Abstract
Cytoplasmic dynein is an ATP-driven motor that transports intracellular cargos along microtubules. Dynein adopts an inactive conformation when not attached to cargo, and motility is activated when dynein assembles with dynactin and a cargo adaptor. It remained unclear how active dynein-dynactin complexes step along microtubules and transport cargos under tension. Using single-molecule imaging, we showed that dynein-dynactin advances by taking 8–32 nm steps towards the microtubule minus-end with frequent sideways and backward steps. Multiple dyneins collectively bear large tension because the backward stepping rate of dynein is insensitive to load. Recruitment of two dyneins to dynactin increases the force generation and the likelihood of winning against kinesin in a tug-of-war but does not directly affect velocity. Instead, velocity is determined by cargo adaptors and tail-tail interactions between two closely packed dyneins. Our results show that cargo adaptors modulate dynein motility and force generation for a wide range of cellular functions.
Introduction
Dynein and kinesin are molecular motors that transport intracellular cargos towards the minus- and plus-end of microtubules (MTs), respectively. While more than 15 different classes of kinesins have been identified in humans, all dynein-related functions in the cytoplasm are associated with a single isoform. Cytoplasmic dynein (‘dynein’ hereafter) is responsible for retrograde transport of intracellular cargos in interphase cells and plays a central role in chromosome positioning, nucleokinesis, and the bipolar assembly of the mitotic spindle during cell division. Defects in dynein function underpin many developmental and neurodegenerative diseases including Parkinson’s, Lissencephaly, and Alzheimer’s disease1, 2.
Dynein is composed of two identical dynein heavy chains (DHCs), a light intermediate chain (LIC) and an intermediate chain (IC) complexed with three light chains (LCs, Fig. 1a)1. The motor domain of DHC comprises a ring of six AAA ATPase subunits (AAA1–6) that connects to an MT through a coiled-coil stalk3-5. The tail domain dimerizes the heavy chains, and recruits LIC and IC6. The tail contacts the AAA+ domains through a linker region, which serves as a mechanical element to drive stepping towards the minus-end7.
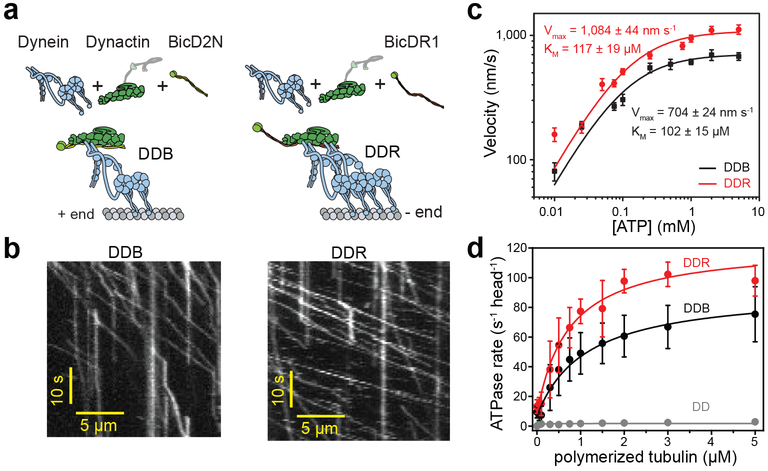
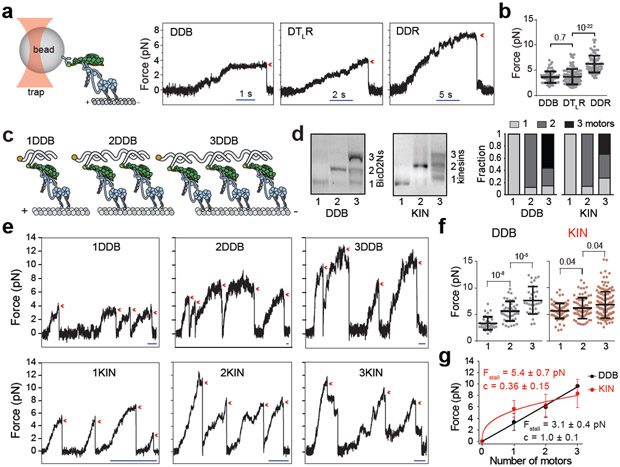
Figure 1. DDR hydrolyzes ATP and moves faster than DDB.
(a) Schematic depiction of the DDB and DDR complexes. The N-terminal coiled-coil domain of BicD2N primarily recruits single dynein, while BicDR1 recruits two dyneins to dynactin. (b) Sample kymographs show the motility of DDB and DDR complexes on MTs (nDDB = 209, nDDR = 223, three independent experiments per condition). (c) Average velocities of DDB and DDR as a function of ATP concentration (mean ± s.e.m., n = 72, 84, 74, 78, 84, 82, 98, 80, 70 motors for DDB; and 70, 75, 72, 78, 106, 86, 100, 92, 98, 80 motors for DDR from left to right, three independent experiments per condition). Data fit well to Michaelis-Menten kinetics with a Hill coefficient of 1 (solid curves, ± s.e. of the fit). (d) MT-stimulated ATPase rates of dynein-dynactin (DD), DDB, and DDR (mean ± s.d.) fit well to Michaelis-Menten kinetics with a Hill coefficient of 1 (solid curves, three independent experiments).
Unlike yeast dynein, which walks processively along MTs, previous studies had shown that mammalian dynein diffuses on an MT, walks only short processive runs, and produces low forces (0.5 – 1.5 pN) in vitro8-11. This contrasted with robust retrograde transport of dynein-driven cargos observed in mammalian cells12, 13. It has been proposed that dynein is able to drive transport only when functioning in large teams and that weak force generation by a single dynein enables fine tuning of the total force by controlling the number of dyneins carrying the cargo10. However, these in vitro studies were performed before it was understood that isolated mammalian dynein is autoinhibited by self-dimerization of its motor domains and is unable to walk processively along MTs6, 11, 14, 15. Therefore, it remains poorly understood how the active dynein machinery steps along MTs and works under tension.
Dynein and its cofactor dynactin form a ternary complex with a cargo-adaptor protein6, 16, 17, in which the motor domains adopt a parallel conformation18, 19, move at velocities similar to that of dynein-driven cargos in cells (Fig. 1a-b)2, 20-22 and produce much higher (4 pN) forces than dynein only23. Recent studies identified several cargo adaptor proteins, such as BicD2 and BicDR1 that link dynein to Rab6 secretory vesicles13, 20. A common structural feature of cargo adaptors is the presence of long coiled-coils2, through which the dynein tail is recruited to dynactin16, 17. Structural studies showed that BicDR1 recruits two dynein dimers to dynactin, while recruitment of two dyneins by BicD2 is less efficient18, 19. Recruitment of a second dynein to dynactin further increases the force generation and velocity of the complex in vitro19, consistent with faster retrograde movement of BicDR1-associated Rab6 vesicles relative to BicD2-associated vesicles in cells13. However, it has remained unclear whether this is due to an increase in dynein copy number or the parallel arrangement of the two motors within the complex18, 19.
In this study, we investigated how different cargo adaptors regulate the velocity, stepping and force production of dynein-dynactin using single-molecule imaging and optical trapping in vitro. We observed that dynein copy number accounts for the increase in force production of dynein/dynactin. However, the motor copy number does not directly account for the faster movement of complexes with two dyneins. Instead, cargo adaptors differentially regulate the velocity of the complex. Through a series of truncations of the second dynein on dynactin, we provide evidence that speed increases via tail-tail interactions, not by a mechanical coupling or ring-ring interactions, between two closely packed dyneins. Finally, we show that complexes with two dyneins more effectively compete against kinesin-1 in a ‘tug-of-war’. These results provide an explanation for how recruitment of two dyneins to dynactin facilitates more robust transport of intracellular cargos towards the MT minus-end.
Results
We investigated how cargo adaptors regulate the velocity and stepping of dynein-dynactin in the presence and absence of load using single-molecule imaging and optical trapping in vitro. In our reconstitution conditions, the N-terminal coiled-coil domain of BicD2 (BicD2N, containing residues 1–400) recruits primarily one dynein to dynactin (DDB), whereas BicDR1 recruits mostly two dyneins (DDR)19. Therefore, we used DDB and DDR as model complexes to study how recruitment of one or two dyneins to dynactin affects motility and force generation (Fig. 1a and Supplementary Video 1). Consistent with our previous observations19, single molecule motility assays showed that DDR moves 50% faster than DDB over a wide range of ATP concentrations (10 μM - 5 mM; Fig. 1b,c, Supplementary Fig. 1). The ATP-dependent increase in velocity of DDB and DDR fits well to a single set of Michaelis-Menten constants with similar affinity for ATP (two-tailed t-test, p = 0.91; Fig. 1c). This contrasts with enzymatic schemes that require the consumption of two ATPs per step24 and is more consistent with a model where stepping is powered by a single ATP hydrolysis event at the AAA1 site25.
