Abstract
Der p 2 is one of the most important allergens from the house dust mite Dermatophagoides pteronyssinus. Identification of human IgE antibody binding epitopes can be used for rational design of allergens with reduced IgE reactivity for therapy. Antigenic analysis of Der p 2 was performed by site-directed mutagenesis based on the X-ray crystal structure of the allergen in complex with a Fab from the murine IgG mAb 7A1 that binds an epitope overlapping with human IgE binding sites. Conformational changes upon antibody binding were confirmed by nuclear magnetic resonance (NMR) using a 7A1-single-chain variable fragment (scFv). In addition, a human IgE antibody construct that interferes with mAb 7A1 binding was isolated from a combinatorial phage display library constructed from a mite allergic patient and expressed as two recombinant forms (single chain Fab -scFab- in Pichia pastoris and Fab in Escherichia coli). These two IgE antibody constructs and the mAb 7A1 failed to recognize two Der p 2 epitope double mutants designed to abolish the allergen-antibody interaction, while preserving the fold necessary to bind antibodies at other sites of the allergen surface. A 10 to 100-fold reduction in binding of IgE from allergic subjects to the mutants additionally showed that the residues mutated were involved in IgE antibody binding. In summary, mutagenesis of a Der p 2 epitope defined by X-ray crystallography revealed an IgE antibody binding site that will be considered for the design of hypoallergens for immunotherapy.
Introduction
House dust mites are one of the most important sources of indoor allergens worldwide. For atopic individuals, exposure to mite allergens can lead to allergic rhinitis, atopic dermatitis and asthma (1-3). Up to 85% of asthmatics are sensitized to house dust mites, and this number may exceed 90% in the tropics (4,5). Currently, thirty-nine mite allergen groups are listed in the official Allergen Nomenclature database of the World Health Organization and International Union of Immunological Societies (WHO/IUIS) (www.allergen.org). Der p 2 is one of the major mite allergens from the species Dermatophagoides pteronyssimus, along with Der p 1 and Der p 23 (6-8). High prevalences of IgE sensitization to Der p 2 (70–90%) have been reported among mite allergic patients (6,9,10). Groups 1 and 2 mite allergens combined bind 50–70% of the IgE that recognizes mite extracts (5). A longitudinal study from birth to 20 years of age showed that, early in life, Der p 1, Der p 2 or Der p 23 initiated IgE responses to mite. These responses increased in IgE prevalence along the first decade of life, with Der p 2 being the allergen with the highest prevalence (10).
The only available treatment for mite allergies consists of administering increasing doses of mite allergen extracts, which can cause secondary side effects to the patient (release of mediators such as histamine and inflammation). These side effects are caused by the interaction of mite allergens with the patient’s immunoglobulin E, IgE, antibodies. The ultimate goal of our studies is to identify IgE antibody epitopes on Der p 2 that can guide the design of mutants with reduced IgE reactivity, also known as hypoallergens. These molecules with lower risk of side effects will eventually be assessed for use in immunotherapy, to mitigate the adverse reactions observed using conventional mite extracts.
Original mapping of IgE epitopes was usually achieved by screening peptides for IgE antibody binding (11). However, many Der p 2 epitopes are defined by the structural features formed from discontinuous regions of the allergen’s primary structure, rather than a continuous peptide sequence (12). Another common approach is the use of site-directed mutagenesis to alter solvent-accessible amino acids and assessing their effect on antibody binding (13,14). While this approach can be used to study conformational epitopes, the low structural resolution of the information provided hinders its utility for rational design. The current study addressed these shortcomings by determining the three-dimensional structure of the allergen in complex with an antibody. In the Protein Data Bank (PDB), only a few structures of allergens in complex with antibodies (mostly IgG antibody Fab or Fab’ fragments) are reported. These include ten allergens in complex with either fragments of monoclonal IgG antibodies (Gal d 4, Bet v 1, Api m 2, Bla g 2, Der p 1, Der f 1, Fel d 1 and Phl p 7) or IgE antibody constructs (Bos d 5, Phl p 2) (15-29). Analysis of the structure of some of these epitopes allowed for the production of mutants with reduced capacity to bind the antibodies (30-33).
Der p 2 has a cylindrical structure with an immunoglobulin-like fold that contains an internal hydrophobic cavity (34,35). X-ray crystal structures of mite allergens in complex with antibody fragments were previously determined for Der p 1 and Der f 1 (24,25). In this study, the first crystal structure of Der p 2 in complex with an antibody (mAb 7A1) that interferes with IgE antibody binding is reported. In addition, a human anti-Der p 2 IgE scFv antibody construct (IgE-scFv) was isolated from a phage display combinatorial library made from a mite allergic individual. This human IgE antibody specifically recognized an epitope overlapping with the epitope recognized by mAb 7A1. Based on the relation of epitopes recognized by this human IgE-scFv and mAb 7A1, the molecular structure of the Der p 2-mAb 7A1 complex was used to design allergen mutants with reduced reactivity to mite allergen group 2-specific human IgE, highlighting the value of high-resolution epitope mapping in the design of novel immunotherapy compounds.
Materials and Methods
Production of purified rDer p 2 and mAb 7A1 Fab
Recombinant allergen (Der p 2.0103) was expressed in Pichia pastoris by methanol induction for 48–96 hours using the vector pPICZαA, and purified from culture media by affinity chromatography using the mAb 7A1. Purity > 95% was assessed on silver stained SDS-PAGE (data not shown).
The mAb 7A1 (isotype IgG1) was produced from the 7A1-H1-G3 cell line and purified by Protein G chromatography, as described previously (36). Its isotype is IgG1. The mAb 7A1 was digested with papain and the resulting Fab were purified using a Pierce Fab preparation kit (Thermo Fisher Scientific, Grand Island, NY) according to the manufacturer’s protocol. Allergen-antibody complex was prepared by mixing the Fab fragment in a 1:1 molar ratio with Der p 2.0103, and incubating at 4ºC for 30 minutes. After incubation, the complex was concentrated, and purified by size exclusion chromatography (SEC) on a Superdex 200 column using 10 mM Tris-HCl and 150 mM NaCl buffer, pH 7.4. Fractions eluted from the SEC column containing Der p 2 in complex with Fab (Der p 2.0103–7A1Fab) were pooled, concentrated to 5 mg/mL, and used for crystallization.
