and Lehner B. (2014). Synonymous mutations frequently act as driver mutations in human cancers. Cell 156, 1324–1335. [DOI] [PubMed] [Google Scholar]
- Thomas LE, Winston J, Rad E, Mort M, Dodd KM, Tee AR, McDyer F, Moore S, Cooper DN, and Upadhyaya M. (2015). Evaluation of copy number variation and gene expression in neurofibromatosis type-1-associated malignant peripheral nerve sheath tumours. Human genomics 9, 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tucker MA, D’Angio GJ, Boice JD Jr., Strong LC, Li FP, Stovall M, Stone BJ, Green DM, Lombardi F, Newton W, et al. (1987). Bone sarcomas linked to radiotherapy and chemotherapy in children. N Engl J Med 317, 588–593. [DOI] [PubMed] [Google Scholar]
- Tukenova M, Guibout C, Hawkins M, Quiniou E, Mousannif A, Pacquement H, Winter D, Bridier A, Lefkopoulos D, Oberlin O, et al. (2011). Radiation therapy and late mortality from second sarcoma, carcinoma, and hematological malignancies after a solid cancer in childhood. Int J Radiat Oncol Biol Phys 80, 339–346. [DOI] [PubMed] [Google Scholar]
- Ullrich RL, Bowles ND, Satterfield LC, and Davis CM. (1996). Strain-dependent susceptibility to radiation-induced mammary cancer is a result of differences in epithelial cell sensitivity to transformation. Radiat Res 146, 353–355. [PubMed] [Google Scholar]
- Ullrich RL, Jernigan MC, Satterfield LC, and Bowles ND. (1987). Radiation carcinogenesis: time-dose relationships. Radiat Res 111, 179–184. [PubMed] [Google Scholar]
- Wei X, Walia V, Lin JC, Teer JK, Prickett TD, Gartner J, Davis S, Program NCS, Stemke-Hale K, Davies MA, et al. (2011). Exome sequencing identifies GRIN2A as frequently mutated in melanoma. Nat Genet 43, 442–446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westcott PM, Halliwill KD, To MD, Rashid M, Rust AG, Keane TM, Delrosario R, Jen KY, Gurley KE, Kemp CJ, et al. (2014). The mutational landscapes of genetic and chemical models of Kras-driven lung cancer. Nature. [DOI] [PMC free article] [PubMed]
- Yong RL, Yang C, Lu J, Wang H, Schlaff CD, Tandle A, Graves CA, Elkahloun AG, Chen X, Zhuang Z, et al. (2014). Cell transcriptional state alters genomic patterns of DNA double-strand break repair in human astrocytes. Nature communications 5, 5799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zack TI, Schumacher SE, Carter SL, Cherniack AD, Saksena G, Tabak B, Lawrence MS, Zhang CZ, Wala J, Mermel CH, et al. (2013). Pan-cancer patterns of somatic copy number alteration. Nat Genet 45, 1134–1140. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.