To test whether the fast velocity of dynein-dynactin is due to a higher ATP hydrolysis rate, we measured the MT-stimulated ATPase activity per dynein in the presence and absence of cargo adaptors. Dynein and dynactin (DD) had very low catalytic activity and MT affinity, consistent with autoinhibition of dynein without a cargo adaptor6. ATPase activity and MT affinity of dynein were dramatically increased in the presence of BicD2N or BicDR1 (Fig. 1d, Supplementary Table 1). In accordance with their velocities, dyneins had a higher ATPase rate in the presence of BicDR1 than in BicD2N (two-tailed t-test; p <0.0001).
Dynein takes variable steps in size and direction
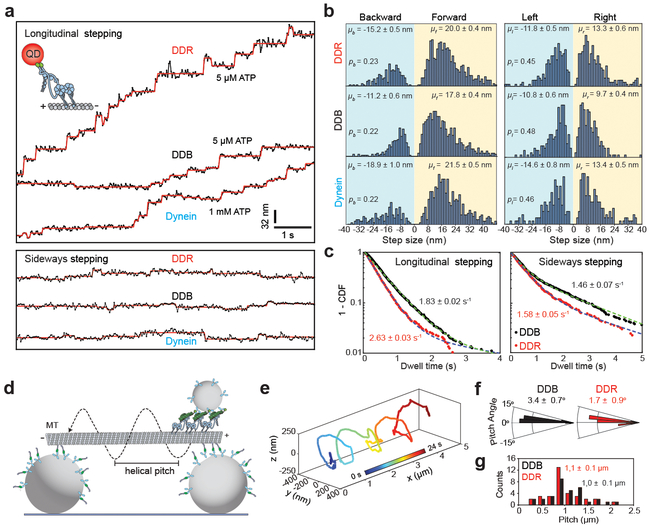
Previous studies on isolated dynein suggested that dynein takes large (32 nm) steps in the absence of load, and that the step size decreases to 16 – 8 nm as the backward load increases10. Another study reported constant 8 nm step size independent of load26. To determine the stepping behavior of active dynein-dynactin motors in unloaded conditions, we labeled dynein with a quantum dot (QD) at the N-terminus and tracked its stepping along MTs in the presence and absence of a cargo adaptor27. Without a cargo adaptor, most dyneins were static on an MT, while others exhibited diffusive movement and frequent release from the MT. On rare occasions, we tracked stepping of motors that exhibited short stretches of slow processive motility in 1 mM ATP (Fig. 2a and Supplementary Fig. 2)23, 28. In the presence of dynactin and a cargo adaptor, dynein motility was robust21, 22, 29. In order to discern individual steps at 30 ms temporal resolution, the ATP concentration was reduced to 5 μM. We observed that DDB and DDR took highly variable (8–32 nm) steps, and exhibited frequent backward and sideways stepping (Fig. 2b) 30, similar to yeast dynein31. DDB and DDR take similar size of steps along the MT long-axis (Fig. 2b), and have similar probabilities to take steps in sideways and backward direction (Fig. 2b, Supplementary Fig. 3). Interestingly, DDR steps at a faster rate than DDB in both longitudinal (2.63 vs. 1.83 s−1 in 5 μM ATP) and sideways directions (Fig. 2c and Supplementary Fig. 3), which primarily accounts for the faster velocity of DDR. We concluded that the recruitment of a second dynein to dynactin increases the stepping rate, but does not alter the size and direction of the steps taken by the complex.
Figure 2. The stepping behavior of mammalian dynein-dynactin in unloaded conditions.
(a) Representative stepping traces of dynein, DDB, and DDR along longitudinal (top) and sideways (bottom) directions. Dynein was labeled with a QD at the N-terminus and its motility was tracked on surface-immobilized MTs at 33 Hz. Horizontal lines represent a fit to a step finding algorithm. n = 21, 19, and 14 for longitudinal and n = 18, 19, and 14 for sideways traces of DDB, DDR, and dynein respectively from 10 independent experiments for each complex. (b) Normalized histograms of longitudinal (left) and sideways (right) steps of dynein, DDB and DDR reveal the average step size in forward (μf), backward (μb), leftward (μl), and rightward (μr) directions (± s.e.m.) and the probability of stepping in backward (pb) and leftward (pl) directions. From top to bottom, n = 963, 1061 and 715 for longitudinal and 534, 463 and 331 for sideways steps. (c) Dwell time distributions of DDB and DDR. Dashed curves represent a fit to a two-exponential decay (decay rate ± s.e.). n = 1430, 963 for longitudinal and 684, 534 for sideways stepping of DDB and DDR, respectively. (d) Schematic of the MT bridge assay (not to scale). MTs are suspended above the coverslip by 2-μm-diameter beads. Motility of 0.5-μm-diameter cargo beads by multiple dynein-dynactin complexes was tracked in three dimensions. (e) An example trajectory of a DDB-driven cargo bead reveals helical motility around the circumference of an MT (n = 18 trajectories). (f) Histograms of helical pitch angles of DDB- and DDR-coated beads (mean ± s.e.m.). n = 32, 34 rotations for DDB and DDR respectively. Positive and negative angles correspond to the right- and left-handed helical movement, respectively. (g) Pitch distributions of DDB- and DDR-coated beads along MT bridges (mean ± s.e.m.). In f and g, n = 32 rotations for DDB (18 beads from eleven independent experiments) and 35 rotations for DDR (16 beads from six independent experiments).
We also tracked unconstrained motion of cargo beads (0.5 μm diameter) driven by multiple dyneins around the circumference of suspended MT bridges in three dimensions (Fig. 2d). We observed that the majority of beads moved with right-handed helical trajectories around MTs (DDB: 84%, DDR: 71%, Fig. 2e-f, Supplementary Fig. 3 and Supplementary Video 2)32, 33. DDB- and DDR-driven beads had similar helical pitches (DDB: 1,047 ± 73 nm, DDR: 1,076 ± 99 nm; mean ± s.e.m., two-tailed t-test, p = 0.81, Fig. 2g), suggesting that sideways stepping of the beads is unchanged by the recruitment of a second dynein to dynactin. We also noticed that helical pitch of these beads is longer than that driven by yeast dynein (580 ± 40 nm)32. Therefore, mammalian dynein more strongly favors motility along the MT long-axis compared to yeast dynein, consistent with dynein’s role in long-range transport in mammalian cells.
Dynein responds asymmetrically to load
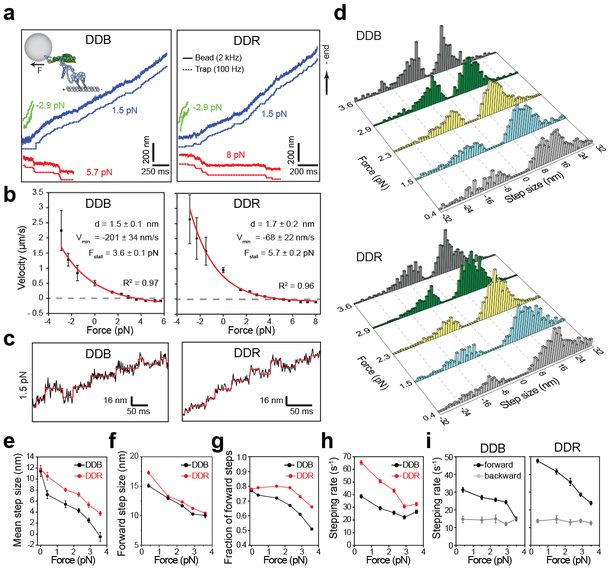
How mammalian dynein-dynactin steps under load has been a subject of considerable debate. While some studies found mammalian dynein to be a surprisingly weak motor (0.5 – 1.0 pN)8-11, other studies reported larger stall forces (4– 7 pN)23, 26, 34. In addition, there is no consensus on whether dynein takes regular 8 nm steps under load26, or takes large steps (24–32 nm) at low load and shorter steps (8 nm) at higher loads8. To address these questions, we exerted constant loads to beads driven by single complexes in the forward and backward direction using optical trap. Similar to yeast dynein35, 36, both DDB and DDR responsed asymmetrically to load, favoring faster movement when pulled forward and resisting backward pull from the trap (Fig. 3a-b). The velocity approached zero at near-stall forces (3.6 ± 0.1 pN for DDB and 5.7 ± 0.2 pN for DDR, mean ± s.e.m.) and both complexes moved slowly towards the plus-end when subjected to super-stall forces. These results contradict the model that dynein forms a catch bond with MTs and is unable to step backward under hindering loads10. Force-velocity (F-V) relationships of DDB and DDR fit well to a simple model that the forward stepping rate increases with assisting load, while the backward stepping rate is load independent36. DDR motility is more resistant to applied load compared to DDB, which might result from sharing of external load by a larger number of dynein heads in DDR36.
Figure 3. Velocity and stepping of mammalian dynein-dynactin under load.