Expression and purification of mAb 7A1 scFv
The DNA encoding the variable regions of mAb 7A1 were amplified with a set of primers from cDNA generated from the 7A1-H1-G3 hybridoma cells using a standard RT-PCR protocol and sequenced using a standard dye-terminator capillary sequencing method (Synbuild, LLC, Tempe, Arizona). The 7A1 heavy and light chain-encoding genes had origins (determined using the IMGT V-QUEST tool (http://imgt.org/IMGT_vquest/vquest)) in the IGHV9–1*02/IGHJ4*01, and IGKV3–4*01/IGKJ1*01 germline genes, respectively. A gene coding for mAb 7A1 scFv (7A1scFv) with a 6X C-terminal poly-histidine purification tag was cloned into the ampicillin resistant pET21b plasmid (Bio Basic, Amherst, NY, USA). The plasmid was co-transformed into E. coli BL21 (DE3) strain together with the pKJE7 (chloramphenicol resistance) plasmid that encodes chaperones DnaK, DnaJ, GrpE (Takara, Mountain View, CA, USA). Plasmid cultures (1 L) were grown at 37 °C to OD600 of 0.8 after which the cultures were cooled to 22 °C, or room temperature, and were induced with 0.4 mM of isopropyl β-D-1-thiogalactopyranoside (IPTG), cooled to 16 °C and grown for 16–18 hours.
Cell pellets obtained after centrifuging the overnight 1 L cultures were resuspended in lysis buffer (20 mM Na2HPO4, 500 mM NaCl, 10 mM imidazole, pH 7.4). After sonication, the cell lysate was centrifuged at 9,000 x g for 10 minutes at 4 °C. The supernatant was purified by immobilized metal affinity using Ni-NTA. After loading the protein onto the column, the column was washed with wash buffer (20 mM Na2HPO4, 500 mM NaCl, 30 mM imidazole and pH 7.4). The protein was eluted using elution buffer (20 mM Na2HPO4, 500 mM NaCl, 250 mM imidazole and pH 7.4). 7A1scFv protein was further purified by gel filtration in elution buffer. After gel filtration, samples containing the protein were dialyzed in buffer containing 20 mM Na2HPO4, 150 mM NaCl, 100 mM EDTA, pH 7.4. The yield was 10 mg/L of culture.
Crystallization, data collection and structure determination
Crystallization of the Der p 2.0103-mAb 7A1 Fab complex (Der p 2.0103–7A1Fab) was performed at 298 K using the vapor diffusion method. The protein complex was mixed at a 1:1 ratio with each crystallization condition. Crystallization conditions that yielded crystals of diffraction quality contained: 100 mM Bis-Tris, 200 mM ammonium acetate, 30–55% 2-methyl-2,4-pentanediol at pH 6.5 (well solution). Data were collected at the Southeast Regional Collaborative Access Team (SER-CAT) 22-ID at the Photon Advanced Source, Argonne National Laboratory (Lemont, IL). Diffraction images were processed using HKL-3000 (37).
Summary for data collection statistics is presented in Table I. The structure was determined using molecular replacement with PDB structures 1KTJ (Der p 2) and 3RVT (Fab) as starting models. The interpretation of the electron density map was facilitated by the knowledge of the sequence of the variable regions of the immunoglobulin, defined as described above. HKL-3000, MOLREP (38) and selected programs from the CCP4 package (39) were used for structure determination. The model was rebuilt using BUCCANEAR (40) and COOT (41), refined using REFMAC (42) and COOT, and validated using MOLPROBITY (43). The final model together with structure factors were deposited to the PDB with accession code 6OY4 (http://www.rcsb.org/structure/6OY4).
Table I.
Data collection, processing and refinement statistics.
| PDB ID | 6OY4 |
| Diffraction source | APS Beamline 22ID |
| Wavelength (Å) | 1.0000 |
| Temperature (K) | 100 |
| Space group | C2 |
| a, b, c (Å) | 180.1, 43.3, 106.0 |
| α, β, γ (°) | 90.0, 125.3, 90.0 |
| Resolution range (Å) | 40.00-2.45 (2.49-2.45) |
| No. of unique reflections | 23826 (1143) |
| Completeness (%) | 94.7 (92.4) |
| Redundancy | 3.0 (2.4) |
| I/σI | 14.5 (2.2) |
| Rmeasure | 0.133 (0.547) |
| Rp.i.m. | 0.074 (0.325) |
| Overall B factor from Wilson plot (Å2) | 47.3 |
| CC1/2 | (0.628) |
| Rcryst | 0.203 (0.257) |
| Rfree | 0.260 (0.312) |
| R.m.s. deviations | |
| Bonds (Å) | 0.011 |
| Angles (°) | 1.4 |
| Average B factors (Å2) | 42.9 |
| Ramachandran plot | |
| Favoured (%) | 97 |
| Allowed (%) | 100 |
Parameters for the highest resolution shell are in parenthesis.
NMR samples and data acquisition
Recombinant Der p 2.0101 (D1S) was expressed in E. coli using the vector pET21a and purified as previously described (44) with the following adjustments: benzonase was added prior to sonication, and the insoluble fraction was incubated with 6 M guanidinium chloride, unbuffered, for 1 h at room temperature with rapid stirring prior to dialysis and refolding (44). Although Der p 2 has been previously studied by NMR (34,45), the shifts were altered in PBS buffer and needed to be reassigned. Assignments were determined using triple resonance NMR techniques with a [U-13C,15N] Der p 2 sample produced in isotopically-labeled media (9). A Der p 2 [U-2H,15N, Ile, Leu, Val (ILV) 1H,13C-methyl] sample was made according to Goto et al. (46), but substituting 2H-glycerol in place of 2H-glucose as the primary carbon source. This technique labels the Ile δ1 methyl and both methyls of Leu and Val with 1H and 13C, while 2H labels the rest of the protein for maximum sensitivity in a high molecular weight complex. For simplicity, this allergen will be referred to as the ‘methyl labeled Der p 2’.