(a) Example traces of bead movement driven by DDB and DDR in a force-feedback controlled trap under assisting (green trace), low hindering (blue trace), and superstall hindering (red trace) loads. (b) Force-velocity curves for DDB and DDR (mean ± s.e.m., n = 17, 21, 19, 22, 27, 27, 30, 25, 26, 24, 17, 15, 15, 19, 20, 20, 22, 24, 24, 23, 30, 21, 18 runs from left to right, three independent experiments per condition). Velocities at 0 pN load were obtained from fluorescence measurements (nDDB = 209, nDDR = 223). Red curves represent a fit to a one-state model, in which the backward stepping is load independent. Uncertainties in the parameters of the fit correspond to s.e. (c) Representative traces of single beads driven by DDB and DDR under 1.5 pN hindering load. Horizontal lines represent a fit to a step-finding algorithm. (d) Normalized step size histograms of DDB and DDR under load. (e) The mean step size, (f) forward step size, (g) the fraction of forward steps, (h) overall stepping rate and (i) the breakdown of forward and backward stepping rates of DDB and DDR under constant hindering loads. In d-i, n = 480, 619, 1025, 709, 352 for DDB and 501, 1263, 1129, 792, 741 for DDR from 0.4 to 3.6 pN (three independent experiments). The values at 0 pN were obtained from fluorescence measurements in Figure 2. Error bars are s.e.m. in e and s.e. from a single exponential decay fit in g-h.
To gain additional insight into the mechanism of mammalian dynein motility, we analyzed the stepping behavior of single complexes under constant hindering load at saturating (1 mM) ATP (Fig. 3c-d). Similar to the unloaded conditions at limiting ATP, dynein takes regular 8 nm, as well as large (16–32 nm) steps both in the forward and backward directions under all loads tested (0.4–3.6 pN). While DDB and DDR had similar stepping properties in the absence of load (Fig. 2b), differences between their stepping became more apparent under load. An increase in hindering load shifted the directional preference of steps taken by DDB and DDR towards the plus-end and reduced the size of the steps taken in forward direction, which led to the reduction of the mean step size (Fig. 3e-f). However, DDR maintained a lower probability of backward stepping, faster stepping rate, and higher velocity than DDB under all loads tested (Fig. 3g-i), suggesting that load sharing between dynein heads increases resistance of the complex to external load.
Analysis of dwell times between consecutive steps at low hindering load (0.4 pN) showed that DDR steps at a faster rate (65 ± 2 s−1; mean ± s.e.) than DDB (39 ± 1 s−1) at saturating ATP (two-tailed t-test; p <0.0001; Fig. 3h and Supplementary Fig. 4). This is consistent with stepping measurements at limiting ATP (Fig. 2c) and accounts for the faster velocity of DDR. Remarkably, we observed that the forward stepping rate decreases by load, while the backward stepping rate remains nearly constant under 0.4 – 3.6 pN load (Fig. 3i), consistent with the concave-up shape of their F-V relationship (Fig. 3b). At near-stall forces of DDB (3.6 pN), the velocity remains close to zero because the motor is equally likely to take steps in the forward and backward directions (pb = 0.5, Fig. 3g-i). Therefore, dynein motility stalls at resistive forces at which backward and forward stepping rates become equal to each other35, 37, not due to complete inhibition of motor stepping.
Tethering multiple dyneins does not change the velocity
We next turned our attention to address how recruitment of a second dynein to dynactin by BicDR1 results in faster movement19. Previous studies on processive motors did not observe a substantial increase in velocity when multiple motors are recruited to a cargo38-40, but this has not been tested for mammalian dynein. To test whether multiple dyneins move faster than a single motor, we linked 1–3 DDBs to a common DNA chassis and measured their velocities (Supplementary Fig. 5a-e and Supplementary Table 2). To eliminate the recruitment of inactive dyneins without dynactin and a cargo adaptor, DDBs were attached to the chassis from BicD2N41. These assemblies walked processively along MTs when incubated with dynein and dynactin (Supplementary Fig. 5f). As more BicD2N were recruited per chassis, the run length increased several-fold, but the velocity remained constant23 (Supplementary Figs. 5 and 6, and Supplementary Video 3). To compare DDB to the plus-end-directed kinesin, we also coupled 1–3 kinesin-1 motors to the same DNA chassis. Similar to dynein, increasing the number of kinesins on the chassis resulted in longer run length, but had little effect on the velocity40 (Supplementary Fig. 5g-h). These results are consistent with previous reports38-40 and show that an increase in motor copy number cannot explain the substantial increase in DDR velocity.
Tail-tail interactions increase the velocity
We next tested whether the specific arrangement of two dyneins bound to dynactin leads to faster movement. Structural studies revealed that the motor domains of two dyneins make contacts at their linker and AAA+ ring domains when they are positioned side by side on an MT18. In addition, the tail domains of two dyneins make extensive contacts (Fig. 4a)19, which can mediate allosteric communication or mechanical coupling between dyneins. If these interactions between two closely packed dyneins facilitate faster movement, we expect elimination of these interactions by truncation of the second dynein at various positions to result in reduction of velocity (Fig. 4a, Supplementary Fig. 7). To eliminate linker-ring interactions while maintaining possible tail-tail interactions, we expressed dynein tail containing residues 1–1,074 of DHC with associated chains (long tail, DynLT)19. To eliminate both interactions, we generated a shorter tail construct (DynST) containing residues 1–481 without the associated chains. We also introduced charge reversal mutations on the second dynein binding site of BicDR1 (BicDR1mut, E165R, and E167K, Supplementary Fig. 7) to eliminate recruitment of a second dynein to dynactin.
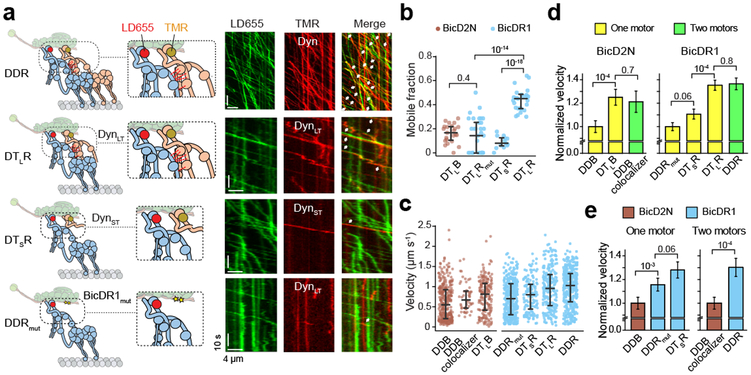
Figure 4. Tail-tail interactions increase the velocity of dynein-dynactin.
(a) (Left) Schematics of DDR assemblies with full-length and truncated dynein constructs labeled with different dyes. Red dashed lines represent tail-tail interactions between two dyneins recruited to dynactin. Yellow stars in BicDR1mut represent point mutations on the recruitment site for the second dynein. See Supplementary Fig. 7c for the schematics of DDB assemblies with full-length and truncated dynein constructs. (Right) Sample kymographs of LD655-dynein and TMR-labeled dynein or dynein tail in the presence of dynactin and BicDR1. White arrows represent colocalization of the processive motility in the TMR and LD655 channels. (b) The ratio of processive TMR-tail to LD655-dynein runs in different dynein-dynactin assemblies (mean ± s.e.m., n = 23, 34, 13, and 27 MTs from left to right, three independent experiments). (c) Velocity comparison of dynein-dynactin complexes assembled with full-length and tail-truncated dyneins (three independent experiments, n = 319, 51 172, 373, 147, 222, and 365 from left to right). The central line and outer edges represent the median and first and third quartiles, respectively. (d) Velocity comparison of complexes assembled with one vs. two dyneins in the presence of BicD2N and BicDR1 (mean ± s.e.m.). (e) Velocity comparison of DDB and DDR complexes assembled primarily with one or two dyneins (mean ± s.e.m.). In b, d, and e, p values are calculated using two-tailed t-test.
To determine how these modifications affect the recruitment of a second dynein, we labeled full-length dynein and dynein tail with different fluorophores and mixed them in the presence of dynactin and a cargo adaptor (Fig. 4a). Because dynein tail lacks the motor domain, its processive run can only be facilitated by its side-by-side arrangement with a full-length dynein on dynactin. We quantified the ratio of processive runs of dynein tail to full length dynein on individual MTs (Fig. 4b). We observed that BicDR1 recruits DynLT as efficiently as full-length dynein as the second motor on dynactin. BicDR1mut reduced the recruitment of DynLT along with dynein by 50%, suggesting that these mutations disrupted the second dynein binding site. Consistent with less efficient recruitment of two dyneins to dynactin by BicD2N19, we observed a lower ratio of DynLT to dynein processive runs using this adaptor. Compared to DynLT, recruitment of DynST to dynactin was less efficient (Fig. 4b), indicating that tail-tail interactions are critical for the stability of second dynein on dynactin.
To determine which dynein truncation leads to lower velocity, we first compared the velocities of complexes assembled with full-length dyneins (Fig. 4a, Supplementary Fig. 7). Consistent with previous observations19, DDB complexes assembled with two dyneins moved 20% faster than other complexes assembled primarily with single dynein, as previously reported19. Similarly, DDR complexes that recruit two dyneins moved 30% faster than DDRmut that recruits mostly single dynein (Fig. 4c-d). We also observed that cargo adaptors differentially affect dynein velocity (Fig. 4c,e). DDR complexes that primarily recruit single dynein (DDRmut) moved 20% faster than DDB. Similarly, DDR complexes assembled with two full-length dyneins moved 30% faster than DDB complexes that contain two full-length dyneins (DDB colocalizers).
Next, we compared the velocities of complexes assembled with one full-length dynein and one dynein tail. Surprisingly, we observed that complexes that contain one full-length dynein and one DynLT moved at a similar velocity to complexes with two full-length dyneins both in the presence of BicDR1 and BicD2N (Fig. 4d). This is in contrast with a previous observation that two full length dyneins are required for faster movement19, and the discrepancy may be related to differences in reconstitution conditions and methods used for quantifying motor velocity. Unlike DynLT, recruitment of DynST to dynactin resulted in slower velocities than complexes assembled with two full-length dyneins (Fig. 4d). Collectively, our results demonstrate that both cargo adaptor identity and the specific arrangement of two dyneins in the complex affect the velocity of dynein-dynactin motility. The increase in velocity does not require stepping and force generation of two full-length dyneins. Instead, side by side arrangement of dynein tail with a single full-length dynein is sufficient for faster movement.
The multiplicity of dynein increases force generation
We next tested whether the recruitment of two dyneins by BicDR1 could account for the observed increase in force generation. To address this question, we first measured the stall force of DDR complexes that contain one full-length dynein and one DynLT (DTLR). We found that DTLR stalls at forces (3.7± 0.1 pN; mean ± s.e.m.) similar to that of DDB (3.6 ± 0.1 pN), and much lower than that of DDR (6.3 ± 0.2 pN, two-tailed t-test, p < 0.0001)19. Because DTLR moves at similar velocities but stalls at nearly one half of DDR stall forces, we concluded that higher force generation, but not the higher velocity, of DDR is due to recruitment of two active dynein motors (Fig. 5a-b).
Figure 5. The multiplicity of dynein-dynactin increases total force generation.
(a) (Left) Schematic of fixed optical trapping assay for measuring the stall force of single DDB tethered from the cargo adaptor (not to scale). (Right) Example traces show typical stall of a bead driven by DDB, DTLR, and DDR. The detachment of the motor from an MT after the stall is denoted by a red arrowhead. (b) Stall forces of DDB, DTLR, and DDR (mean ± s.d., n = 67 runs from 16 beads from three independent experiments for DDB, 111 runs from 24 beads from four independent experiments for DTLR, and 84 runs from 21 beads from three independent experiments for DDR). The line and whiskers represent the mean and s.d., respectively. (c) Schematic shows the assembly of 1–3 DDBs on a DNA chassis. (d) Quantification of DNA chassis labeled with 1, 2 and 3 BicD2Ns or kinesin-1 motors (right) from a native agarose gel (left) from three independent experiments. Full gels can be found at Supplementary Fig. 8. (e) Example traces show typical stalls of beads driven by single 1–3 DDB and KIN chassis in a fixed trap assay. The detachment of the motor from an MT after the stall is denoted by a red arrowhead. Scale bars are 1 s. (f) Stall forces of 1–3DDB and 1–3KIN chassis. From left to right, n = 35 from 12 beads, 42 from 15 beads, 32 from 10 beads, 83 from 19 beads, 72 from 17 beads and 143 from 26 beads (three independent experiments per condition). The line and whiskers represent the mean and s.d., respectively. (g) Calculated stall forces of the DNA chassis assembled with one, two and three motors after accounting for incomplete complexes (see Methods). Error bars represent s.d. Solid curves represent a fit to the power law (y = Fstall.xc) where Fstall is the stall force of one motor and c is the scaling exponent (mean ± s.e.). In b and g, p values are calculated using two-tailed t-test.
We also measured the stall forces of DNA chassis assembled with multiple DDBs to determine how overall force generation increases by the DDB copy number (Fig. 5c, Supplementary Fig. 5). We estimated that 82% of the DNA chassis that contains two complementary sites to BicD2N-DNA (2DDB) recruits two BicD2Ns and 57% of the chassis with three complementary sites (3DDB) recruit three BicD2Ns (Fig. 5d). As the number of motors on the chassis was increased, we observed a substantial increase in DDB stall force (Fig. 5e-f), demonstrating that dynein copy number contributes to collective force generation of the complex42, 43. Unlike dyneins, the coupling of multiple kinesins using the same DNA chassis resulted in a modest increase in stall force40, 44 (Fig. 5e-f). Stall forces of 1–3 DDB assemblies are additive with the number of dynein motors42, 45, while the stall force of kinesin assemblies increases sub-proportionally with the number of kinesins46 (Fig. 5g).
Two dyneins win against kinesin in a tug-of-war
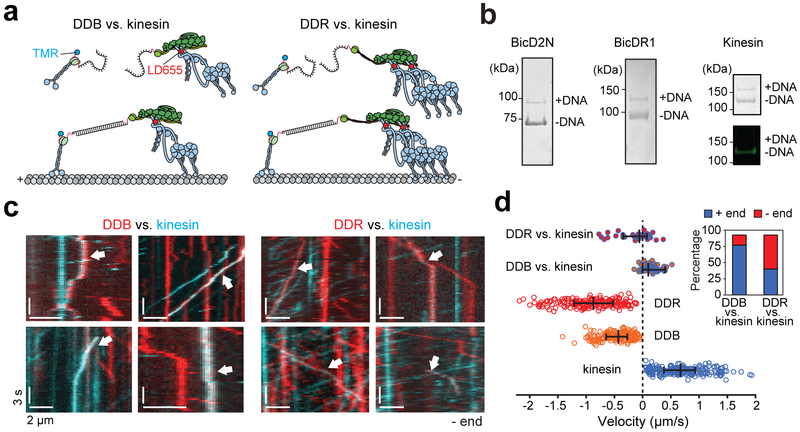
We previously showed that DDB is a strong opponent to kinesin in a one-to-one tug-of-war in vitro23. DDB slows down the overall velocity of kinesin by ~20-fold, but kinesin still wins a majority (80%) of these competitions and moves dynein towards the MT plus-end, presumably because it exerts a higher force (6 pN) than DDB. We asked whether coupling of two dyneins by BicDR1 would enable dynein-dynactin to compete more effectively against kinesin. We pitted one DDR or DDB against one kinesin using a DNA scaffold. The motors were labeled with different colored fluorophores and the direction and velocity of the colocalizers were analyzed as individual complexes moved on MTs (Fig. 6a-b and Supplementary Video 4). We observed that 57% of the kinesin-DDR colocalizers moved towards the minus-end, and the median velocity of all colocalizers was 59 nm s−1 towards the minus-end (Fig. 6c-d). In comparison, only 17% of the kinesin-DDB colocalizers moved towards the minus-end, and the median velocity of all colocalizers was 94 nm s−1 towards the plus-end (Fig. 6c-d). Tsshese results show that the recruitment of a second dynein favors dynein-dynactin to win the tug-of-war.
Figure 6. Recruitment of a second dynein shifts the balance towards the MT minus-end in a tug-of-war.
(a) Kinesin and BicDs were labeled with complementary DNA oligos and different-colored fluorescent dyes. Kinesin and dynein assemblies were connected by DNA hybridization. (b) (Left) The fraction of BicDs labeled with DNA was estimated from a denaturing gel. (Right) Kinesin was sub-stoichiometrically labeled with DNA (top). Fluorescence image of the same gel shows that DNA-labeled kinesin does not get labeled with TMR (bottom). n = 3 independent experiments. Full gels can be found at Supplementary Fig. 5c-d. (c) Kymographs represent the motility of TMR-kinesin (cyan), LD655-dynein (red), or kinesin-dynein assemblies (white arrows). (d) Velocity distribution of individual motors and kinesin-dynein assemblies. Negative velocities correspond to the movement towards the MT minus-end (n = 36, 46, 193, 179, and 210 from top to bottom, three independent experiments per condition). The line and whiskers represent the median and 65% c.i., respectively. The inset shows the percentage of plus- and minus-end directed runs when DDB or DDR pitted against kinesin.
Discussion
Our results showed that cargo adaptors regulate the velocity and stepping of dynein-dynactin along MTs. Mammalian dynein-dynactin is a robust motor that has similar stepping and load-bearing properties to dynein from yeast31. Contrary to earlier reports of regular 8 nm26 or larger (24–32 nm) steps8, dynein takes variable (8–32 nm) steps (Fig 2b and 3d). Unlike kinesin-1 that predominantly follows a single MT protofilament towards the plus-end47, dynein frequently takes sideways and backward steps on an MT. This may be due to the flexible and elongated structure of DHC, which allows a large diffusional search of a stepping head for the next tubulin binding site.