Complexes were made by mixing 7A1scFv with an excess of methyl labeled Der p 2 and incubating the mixture for at least 30 minutes at room temperature before applying the sample to a 26–600 S75 Superdex column flowing at 0.3 mL/min at 4°C with PBS. The Der p 2–7A1scFv fractions were exchanged into 2H-PBS and concentrated to 500 μL. Data was acquired on an Agilent 800 MHz spectrometer with a cryogenically cooled probe using the 1H-13C heteronuclear multiple quantum coherence (HMQC) pulse sequence for 12 hours at 37 °C.
Site directed mutagenesis of the mAb 7A1 epitope on Der p 2 and expression and purification of allergens
Site-directed mutagenesis of the mAb 7A1 epitope on Der p 2 was performed using the QuikChange Site-Directed Mutagenesis kit (Agilent Technologies, Santa Clara, CA). The sequences of the mutated DNA in the vector pPICZαB were confirmed before linearization and transformation into the P. pastoris.
Recombinant Der p 2.0103 wild type and mutants were expressed in a methanol inducible P. pastoris system, and purified over a mAb 7A1 or 1D8 affinity chromatography column, respectively. The allergens were eluted with 0.1 M glycine, 0.15 M NaCl, pH 2.5, concentrated and dialyzed in PBS. The final protein concentration was determined by Advanced Protein Assay (Cytoskeleton Inc., Denver, CO).
Comparison of mAb 7A1 and IgE antibody binding to epitope mutants versus wild type Der p 2
Direct binding of mAbs 7A1 and 1D8 to Der p 2 wild type and mutants was assessed using a direct antibody binding immunoassay. The first wells of a microtiter plate were coated with equal amounts of Der p 2.0103 and the mutants thereof, diluted ½ across the plate and incubated overnight at 4 °C. The plate was washed, blocked for 30 minutes with phosphate buffered saline, pH 7.4, containing 0.05% Tween 20 (PBS-T) with 1% bovine serum albumin, incubated first for 1 h with biotinylated mAb 7A1 or 1D8 (1:1,000 dilution) and then streptavidin-peroxidase for 30 min. The plate was developed using 2,2’-azino-bis(3-ethylbenzothiazoline-6-sulphonic acid (ABTS) substrate. Absorbance was read at 405 nm on a Bio-Tek EL800 Microplate Reader (Bio-Tek Instruments, Inc., Winooski, VT).
This immunoassay also allowed to confirm that the structure of Der p 2 was conserved in the wild type and the two mutants, by testing their ability to bind mAb 1D8, an antibody whose epitope lies distal from that of mAb 7A1 (45). In addition, the two Der p 2 mutants and Der p 2.0103 were assessed for protein folding by NMR (1D-NOESY), 100 ms mixing time and watergate-3919 water suppression. Each non-labelled mutant and wild type allergen (500 μg of each mutant, and 4.5 mg of WT) were examined in PBS buffer, 10% D2O, and 30 μM DSS. To compare to an unfolded state, 6 M urea and 1 mM DTT were added to the wild type, and boiled for 10 minutes.
The ability of mAb 7A1 epitope mutants and wild type rDer p 2.0103 to inhibit binding of murine IgG mAbs or human IgE Abs to Der p 2 was assessed using ELISA. The wild type and mutant allergens (at 0.001, 0.01, 0.1, 1, 10, 100 μg/mL) were preincubated for 1 h with either biotin-labeled mAb 7A1 (1/1,000 dilution; 4.8 μg/mL) or human plasma containing anti-Der p 2 IgE (1/2 or 1/4 dilution) and subsequently added to microtiter plates coated overnight with 10 μg/mL rDer p 2.0103, and blocked for 1 h as described above. The plates were incubated for 3 hours, washed and incubated for 30 min with streptavidin-peroxidase or 1 h with peroxidase labeled goat anti-human IgE, respectively, and developed as described above.
Plasma were obtained from PlasmaLab International (Everett, WA) which operates in full compliance with Food and Drug Administration regulations. Informed donor consent was obtained from each individual prior to the first donation. Five plasma samples from mite allergic patients were selected based on previous in-house multiplex assays results with an average of IgE reactivity to Der p 2 of 147 ± 95 IU/L (range 47–262 IU/L) and total IgE of 3,555 ± 6,390 IU/L (range 535–14981 IU/L) measured by multiplex technology.
Inhibition of IgE antibody binding to Der p 2 by IgG mAb 7A1
The overlap between mAb 7A1 and serum IgE antibody binding was investigated by measuring the inhibition of IgE antibody binding to Der p 2 by three murine IgG mAbs: 7A1, 1D8 and αDpX by immunoassay. The inhibitors (allergens Der p 1 or Der p 2 to test specificity of the inhibition, or mAbs 7A1, 1D8 and αDpX; each to a final concentration of 100 μg/mL) were preincubated for 1 h with human plasma containing anti-Der p 2 IgE (1/2 or 1/4 dilution) and subsequently added to microtiter plates coated overnight with 10 μg/mL rDer p 2.0103, and blocked for 1 h as described above. The plates were incubated for 3 hours, washed, incubated for 1 h with peroxidase labeled goat anti-human IgE, and developed as described above.
Construction of phage display libraries from RNA isolated from mite allergic subjects
The identification of anti-Der p 2 IgE from phage display library was performed as previously described (47). Blood was obtained from two subjects who were ImmunoCAP Class 4 and Class 5 for dust mite (Der p) sIgE. Written informed consent was obtained and research was approved by the University of Virginia Human Investigation Committee (Protocol #13166). For each subject, 50 mL of heparinized venous blood were collected, from which peripheral blood mononuclear cells (PBMCs) were prepared by Ficoll gradient centrifugation (48,49). RNA was extracted from 10 million PBMCs using the RNeasy Mini Kit (Qiagen, Hilden, Germany). RNA was then quantitated and analyzed by agarose gel to check for degradation.
The QuantiTect Reverse Transcription Kit (Qiagen, Hilden, Germany) was used for first strand cDNA synthesis. Two first strand synthesis reactions for each subject RNA sample were prepared, one with an oligo-dT primer and one with an IgE-specific primer (5’-GCA CTG TAA ACT AGT CAC GGT GGG CGG GGT G-3’). For each reaction, 1 μg of RNA was mixed with gDNA Wipeout Buffer and RNase-free water, then incubated at 42 °C for 2 minutes before the reverse transcriptase, RT Buffer, and primer were added. This was followed by a 30 minute incubation step at 42 °C and a 3 minute inactivation step at 95 °C.
A scFv-encoding phagemid library was made from each subject essentially as described by Levin et al. (50). In summary, genes encoding the H chain and κ chain variable (V) domains were amplified in a series of PCR steps using the first-strand cDNA synthesized as described above. Genes encoding Vλ domains were similarly amplified from cDNA generated from an unrelated donor. H chain V domain-encoding genes were amplified (Pfu Ultra II Fusion HS DNA polymerase (Agilent, Santa Clara, CA) to introduce a 5’ restriction site (SfiI) and 3’-linker sequence while κ and λ V domain encoding sequences were similarly amplified to introduce a 5’-linker region and the 3’ restriction site (NotI). Finally, overlap-extension PCR was performed to generate the VH-linker-Vκ/Vλ-encoding gene product. Primers used in these PCR reactions were those described by Andréasson et al. (51). The final PCR products were cloned into a modified (52) version of the pFAB5c.His vector (53) and transformed by electroporation into One Shot E. coli Top 10F’ electrocompetent cells (Invitrogen, Carlsbad, CA) using a BTX Electroporation system (BTX, Holliston, MA, USA). The phage library displaying scFv was produced essentially as previously described (52) using M13 K07 helper phage (New England Biolabs, Ipswich, MA, USA). The phage stock was precipitated once with PEG/NaCl and dissolved in PBS supplied with 0.1% BSA.
Selection of anti-Der p 2 antibody constructs from a phage display library of a mite allergic subject
Selection of Der p 2-specific scFv was performed using natural Der p 2 purified from dust mite extracts by affinity chromatography that had been biotinylated with EZ Link Sulfo-NHS-LC-Biotin (NHS is Hydroxysuccinimide) (Thermo Scientific, Rockford, IL, USA). Phage stocks were, prior to incubation with allergen-carrying beads, incubated with M280-Streptavidin Dynabeads (Invitrogen, Carlsbad, CA) to remove phages with unwanted binding properties. Two rounds of selection (0.5 mL total volume, each) were performed using 50 and 25 pmol biotinylated Der p 2, respectively, coated onto M280-Streptavidin Dynabeads. Selection was performed in PBS/0.05% Tween 20 containing 0.5% BSA (first round of selection) or 0.5% fish gelatin (Sigma, St. Louis, MO, USA) (second round of selection).
After two rounds of selection, scFv-encoding genes were re-cloned into the pHP2–15 vector (52) that allows for the production of soluble scFv (IgE-scFv) carrying a FLAG tag and a hexahistidine tag. Ligated DNA was transfected into One Shot™ TOP10 chemically competent E. coli (Invitrogen, Carlsbad, CA) according to the manufacturer’s instructions. Individual colonies were picked and scFv was produced in 96-well deep-well plates and tested for binding to the Der p 2 antigen by ELISA. Briefly, 96-well microtitre plates were coated with 1 μg/well streptavidin (Invitrogen, Carlsbad, CA), and biotinylated natural Der p 2 protein (1.25 μg/well) was subsequently added (wells without added allergen were used as negative control). Bacterial supernatant diluted in PBS containing 0.5% BSA and 0.05% Tween was added. Bound scFv was detected by Monoclonal anti-FLAG® M2-Peroxidase (Sigma, St Louis, MO) and detected using 1-Step™ Ultra TMB-ELISA Substrate Solution (Thermo Fisher Scientific, Waltham, MA).
Clones producing allergen-binding scFv were picked, after which soluble scFv was again produced in 96-well plates and sterile filtered. The binding of soluble scFv to allergen was assessed using surface plasmon resonance (SPR) technology using a MASS-16 instrument (Sierra Sensors, Hamburg, Germany). Monoclonal anti-FLAG® M2 antibody (Sigma, St Louis, MO) was immobilized (30 μg/mL during 7 min at pH 4) on a High Capacity Amine Sensor (Sierra Sensors GmbH, Bruker Corp., Munich, Germany) chip. Measurements were performed in Dulbecco’s PBS (HyClone, South Logan, UT) containing 0.01% Tween 20. Briefly, 30 μL diluted scFv fragments (dilution selected to produce a binding of scFv in the range 50–250 RU) was injected over the surface (10 μL/min) followed by injection of 50 μL, 120 nM Der p 2 antigen at 25 μL/min, and a dissociation time of 200 s. The reference channel carried no immobilized scFv. After each cycle the surface was regenerated with 15 μL 10 mM glycine buffer pH 2.2 containing 0.5M NaCl (15 μL/min). DNA encoding scFv that bound the antigen in SPR analysis were sequenced using Sanger sequencing technology (GATC, Konstanz, Germany). Additional SPR analysis of the binding properties of sequence-unique clones was performed using 30 nM and 120 nM of antigen.