The velocity and step size of dynein were strongly affected by external load. Dynein moves faster when subjected to assisting load and moves slowly backward when the hindering load exceeds the stall force. DDR was more resistant to hindering load than DDB, presumably because the external load is shared by a larger number of dynein monomers in DDR23. Consistent with earlier reports, multiple dynein-dynactins stand against larger backward loads more effectively than kinesins, and the overall stall force is additive with the dynein copy number10, 23. A previous study10 proposed that this is due to catch-bond formation of dynein with an MT, which prevents the motor to step backward under high hindering loads. In addition, load-dependent reduction of dynein step size from regular 32 nm steps to 8 nm steps enable trailing motors to catch up with load-bearing motors in the lead for clustering on a cargo. Our results are not fully consistent with this view, because we observed that dynein takes variable steps under all loads tested. While the motor took less frequent large forward steps under high load, the reduction in the mean step size under high load was mostly due to the increase in the proportion of backward steps (Fig. 3e-g). Additionally, we found that dynein steps backward under superstall loads (Fig. 3b). We propose that stall forces of a few dyneins are additive because individual motors remain strongly attached to the MT for an extended period of time when pulled backwards35, 36, 48, 49 and the backward stepping rate of dynein is independent of 1–4 pN hindering load (Fig. 3i). In comparison to dynein, kinesin dissociates within a few seconds after a stall and prematurely releases from the MTs at sub-stall forces44, which limits the ability of multiple kinesins to bear large tension46. This implies that recruiting a larger number of kinesin and dynein on a cargo while maintaining their ratios constant would shift the transport towards the retrograde direction10.
Our results also provide an explanation for how recruitment of a second dynein increases the velocity of dynein-dynactin. We did not observe substantial differences in the size and direction of steps taken by DDB and DDR in the absence of load. Therefore, the tight-packing of two dyneins do not restrict the inherent backward and sideways steps, and favor a more direct and faster route toward the MT minus-end19. Instead, DDR moves faster because it hydrolyzes ATP at a faster rate and steps more frequently than DDB (Fig. 2c). We showed that cargo adaptors differentially regulate dynein velocity, which may be due to slight differences in structural arrangements and contacts between dyneins when recruited to dynactin by these proteins18, 19. Recruitment of a second dynein to dynactin increases the velocity of both DDB and DDR due to the interactions between the tails of two dyneins positioned side-by-side. It is possible that extensive contacts between two dyneins allosterically modulate the catalytic activity of each motor, or the presence of dynein tail prevents dynein motor domains from reorienting into conformations that impair stepping along MTs. Higher resolution structures of dynein-dynactin assemblies on MTs are required to dissect how cargo adaptors regulate the structural arrangement of dyneins in the complex.
Contrary to regulation of dynein velocity, the number of dyneins bound to dynactin determines the overall force generation of the complex43. We showed that kinesin-1 is more likely to win tug-of-war and move the cargo assemblies towards the MT plus-end when pitted against DDB, which primarily recruits one dynein. Recruitment of a second dynein by BicDR1 shifts the force balance towards dynein and facilitates faster minus-end-directed transport of cargo assemblies in vitro (Fig. 6). This result is consistent with studies of Rab6 vesicle transport that overexpression of BicDR1 during the early phase of neural development restricts anterograde transport by kinesin-3 and accumulates secretory vesicles around the centrosome13. It remains to be tested whether this is because DDR is more processive, moves faster or produces higher forces than DDB13.
Our results challenge the current view that dynein is able to drive transport when functioning in large teams or clustering on a cargo, and that weak force generation of single dynein enables fine tuning of the total force by controlling the number of dyneins carrying the cargo10, 50. Instead, we propose that intracellular cargos are transported retrogradely by a small number of dynein motors over long distances along MTs. In addition, dynein’s resistance to large hindering load may enable multiple motors to efficiently perform ‘heavy-duty’ cellular functions, such as transporting nuclei in migrating neurons and positioning the mitotic spindle during mitosis1.
Methods
BG activation of LD655.
LD655-NHS (2 mmol, Lumidyne Technologies) was dissolved in 1 mL dry DMF, and mixed with 200 μL DIEA and 2 mg BG-NH2 (NEB). The reaction mixture was vortexed and then incubated in the dark at room temperature for 15 mins. The reaction solution was then poured into 30 mL of ethyl acetate and centrifuged. The precipitated residue was dissolved in 2 mL of ddH2O and purified using the C18 T3 column with a 10 mM TEAA pH 6.5 buffer mobile phase in a gradient of 10‐90% acetonitrile. After evaporation of acetonitrile, the product was concentrated, buffer exchanged over a Sep‐Pak C18 column and eluted with methanol and dried by a rotary evaporator. In electrospray ionization mass spectrometry (ESI-MS), the mass divided by charge number (m/z) for C60H69N9O15S4 [M-H]− was 1288.6, similar to the calculated value of 1288.4.
Cloning and plasmid generation.
The constructs that express native human dynein (SNAP-DYNC1H1), phi dynein mutant (SNAP-DYNC1H1E1518K/R1567K) in a pACEBac1 vector backbone; and mouse BICD2–400-GFP, BICD2–400-SNAPf, and BICDR1-GFP in a pOmniBac vector backbone were provided by A. P. Carter19, 22. BicDR1-SNAPf was sf9-codon optimized and cloned into a pOmniBac-derived vector. Dynein tail constructs containing residues 1–481 (DynST) and 1–1,074 (DynLT) of DYNC1H1 were fused with an N-terminal SNAP-tag and a C-terminal GST tag into the pOmniBac vector. The BicDR1E165R/E167K-GFP construct was generated by mutagenesis of BICDR1-GFP in the pOmniBac vector. All constructs contain His6-ZZ tag followed by a TEV protease cleavage site for protein purification. Except for DynST, all dynein heavy chain constructs were coexpressed by fusing to the pDyn2 plasmid that contain genes from IC2C, LIC2, TCTEX1, LC8, and ROBL1, as described22.
A cDNA for a full-length human kinesin-1 (KIF5B; clone id: 8991995) was obtained from GE Dharmacon. SNAPf-GFP was fused to the C-terminus of tail truncated human kinesin (1–560 amino acids) and cloned into the pET17b vector containing a His6 tag at the C-terminus. The list of constructs used for each dataset is given in Supplementary Table 3.
Protein expression and purification.
Dynein, BicD2N and BicDR1 constructs were expressed in sf9 cells. SNAP-tagged proteins were labeled with BG-functionalized TMR (NEB) or LD655 dyes, or biotin (NEB), and purified as described22. Native dynactin was purified from pig brains using the large scale-SP-sepharose protocol and anion exchange chromatography using a MonoQ column (GE)29. Human kinesin-1 (K560-SNAPf-GFP) was expressed in BL21DE3 cells and purified by Ni-NTA chromatography, as described previously51. Protein concentration was determined using the Bradford colorimetric assay.
ATPase assays.
MTs were polymerized by adding 5 mg ml−1 pig brain tubulin to BRB80 buffer (80 mM PIPES pH 6.8, 1mM MgCl2, 1 mM EGTA) supplemented with 1 mM GTP,10 μM taxol and 10% Dimethyl sulfoxide (DMSO) for 1 hour at 37°C. Taxol-stabilized MTs were pelleted at 50,000 g, resuspended in DLB (30 mM HEPES, pH 7.2, 2 mM MgCl2, 1 mM EGTA and 10% glycerol) supplemented with 10 μM taxol and stored in dark at room temperature. ATPase assays were carried out using the EnzCheck Phosphate Assay Kit (Life Technologies). In order to maximize the assembly of dynein to dynactin, 1:9:16 molar ratio of dynein, dynactin, and BicD2N or BicDR1 were incubated on ice for 10 min followed by an immediate use. We also used a full-length dynein mutant (phi-dynein) that interacts more stably with dynactin but moves at wild-type dynein velocities6.
The fraction of dynein assembled with dynactin and a cargo adaptor was estimated by GFP pull down experiments. Briefly, we mixed dynein, dynactin and a GFP-tagged cargo adaptor using the same concentrations as in the ATPase assays. The mixture was mixed with antiGFP-beads and pelleted at 13,000 g for 5 min. The pellet and supernatant were run on a denaturing gel and the ratio of dynein pelleted with beads was calculated from gel pictures in ImageJ. Nonspecific binding of dynein and dynactin to beads in the absence of a cargo adaptor was accounted for in control assays. The motors pelleted with beads were assumed to be the active fraction, whereas the rest of the motors in solution were not associated with dynactin and a cargo adaptor. We estimated that 9% of dynein is associated with dynactin under these conditions. Because dynein not associated with dynactin exhibits extremely low catalytic activity under the same tubulin concentrations, the ATPase rate of the 90% unbound fraction was low and it was accounted for when calibrating the ATPase activity of fully assembled complexes.