Relative epitope mapping of scFv
To examine if the identified IgE scFv bound to the same epitope as murine IgG mAbs 7A1 or 1D8, an epitope mapping was performed by ELISA. Streptavidin (Sigma, St Louis, MO) was coated in 96-well microtitre plates overnight and biotinylated natural Der p 2 protein (1.25 μg/well) was added. After 1 hour, the plate was washed and mAb 7A1 or mAb 1D8 was added at 0.2 μg/mL or 1 μg/mL, respectively, in PBS containing 0.5% BSA and 0.05% Tween. After 1 hour of incubation, scFv-containing culture supernatant was added and incubated for an additional hour at room temperature. Finally, the plate was washed and bound scFv was detected as described above.
Expression and purification of recombinant anti-Der p 2 IgE antibody constructs based on the isolated scFv
Recombinant Fab expressed in Escherichia coli
The Fab was designed from the construct obtained from a phage display library as described above and was expressed using a dual expression single vector that features two promoters for simultaneous expression of the heavy and light chain of the Fab. A pMAL-c4x plasmid containing a cassette (kindly provided by Levi Blazer, Toronto, Canada) coding for an anti-Der p 2 Fab was transformed into E. coli (BL21-T1R strain) (New England BioLabs, Inc., Ipswich, MA). A plasmid culture (1 L) was grown at 37 °C by shaking at 250 rpm until OD600 reached a value between 0.6 and 1.0. Then, Fab expression was induced by adding 0.5 mM IPTG and the culture continued shaking at 30 °C for 4 hours. The Fab was purified from culture supernatants through a C-terminal 6x poly-histidine tag by nickel-agarose affinity chromatography.
Recombinant scFab expressed in Pichia pastoris
The IgE construct derived from a PBMC library was also expressed in P. pastoris as a scFab (PBMCD2) with a 60 amino acid linker that connects the C-terminus of the variable plus constant regions of the light chain with the N-terminus of the variable plus constant regions of the heavy chain sequences (54). The pPICZαB vector with a DNA insert encoding for the scFab was electroporated into KM71 cells and cells were grown in a methanol inducible system for 48–72 hours. Expressed scFab was purified by nickel-agarose affinity chromatography.
Assessment of binding of IgE antibody constructs Fab and scFab to the mutants
A microtiter plate was coated overnight with mAb 1D8. The plate was washed and incubated with Der p 2.0103 wild type or mutants at 100 ng/mL for 1 h. Supernatants from P. pastoris and E. coli cultures expressing the Der p 2-specific construct were then added, diluted 1:2 across the plate and incubated for 1 h. Incubations with biotinylated anti-His Tag Ab (1:500) for 1 h and streptavidin peroxidase (1:1,000) for 30 minutes followed, and the plate was developed as described above.
Results
Crystal structure of Der p 2 in complex with Fab from mAb 7A1
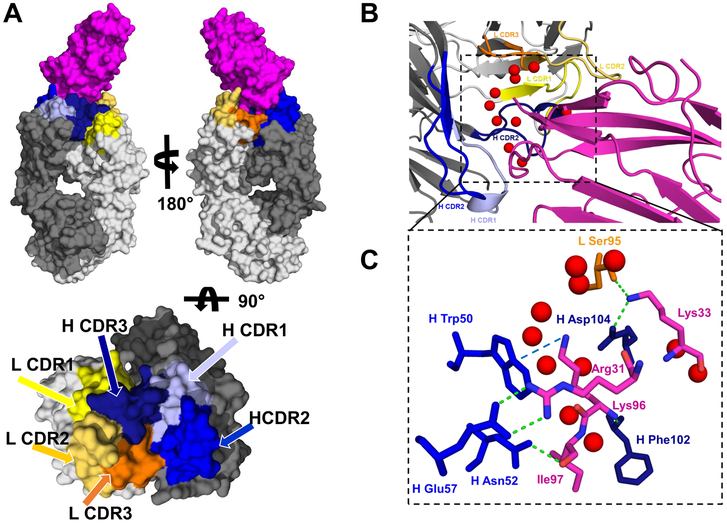
The structure of Der p 2.0103 in complex with the Fab of mAb 7A1 (Der p 2.0103–7A1Fab complex) was determined at 2.45 Å resolution (Table I). The complex crystallized in the C2 space group with one complex copy per asymmetric unit (Fig. 1A). The epitope has an area of approximately 750 Å2, and 64% of this area corresponds to a region that interacts with the variable fragment of the antibody heavy chain (Fig. 1B). Most of the H-bond interactions between residues forming the epitope and the paratope are formed by two fragments of Der p 2.0103 (Arg31-Gly32-Lys33-Pro34 and Lys96-Ile97) that interact mostly with amino acids from complementarity-determining regions 2 and 3 (CDR2 and CDR3) of the heavy chain and a residue from L CDR3 (CDR3 from light chain) (Table II, Fig. 1C). One of the H CDR2 interactions (H chain Glu57 with Der p 2 Arg31) involved a residue (Glu57) likely not encoded by the H chain germline gene (Fig. 1C, Supplemental Fig. 1), but introduced as a consequence of codon insertion (55,56) that occurred during the somatic hypermutation process that was part of the evolution of this antibody specificity. Direct contacts between allergen and antibody are not only mediated by H-bonds, but there are several residues, such as Pro34, Pro95, Ile97 and Pro99 that participate in hydrophobic interactions. In addition, there is a cation-π interaction involving Lys96 (Der p 2) and Trp50 (H CDR2) with the ~3.5Å distance between the interacting side chains (Fig. 1C, Supplemental Fig. 1). The structural analysis shows that Lys33 and Pro34 are responsible for a significant fraction of the epitope area that is in contact with the light chain of mAb 7A1, while Lys96 and Ile97 play a similar role in the contact with the heavy chain. Ile97 is wedged against CDR2, CDR1 and indirectly against CDR3, but in the last case the interaction is mediated by a water molecule. The epitope-paratope interactions are not only mediated by direct contacts, but also by indirect contacts through water molecules. In fact, the crystal structure revealed 11 water molecules (Fig. 1B and 1C), which were trapped between the allergen and antibody and participate in mediating contacts between these two molecules.
Figure 1. Structure of Der p 2.0103-mAb 7A1 (Fab) complex.
A) Space filling representation of the complex (top). Der p 2 is shown in magenta, while light and heavy chain of the antibody are marked in light and dark grey, respectively. CDRs are mapped on mAb 7A1 molecular surface and marked with different colors (bottom). B) Interaction between the epitope and the paratope. Positions of water molecules buried between allergen and the antibody are marked with red spheres. For clarity of the presentation, only positions of water oxygen atoms are shown. C) The inset shows the residues from epitope and paratope that are involved in formation of H-bonds (green dashed lines) and a cation-π interaction (blue dashed line), presented in stick representation. The color scheme is the same as in the other panels.
Table II.
H-bonds formed between Der 2.0103 and mAb 7A1.
| Der p 2.0103 | 7A1 | CDR | Distance (Å) |
|---|---|---|---|
| Arg31 [Nη1] | H Glu57 [Oε2] | H CDR2 | 2.8 |
| Arg31 [Nη2] | H Glu57 [Oε1] | H CDR2 | 2.9 |
| Gly32 [O] | H Asp104 [N] | H CDR3 | 2.9 |
| Lys33 [Nξ] | H Asp104 [Oδ2] | H CDR3 | 2.6 |
| Lys33 [Nξ] | L Ser95 [O] | L CDR3 | 2.9 |
| Lys96 [N] | H Phe102 [O] | H CDR3 | 3.0 |
| Ile97 [O] | H Asn52 [Nδ2] | H CDR2 | 2.8 |
Conformational changes upon mAb 7A1 Fab binding to Der p 2 by X-ray crystallography
In the crystal structure of the Der p 2.0103–7A1Fab complex the majority of the symmetry-related contacts are formed by the Fab fragment. Therefore, the structure provided an opportunity to compare the conformation of the antibody bound Der p 2.0103 with the conformation of the free allergen (Der p 2.0101; 1KTJ). The superposition of these two structures resulted in a rmsd value of 1.0 Å (chain A of 1KTJ, 122 superposed Cα atoms), which is similar to the rmsd value obtained after superposition of the antibody bound Der p 2.0103 with the structure of Der f 2 (PDB codes: 1XWV and 2F08). The antibody binding induces several localized perturbations to Der p 2.0103, with notable regions including the Glu62-Gly67 fragment (Fig. 2A-C, small oval), along with a discontinuous sequence of the allergen that is distal from the epitope (Fig. 2A-C, large oval). In the former, residues 63 and 64 showed the greatest variation, with displacement distances of 2.3 and 3.8 Å, respectively, between the two structures (Fig. 2E; amplified view of the “small oval” in Figure 2C). Despite its proximity, the actual epitope region experienced significantly reduced conformational changes relative to this region (Fig. 2F). The distal (big oval) region encompassed a larger number of residues, potentially contributing to the increased disorder of the region, particularly in the vicinity of the Cys73-Cys78 disulfide bridge. However, this disorder, represented by the B-factor values in Fig. 2A, prevented the accurate modeling of residues 48–49, 75–79 and 109–112 in the bound structure, precluding further structural comparison with the free form.
Figure 2. Mobility of Der p 2 in the structure with the mAb 7A1 complex (A) compared with Der p 2 alone (B).
Comparison of B-factors between mAb 7A1 bound (A) and free (PDB code: 1KTJ) Der p 2.0101 (B). Red color and the larger diameter of ribbon correspond to the highest mobility/displacement of residues, blue color and the smallest diameter correspond to regions with the lowest mobility. Other colors and diameters of the ribbon indicate intermediate mobility of the residues. C) Superposition of main chains for mAb 7A1 bound (magenta) and free (gray) Der p 2.0103. Ovals mark regions that display the most pronounced conformation differences. D) Display of Der p 2 residues in X-ray crystal structures, associated with conformational changes of methyl residues in Der p 2 observed by NMR (see Figure 3). The ribbon diagram of Der p 2 alone (1KTJ) is colored green for residues that would interact with mAb 7A1, grey for other residues, and yellow sticks are shown for Ile28, Leu61, Val63, and Ile97. Der p 2 in the mAb 7A1 Fab complex is colored black and Ile28, Leu61, Val63, and Ile97 are rendered with purple sticks. The Fab is rendered in dark red with a ribbon and mesh surface. E, F) Structural changes of specific Der p 2 areas upon antibody binding. Magenta – antibody bound Der p 2.0103, Grey – free Der p 2.0103 (PDB code: 1KTJ). E) Zoom on a region that was marked on panel 2C with the smaller oval. F) Comparison of conformation of the amino acids forming the mAb 7A1 epitope.