Assay reaction consisted of dynein motility buffer (DMB: 30 mM HEPES pH 7.0, 5 mM MgSO4, 1 mM EGTA, 1 mM tris(2- carboxyethyl)phosphine (TCEP) supplemented with 25 mM KCl, 2.5 nM dynein, 22.5 nM dynactin, 40 nM BicD2N or BicDR1, 200 nM methylthioguanosine, 1 U ml−1 purine nucleoside phosphorylase, 2 mM DTT, taxol-stabilized MTs, and 2 mM ATP. The total reaction volume was 100 μL per sample. The rate of inorganic phosphate generation was measured using a 96-well plate reader (μQuant, BioTek Instruments) set to record absorbance at 360 nm at 60 s intervals for 30 min against a buffer-only blank. All reactions were performed in triplicate. ATPase activity for DDB and DDR were calculated from the raw data by accounting for the fraction of fully assembled complexes in the mixture.
Construction of the DNA chassis.
DNA oligos were designed by the oligoAnalyzer tool (IDT DNA) to minimize the formation of secondary structures. To generate the desired DNA chassis that can engage one to three motors, DNA fragments were joined using short complementary DNA splints (Supplementary fig. 5 and Supplementary Table 2). These fragments were annealed by heating to 95 °C followed by slowly cooling down to ambient temperature in 30 mM Tris pH 7.4, 150 mM NaCl. The annealed DNA fragments were then phosphorylated and ligated by incubating with T4 polynucleotide kinase (PNK) and T4 DNA ligase for 4 h at ambient temperature in T4 DNA ligase reaction buffer (NEB). Excess DNA oligonucleotides and incomplete DNA products were removed and the DNA chassis were gel purified using 5% TBE-urea denaturation polyacrylamide gel electrophoresis. The DNA fragments were extracted from the gel and their concentration was quantified from the absorbance at 260 nm.
Labeling with DNA oligonucleotides.
For labeling cargo adaptors and kinesin with DNA, 25 μM DNA oligos with a 6-carbon spacer and amino group modification at their 5’-ends were functionalized with 4 mM BG-GLA-NHS (dissolved in anhydrous DMSO) in 50 mM HEPES pH 8.5 buffer containing 50% DMSO. The mixture was reacted overnight in dark at room temperature. The excess ligand was removed by ethanol precipitation. Isolated BG-DNA was dissolved in DMB and its concentration was quantified from the absorbance at 260 nm.
BicD2N- or BicDR1-SNAPf was mixed with BG-DNA in DMB for 1 h at 4 °C. Excess unreacted DNA was removed using TSKgel G4000SWXL size exclusion column (Tosoh). For labeling kinesin with DNA, K560-SNAPf was mixed with BG-DNA in DMB for 1 h at 4 °C. The protein was then labeled by mixing with 10-fold excess BG-TMR-Star (NEB) for 30 min at 4 °C. Excess unreacted DNA and fluorophores were removed via the MT bind and release assay. The DNA labeling efficiency was quantified by comparing the intensities of the DNA-labeled and unlabeled protein bands on a denaturing gel. The DNA concentration was adjusted to reduce the labeling efficiency to ~30% in order to minimize the likelihood (<9%) of labeling a single dimeric protein with two DNA oligonucleotides.
To link multiple DDBs to a DNA chassis, dynein, dynactin and BicD2N-DNA were mixed at 3:5:1 molar ratio in DMB supplemented with 1 mg/ml BSA. BicD2N was kept limiting to minimize binding of free BicD2N to the DNA chassis. The protein mixture was incubated on ice for 10 min, and then mixed with the DNA chassis at 200:1 molar ratio in DMB supplemented with 1 mg/ml BSA and 50 mM NaCl for 30 min at room temperature. For linking multiple kinesins to DNA chassis, DNA-labeled kinesin was mixed with the DNA chassis at 200:1 molar ratio in BRB80 supplemented with 50 mM NaCl for 30 min at room temperature. Gel shift assays were performed on 1.5% agarose gel in 1x TBE buffer and run for 3 h at 4 °C. To quantify the fractions of DNA chassis labeled with 1, 2 or 3 motors, the gel image was background subtracted and the ratio of the signal detected from an integrated area on a given band over the total signal from all bands was calculated in ImageJ.
Functionalization of beads and QDs.
860 nm and 500 nm diameter carboxy latex beads (Life Technologies) were washed twice in activation buffer (10 mM MES pH 6.0, 100 mM NaCl) by pelleting at 10,000 g. The resuspended beads were then activated by mixing with 1 mg each of 1-Ethyl-3-(3-dimethylaminopropyl) carbodiimide (EDC) and N-hydroxysulfosuccinimide (Sulfo-NHS) crosslinkers (Pierce) dissolved in 100 μl of dimethylformamide (DMF) for 30 min. The beads were then allowed to react with rabbit polyclonal αGFP antibodies (Covance) in coupling buffer (100 mM sodium phosphate, pH 7.4) by shaking for 2 h at room temperature. The beads were then passivated by adding 10 mg/ml bovine serum albumin (BSA) for 6 h at 4 °C, washed three times with phosphate-buffered saline (PBS), pH 7.4, and stored at 4 °C in PBS supplemented with 0.5 mg/ml BSA and 0.1% sodium azide. 200 nM amine-coated QDs emitting at 655 nm (ThermoFisher) was mixed with 1 mM BG-GLA-NHS (NEB) in 100 mM sodium borate buffer, pH 8.0 for 2 h in dark at room temperature. The excess unreacted ligand was filtered and the functionalized QDs were spin-concentrated using 30,000 MWCO centrifugal filter units (Amicon).
Motility assays.
Biotinylated MTs were prepared by mixing unlabeled pig tubulin and 2% biotinylated tubulin in BRB80 supplemented with 2 mM GTP and 20% DMSO for 30 min at 37°C. 100 nM taxol was added to the mixture and the reaction was kept for an additional 60 min. Unpolymerized tubulin was removed by pelleting at 20,000 g for 12 min and resuspending MTs in BRB80 containing 100 nM taxol. For MT-bridge assays, MTs were polymerized using Cy5-labeled tubulin instead of biotinylated tubulin. To immobilize biotinylated MTs to the surface of a flow chamber, 1 mg/ml BSA-biotin (Sigma) was introduced into the flow chamber and then washed with BRB80 buffer supplemented with 1.25 mg/ml casein (Sigma). The chamber was incubated with 1 mg/ml streptavidin (NEB) and washed with BRB80-casein.
For single-molecule DDB and DDR motility experiments, fluorescently-labeled dynein was used for imaging. Dynein, dynactin and a cargo-adaptor (BicD2N or BicDR1) were mixed at 1:5:20 molar ratio in DMB supplemented with 1 mg/ml BSA. For experiments with dynein tail, TMR-labeled tail was added to the mixture at a 4-fold molar excess of LD655-labeled dynein. The mixture was incubated on ice for 10 min, diluted in DMB supplemented with 1.25 mg/ml casein (DMB-C) and then flowed into the chamber. After 2 min, the chamber was washed with 40 μl DMB-C twice. Finally, the chamber was washed with 20 μl dynein stepping buffer (DMB-C supplemented with 2.5 mM PCA (protocatechuic acid), 35 μg/ml PCD (protocatechuate-3,4-dioxygenase), 1 mM Mg.ATP) and motility was recorded for 5 min.
For multiple DDBs on a DNA chassis, the protein-DNA mixture was diluted in DMB-C and flown into the chamber. The chamber was washed with DMB-C and dynein stepping buffer. For imaging single or multiple kinesins, the complexes were diluted in BRB80 supplemented with 1.25 mg/ml casein (BRB-C). The chamber was washed with BRB-C and then with stepping buffer (BRB-C supplemented with 2.5 mM PCA, 35 μg/ml PCD and 1 mM Mg.ATP), and imaged immediately. Motility of individual DDB and kinesin chassis was monitored by tracking the TMR label on the chassis.
For DDB and DDR stepping measurements, SNAP-dynein was assembled with dynactin and a cargo adaptor at 1:5:20 molar ratio in DMB supplemented with 1 mg/ml BSA for 10 min on ice, mixed with 1000x excess BG-coated QD-655 and left for another 5 min on ice. For dynein stepping, dynein and cargo adaptor were not added to the mixture. The mixture was then diluted in DMB-C, flown into the chamber, and incubated for 2 min. The motility chamber was washed with DMB-C to remove unbound protein and QDs. The sample was imaged in dynein stepping buffer supplemented with 1 mM Mg.ATP for dynein only and 5 μM Mg.ATP for DDB and DDR.