Conformational changes by NMR in Der p 2 upon complexation with mAb 7A1 scFv
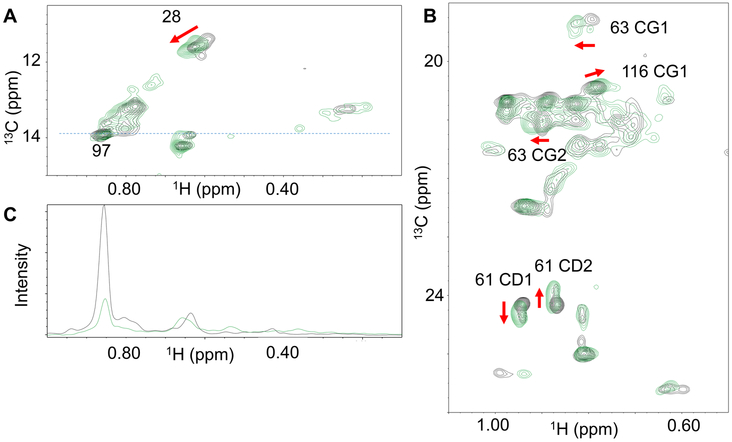
The B-factor values obtained from the crystal structures suggest that binding of mAb 7A1 significantly alters Der p 2 structure and dynamics, particularly in the region distal from the epitope site discussed above. To explore this possibility, 1H-13C-NMR was used to monitor conformational changes in 13C- methyl labeled ILV side chains, a labeling scheme which is particularly suited to maintain high sensitivity even in high molecular weight systems (57). The resulting HMQC spectra of ILV-methyl labeled Der p 2 in both its free form, and in complex with the 7A1scFv is shown in Fig. 3. Here, chemical shift perturbations were observed for I28, L61, V63 and V116 (Fig. 3A and B), along with a significant reduction in NMR signal intensity of Ile97 upon formation of the mAb 7A1 complex. The reduction in intensity of Ile97 may be due to conformational exchange upon binding, or due to the fact that the scFv is fully 1H labeled, which could be a source of relaxation reducing the signal intensity as seen by Hamel and Dahlquist in a similar situation (58). These findings are consistent with the crystal structures, in which all the aforementioned residues that show shift changes in the NMR spectrum display conformation changes upon mAb 7A1 binding (Fig. 2C and 2D). Additionally, the slight shift in the peak for Val116 1H-13C in Fig. 3B, mirrors the distal conformational changes shown in Fig. 2A and C, further validating the structural rearrangements observed in the crystal structures.
Figure 3. NMR HMQC spectra of methyl labeled Der p 2.
NMR HMQC spectra of Der p 2 alone is shown in black, while Der p 2 in complex with 7A1scFv is shown in green. Panel A shows the Ile region and Panel B shows the Val-Leu region. Panel C is a slice through panel A at 13.9 ppm (blue dashed line, panel A) comparing relative intensities. Arrows indicate shift changes and methyl resonances with changes are labeled. Stereo assignments of the Val and Leu were not made so the labeling of stereochemistry is arbitrary.
Epitope analysis for rational design of mAb 7A1 epitope mutants
The crystal structure of the Der p 2.0103–7A1Fab complex along with the 1H-13C-NMR studies allowed for the identification of several allergen residues that are critical for interaction with the antibody. Based on these insights, two double mutants (Arg31Ala-Lys33Ala and Lys96Glu-Ile97Glu) of the allergen were designed to decrease their ability to bind mAb 7A1. The first set of mutations was designed to affect the interaction with the light chain (Arg31Ala-Lys33Ala) through replacing long and positively charged side chains with methyl groups to reduce the number of H-bonds in the epitope-paratope interface (Table 2). Arg31 is responsible for a strong interaction with Glu57 from H CDR2, while Lys33 participates in formation of two hydrogen bonds with Ser95 from L CDR3 and Asp104 from H CDR3. Therefore, mutation of these two residues to alanine was expected to completely abolish their ability to form the network of H-bonds that involved their side chains (Fig. 1C). The second set of mutations (Lys96Glu-Ile97Glu) targeted the interaction with the heavy chain. Here, a stretch of residues (Val94-Pro99) was identified in the structure that lined the complex interface and might therefore play a role in antibody binding, from which Lys96 and Ile97 were selected to be substituted (Fig. 1C). Lys96 forms a hydrogen bond through a Phe102 (H CDR3 – CDR3 from heavy chain) main chain oxygen atom and is also involved in interactions with a water molecule that is in the allergen-antibody interface. This water molecule is bound through hydrogen bonds with two neighboring water molecules, as well as with Asp98 (L CDR3). Moreover, Lys96 is close (3.7 Å) to the carboxyl group of Asp104 (H CDR3). Substitution to Glu at position 96 was expected to perturb this hydrogen bond network, while also giving rise to unfavorable electrostatic repulsion with the negatively charged carboxylate groups. Ile97 also forms a hydrogen bond through main chain atoms with amide group of Asn52, which is not expected to be affected by the mutation (H CDR2; Table 2), and in addition interacts through the side chain and hydrophobic interactions with Asn31 (H CDR1) and Tyr54 (H CDR2). The Ile97Glu mutation also eliminated the large aliphatic side chain of Ile97 that was wedged against H CDR1 and H CDR2, and was expected to significantly reduce the ability of this residue to form extensive hydrophobic contacts. In summary, the mutation did not only result in substitution of a hydrophobic amino acid with a hydrophilic one, but also changed the charge from neutral to negative.
Effect of epitope mutations on mAb 7A1 binding
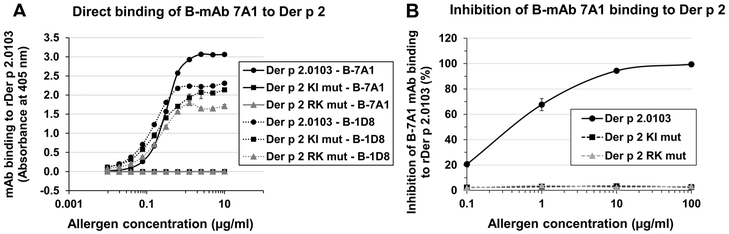
The capacity of the two Der p 2 mutants to bind the mAb 7A1 was completely abolished by the amino acid substitutions discussed previously, as shown by direct and inhibition binding assays (Fig. 4). However, the ability of mAb 1D8 (whose epitope lies opposite from the mAb 7A1 epitope) to bind the mutants was preserved (Fig. 4A), suggesting that the overall fold of Der p 2 mutants was conserved. Similar folding of the mutants versus the wild type was confirmed by NMR. In a 1D experiment, both mutants showed 1H dispersion from 10.5 ppm to 6.2 ppm in the amide region with overlapping peaks (Supplemental Fig. 2) and methyl peaks approached 0 ppm typical of well-folded proteins. Unfolded proteins have a narrow amide dispersion centered around 8.2 +/− 0.5 ppm, and typically no methyl peaks below 0.85, as can be seen when Der p 2 is treated with 6 M urea and 1 mM DTT (Supplemental Fig. 2). These results suggest that the structure was largely maintained in both mutants.