For tug-of-war experiments, BicD2N or BicDR1 labeled with a DNA oligo were mixed with LD655-labeled dynein and dynactin at 2:5:1 molar ratio in DMB supplemented with 1 mg/ml BSA and incubated on ice for 10 min. Kinesin labeled with a complementary DNA oligo and TMR was then added to the mixture at an equimolar ratio to the cargo adaptor. 50 mM NaCl was added for DNA annealing, and the mixture was incubated for 10 min on ice. The mixture was then flown into the chamber, incubated for 2 min, and washed with DMB-C. The sample was washed with dynein stepping buffer and imaged immediately.
Microscopy.
Fluorescence imaging experiments were performed using a Nikon multicolor TIRF microscope equipped with a Nikon Ti-E microscope body, a 100× 1.49 N.A. apochromat oil-immersion objective (Nikon), perfect focusing system and an electron-multiplied charge-coupled device (EM-CCD) camera (Andor, Ixon EM+, 512×512 pixels) with an effective pixel size of 160 nm after magnification. Cy3 and Cy5 signals were filtered through 585/40 nm and 655/40 nm bandpass emission filters (Semrock), respectively. Multi-color fluorescence imaging was performed using the time-sharing mode. GFP, TMR and LD655 were excited with 0.05 kW cm−2 488 nm, 561 nm, and 633 nm laser beams (Coherent), respectively and movies were recorded at 150–300 ms per frame. QDs were excited with 1 kW cm−2 488 nm beam and movies were recorded at 30 ms per frame.
Data analysis.
Movies were analyzed using ImageJ. Kymographs were generated by plotting segmented lines along the MTs. The processive movement was defined and analyzed as described previously23. In velocity analysis, complexes that exhibit diffusive movement, run shorter than 200 nm and paused for more than 1 s were excluded. For two-color imaging, the fluorescent channels were overlaid in ImageJ to generate a composite image. Co-localization events were manually scored in kymographs. For dynein-tail two color imaging, tail spots that did not colocalize with the dynein channel were included in the velocity analysis. For stepping analysis, fluorescent spots of single QDs were localized using a 2D Gaussian fitting tracker algorithm, and trajectories were fitted by a custom-written step finder algorithm based on Schwartz Information Criterion49.
MT bridge assays.
MT bridges were assembled using taxol-stabilized MTs, as previously described32. 2 μm diameter carboxyl-latex beads were functionalized with GFP-antibody using EDC-NHS crosslinking. The beads were incubated with GFP-SRS-MTBD, which contains MT binding domain and stalk of mouse cytoplasmic and tightly binds to an MT52, for 10 min in DMB buffer. Excess protein was removed by pelleting and resuspending beads at 15,000 g. 0.5 μm diameter GFP-antibody coated latex beads (cargo beads) were incubated with DDB or DDR complexes assembled using BicD2N-GFP or BicDR1-GFP on ice for 10 min. Unbound protein was removed by pelleting and resuspending the beads at 8,000 g for 3 min. The beads were resuspended in 15 μl DMB supplemented with 1 mM ATP, 1 mg/ml casein, 10 μM taxol, 2 mM PCA and 50 nM PCD.
GFP-SRS-MTBD coated beads were nonspecifically adsorbed to the surface of a flow chamber for 10 min. The sample was washed twice with 30 μl DMB-C. Cy5-MTs were flowed into the chamber and incubated for 10 min. Unbound MTs were washed twice with 30 μl DMB-C. Finally, 0.5 μm cargo beads coated with DDB or DDR were flown into the chamber. Cy5-MTs were excited with 632 nm beam in the epifluorescence mode, and the signal was detected by the CMOS camera (Hamamatsu Orca v2, 109 nm effective pixel size) at 10 Hz. The sample was scanned to identify a stable MT bridge that is 10–30 μm long between two large beads and does not exhibit oscillations more than 200 nm. Freely diffusing cargo beads were trapped using a custom-built optical trap (see below), brought near an MT bridge and released for motility. The sample was excited with brightfield using a halogen lamp and a Nikon 1.40 NA oil condenser and the transmitted light was collected using a Nikon 1.49 NA oil objective and a monochrome CCD camera. Movies were recorded at 100 ms per frame. The xy position of the bead was determined by a 2D Gaussian fitting. The z position was determined from the bead intensity and calibrated from the radius of rotations in the y-axis. 3D traces were smoothed using a moving average of ten consecutive frames.
Optical trapping assays.
Optical trapping experiments were performed on a custom-built optical trap microscope setup36. Briefly, motor coated beads were trapped with a 2 W 1064 nm laser beam (Coherent) focused on the image plane using a 100 × 1.49 N.A. apochromat oil-immersion objective (Nikon). Cy5-labeled axonemes were excited with a 633-nm laser (Coherent), imaged using a monochrome camera (The Imaging Source) and moved to the center of the field of view using a locking XY stage (M-687, Physik Instrumente). The trap was steered with a pair of perpendicular acousto-optical deflectors (AODs, AA Opto-Electronic). The trapped bead was lowered to the surface of the axonemes using a piezo flexure objective scanner (P-721 PIFOC, Physik Instrumente). Bead position relative to the center of the trap was monitored by imaging the back-focal plane of a 1.4 N.A. oil-immersion condenser (Nikon) on a position-sensitive detector (First Sensor Inc.). For calibrating the detector response, a trapped bead was rapidly raster-scanned by the AOD and trap stiffness was derived from the Lorentzian fit to the power spectrum of the trapped bead.
For single motor experiments, 860 nm diameter anti-GFP coated latex beads were incubated with DDB and DDR assembled with BicD2N-GFP or BicDR1-GFP for 10 min on ice. For DTLR stall force experiments, dynactin and BicDR1 were assembled with un-tagged full-length dynein and biotin-labeled DynLT and mixed with 800 nm diameter streptavidin-coated beads (Spherotech) to ensure that complexes are attached to the beads through DynLT. For DNA chassis experiments, the chassis were labeled with biotin and the DNA-protein mixture was mixed with streptavidin-coated beads for 10 min on ice. In all experiments, the protein and DNA were diluted such that less than 30% of the trapped beads exhibit motility within 1 min of contact with the axonemes to ensure that 95% of the tested beads are driven by a single complex.
Forces generated by DNA chassis that contains one, two and three motors in Fig. 5g were calculated from stall force measurements (Fig. 5f) and the fractions of fully-assembled complexes and subcomplexes (Fig. 5d), as follows.
where Fn is the average stall force measured using a DNA chassis with n binding sites, fi is the force generation of i motors (i ≤ n), and si is the fraction of complexes with i motors bound to a DNA chassis with n binding sites.
For fixed trap assays, the trap stiffness was adjusted to allow beads to travel the linear range of the detector (100 – 150 nm) before stalling. To generate stall force histograms, position data from trap recordings (5 kHz) were down-sampled to 500 Hz and traces were then manually scored. A stall event is defined as a bead position to reach a plateau and remain stationary for at least 100 ms before rapidly (<2 ms) moving back to the trap center.
For force clamp assays, beads that walk for at least 100 nm against the trap were subjected to a force feedback control. Bead signal was acquired at 5 kHz and position feedback was performed at 100 Hz. Bead position was downsampled to 500 Hz and fit to a step-finding algorithm. Runs shorter than 200 nm in length or runs that contain sudden jumps larger than 50 nm were excluded from the velocity analysis. Velocities were determined from the slope of the run.
F-V curves were fitted to a one-state model with a fixed backward stepping rate and a force-dependent forward stepping rate36.
where F is the external force, d is the characteristic distance, Fstall is the stall force, Vmin is the minimum velocity, kB is the Boltzmann constant and T is absolute temperature.
Statistics and reproducibility.
At least three independent repetitions were performed to obtain any result. Number of replicates (n) and statistical analysis methods are clearly stated in the main text or in the Figure legends. Representative data are shown from independently repeated experiments with similar results.
Reporting Summary.
Further information of research design is available in the Nature Research Reporting Summary linked to this article.
Data availability.
All data that support the conclusions are available from the authors on request.
Code availability.
Code used in this paper is available from the corresponding author upon request.
Supplementary Material
Acknowledgments
We are grateful to the members of the Yildiz laboratory for helpful discussions, A. P. Carter, L. Urnavicius, S. Lacey (MRC, Cambridge) for providing plasmids and helping with protein expression and purification, and D. Drubin and C. Kaplan for multi-color TIRF microscopy. This work was funded by grants from the NIH (GM094522), and NSF (MCB-1055017, MCB-1617028) to A.Y, NIH (GM098859) to S.C.B. and NIH F31 fellowship to L.F.
Footnotes
Competing Interests
S.C.B. holds an equity interest in Lumidyne Technologies. All other authors declare no competing interests.
References
- 1.Roberts AJ, Kon T, Knight PJ, Sutoh K & Burgess SA Functions and mechanics of dynein motor proteins. Nature reviews. Molecular cell biology 14, 713–726 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Reck-Peterson SL, Redwine WB, Vale RD & Carter AP The cytoplasmic dynein transport machinery and its many cargoes. Nature reviews. Molecular cell biology 19, 382–398 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Carter AP, Cho C, Jin L & Vale RD Crystal structure of the dynein motor domain. Science 331, 1159–1165 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kon T et al. The 2.8 A crystal structure of the dynein motor domain. Nature 484, 345–350 (2012). [DOI] [PubMed] [Google Scholar]
- 5.Schmidt H, Gleave ES & Carter AP Insights into dynein motor domain function from a 3.3-A crystal structure. Nature structural & molecular biology 19, 492–497, S491 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhang K et al. Cryo-EM Reveals How Human Cytoplasmic Dynein Is Auto-inhibited and Activated. Cell 169, 1303–1314 e1318 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Can S, Lacey S, Gur M, Carter AP & Yildiz A Directionality of dynein is controlled by the angle and length of its stalk. Nature 566, 407–410 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mallik R, Carter BC, Lex SA, King SJ & Gross SP Cytoplasmic dynein functions as a gear in response to load. Nature 427, 649–652 (2004). [DOI] [PubMed] [Google Scholar]
- 9.McKenney RJ, Vershinin M, Kunwar A, Vallee RB & Gross SP LIS1 and NudE induce a persistent dynein force-producing state. Cell 141, 304–314 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rai AK, Rai A, Ramaiya AJ, Jha R & Mallik R Molecular adaptations allow dynein to generate large collective forces inside cells. Cell 152, 172–182 (2013). [DOI] [PubMed] [Google Scholar]
- 11.Torisawa T et al. Autoinhibition and cooperative activation mechanisms of cytoplasmic dynein. Nature cell biology 16, 1118–1124 (2014). [DOI] [PubMed] [Google Scholar]
- 12.Moughamian AJ & Holzbaur EL Dynactin is required for transport initiation from the distal axon. Neuron 74, 331–343 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schlager MA et al. Bicaudal d family adaptor proteins control the velocity of Dynein-based movements. Cell reports 8, 1248–1256 (2014). [DOI] [PubMed] [Google Scholar]
- 14.Trokter M, Mucke N & Surrey T Reconstitution of the human cytoplasmic dynein complex. Proceedings of the National Academy of Sciences of the United States of America 109, 20895–20900 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Toropova K, Mladenov M & Roberts AJ Intraflagellar transport dynein is autoinhibited by trapping of its mechanical and track-binding elements. Nature structural & molecular biology 24, 461–468 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Urnavicius L et al. The structure of the dynactin complex and its interaction with dynein. Science 347, 1441–1446 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chowdhury S, Ketcham SA, Schroer TA & Lander GC Structural organization of the dynein-dynactin complex bound to microtubules. Nature structural & molecular biology 22, 345–347 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Grotjahn DA et al. Cryo-electron tomography reveals that dynactin recruits a team of dyneins for processive motility. Nature structural & molecular biology 25, 203–207 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Urnavicius L et al. Cryo-EM shows how dynactin recruits two dyneins for faster movement. Nature 554, 202–206 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Splinter D et al. BICD2, dynactin, and LIS1 cooperate in regulating dynein recruitment to cellular structures. Molecular biology of the cell 23, 4226–4241 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.McKenney RJ, Huynh W, Tanenbaum ME, Bhabha G & Vale RD Activation of cytoplasmic dynein motility by dynactin-cargo adapter complexes. Science 345, 337–341 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schlager MA, Hoang HT, Urnavicius L, Bullock SL & Carter AP In vitro reconstitution of a highly processive recombinant human dynein complex. The EMBO journal 33, 1855–1868 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Belyy V et al. The mammalian dynein-dynactin complex is a strong opponent to kinesin in a tug-of-war competition. Nature cell biology 18, 1018–1024 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ross JL, Wallace K, Shuman H, Goldman YE & Holzbaur EL Processive bidirectional motion of dynein-dynactin complexes in vitro. Nature cell biology 8, 562–570 (2006). [DOI] [PubMed] [Google Scholar]
- 25.DeWitt MA, Cypranowska CA, Cleary FB, Belyy V & Yildiz A The AAA3 domain of cytoplasmic dynein acts as a switch to facilitate microtubule release. Nature structural & molecular biology 22, 73–80 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Toba S, Watanabe TM, Yamaguchi-Okimoto L, Toyoshima YY & Higuchi H Overlapping hand-over-hand mechanism of single molecular motility of cytoplasmic dynein. Proceedings of the National Academy of Sciences of the United States of America 103, 5741–5745 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.DeWitt MA, Chang AY, Combs PA & Yildiz A Cytoplasmic dynein moves through uncoordinated stepping of the AAA+ ring domains. Science 335, 221–225 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Duellberg C et al. Reconstitution of a hierarchical +TIP interaction network controlling microtubule end tracking of dynein. Nature cell biology 16, 804–811 (2014). [DOI] [PubMed] [Google Scholar]
- 29.King SJ & Schroer TA Dynactin increases the processivity of the cytoplasmic dynein motor. Nature cell biology 2, 20–24 (2000). [DOI] [PubMed] [Google Scholar]
- 30.Elshenawy MM et al. Lis1 activates dynein motility by pairing it with dynactin. bioRxiv (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Reck-Peterson SL et al. Single-molecule analysis of dynein processivity and stepping behavior. Cell 126, 335–348 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Can S, Dewitt MA & Yildiz A Bidirectional helical motility of cytoplasmic dynein around microtubules. eLife 3, e03205 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mitra A, Ruhnow F, Nitzsche B & Diez S Impact-Free Measurement of Microtubule Rotations on Kinesin and Cytoplasmic-Dynein Coated Surfaces. Plos One 10, e0136920 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nicholas MP et al. Control of cytoplasmic dynein force production and processivity by its C-terminal domain. Nature communications 6, 6206 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gennerich A, Carter AP, Reck-Peterson SL & Vale RD Force-induced bidirectional stepping of cytoplasmic dynein. Cell 131, 952–965 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Belyy V, Hendel NL, Chien A & Yildiz A Cytoplasmic dynein transports cargos via load-sharing between the heads. Nature communications 5, 5544 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nishiyama M, Higuchi H & Yanagida T Chemomechanical coupling of the forward and backward steps of single kinesin molecules. Nat. Cell Biol. 2002. 4(10):790–7 (2002). [DOI] [PubMed] [Google Scholar]
- 38.Lu H et al. Collective dynamics of elastically coupled myosin V motors. The Journal of biological chemistry 287, 27753–27761 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Derr ND et al. Tug-of-war in motor protein ensembles revealed with a programmable DNA origami scaffold. Science 338, 662–665 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Furuta K et al. Measuring collective transport by defined numbers of processive and nonprocessive kinesin motors. Proceedings of the National Academy of Sciences of the United States of America 110, 501–506 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Driller-Colangelo AR, Chau KW, Morgan JM & Derr ND Cargo rigidity affects the sensitivity of dynein ensembles to individual motor pausing. Cytoskeleton 73, 693–702 (2016). [DOI] [PubMed] [Google Scholar]
- 42.Mallik R, Petrov D, Lex SA, King SJ & Gross SP Building complexity: an in vitro study of cytoplasmic dynein with in vivo implications. Current biology : CB 15, 2075–2085 (2005). [DOI] [PubMed] [Google Scholar]
- 43.Vershinin M, Carter BC, Razafsky DS, King SJ & Gross SP Multiple-motor based transport and its regulation by Tau. Proceedings of the National Academy of Sciences of the United States of America 104, 87–92 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Jamison DK, Driver JW & Diehl MR Cooperative responses of multiple kinesins to variable and constant loads. The Journal of biological chemistry 287, 3357–3365 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Soppina V, Rai AK, Ramaiya AJ, Barak P & Mallik R Tug-of-war between dissimilar teams of microtubule motors regulates transport and fission of endosomes. Proceedings of the National Academy of Sciences of the United States of America 106, 19381–19386 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Shubeita GT et al. Consequences of motor copy number on the intracellular transport of kinesin-1-driven lipid droplets. Cell 135, 1098–1107 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ray S, Meyhofer E, Milligan RA & Howard J Kinesin follows the microtubule’s protofilament axis. J Cell Biol 121, 1083–1093 (1993). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Nicholas MP et al. Cytoplasmic dynein regulates its attachment to microtubules via nucleotide state-switched mechanosensing at multiple AAA domains. Proceedings of the National Academy of Sciences of the United States of America 112, 6371–6376 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Cleary FB et al. Tension on the linker gates the ATP-dependent release of dynein from microtubules. Nature communications 5, 4587 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rai A et al. Dynein Clusters into Lipid Microdomains on Phagosomes to Drive Rapid Transport toward Lysosomes. Cell 164, 722–734 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
Methods references
- 51.Dogan MY, Can S, Cleary FB, Purde V & Yildiz A Kinesin’s front head is gated by the backward orientation of its neck linker. Cell reports 10, 1967–1973 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Gibbons IR et al. The affinity of the dynein microtubule-binding domain is modulated by the conformation of its coiled-coil stalk. The Journal of biological chemistry 280, 23960–23965 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data that support the conclusions are available from the authors on request.