Figure 4. Mutations abolished mAb 7A1 binding to Der p 2.
A) Direct binding assay showing that the two Der p 2 mutants in the epitope for mAb 7A1 did not bind B-mAb 7A1, whereas both bound B-mAb 1D8 (positive control). Plot represents an average of two experiments. B) Results of inhibition assays showing that the two Der p 2 mutants did not bind the mAb 7A1. Plot represents an average of three experiments (each run in duplicate).
Relevance of the mAb 7A1 epitope regarding subjects’ IgE Ab binding to Der p 2
To assess the overlap between mAb 7A1 and IgE binding, inhibition of plasma IgE antibody binding to Der p 2 by mAb 7A1 and two other murine IgG mAb was measured. The three mAbs (7A1, 1D8 and αDpX) inhibited binding of IgE antibodies in different degrees depending on the individual (n = 5), reaching maximum inhibitions of up to 39.0%, 49.7 and 42.7%, respectively (Fig. 5A). The specificity of the IgE inhibition to Der p 2 was shown in the same experiment using natural Der p 1 as a negative control (Fig. 5B).
Figure 5. Inhibition of IgE antibody by IgG mAbs and allergens.
A) Overlap of IgE and murine mAb IgGs. Plots show maximum inhibitions (using inhibitor at 100 μg/mL). Percentage of inhibition varies by patient (n = 5). B) Specificity of inhibition of IgE antibody binding to Der p 2 using natural Der p 1 as negative control (allergen concentrations are 100 μg/mL). For both panels data are average of duplicates ± SD. For panel B long and short horizontal lines indicate geometric means and 95% CIs, respectively.
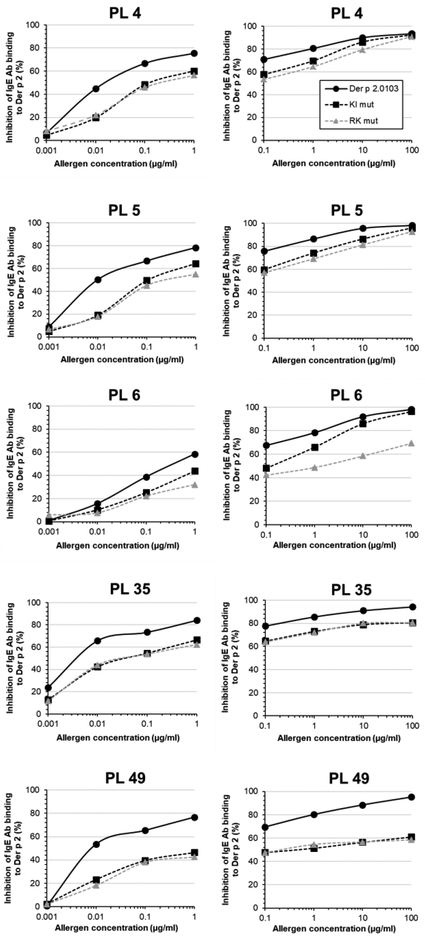
In addition, IgE binding of the Der p 2 mutants compared to wild type was also tested using patient blood plasma. The mutants displayed a reduction in IgE Ab binding relative to the wild type for all individual plasma tested (n = 5) in inhibition assays. However, the ability of the mutants to inhibit binding of patient’s IgE antibodies versus the wild type varied depending on the patient, and can be summarized by the following scenarios: a) both mutants inhibited plasma IgE antibody binding to a similar degree and reached 100% inhibition at high allergen concentration, with an approximately 10-fold higher IC50 than the wild type (as shown by inhibition curves of patients 4 and 5 that were displaced to the right versus the wild type curves), b) the two mutants similarly inhibited plasma IgE antibody binding, but failed to inhibit all IgE binding even at high protein concentration, whereas wild type allergen was able to fully inhibit IgE binding to the immobilized allergen (patients 35 and 49), c) the mutants behaved differently in that the Lys96Glu-Ile97Glu mutant, although less effective than the wild type allergen, had about a 10-fold higher capacity to inhibit IgE than the Arg31Ala-Lys33Ala mutant (that did not reach 100% inhibition) (patient 6). The differences in the ability to inhibit plasma IgE antibody binding to wild type Der p 2 between mutants and wild type were from 10-fold (patients 4, 5 and 35) up to 100-fold (mutant Arg31Ala-Lys33Ala in patient 6 and both mutants for patient 49) (Fig. 6). In summary, there is a subject-specific epitope profile detected by IgE-specific for Der p 2.
Figure 6. Reduced capacity to inhibit patient’s IgE antibody binding by Der p 2 mutants versus the wild type.
A reduction of the IgE Ab binding in different degrees by the 2 mutants was seen for all patients (n = 5). Left and right plots are results from two experiments per patient using a different range of inhibition concentrations. Data are averages of duplicates ± SD.
Selection and reactivity profile of a human monoclonal anti-Der p 2 IgE fragment
Two phage display libraries were generated from IgE H chain V domain-encoding transcripts of highly mite-allergic subjects, and a diversity of light L chain V domain-encoding transcripts. The two libraries were screened for Der p 2 binding constructs, and Der p 2-specific scFvs were selected from one of the two combinatorial libraries, which was constructed from a subject with ImmunoCAP class 4 for dust mite sIgE. Four isolated Der p 2-specific scFv had clonally related heavy chain variable domains (as evidenced by the presence of an identical, 15-residue long H CDR3 sequence) and light chain variable domains differing only in a few residues. Since H CDR3 has a dominant role in antibody recognition and has been reported to be sufficient for most antibody specificities (59), and the four clones had identical H CDR3, only one clone was selected for expression. The selected clone, H04_B04, bound both to immobilized Der p 2 as demonstrated by ELISA, and to soluble Der p 2 as demonstrated by real-time binding assays with surface plasmon resonance-based detection (Fig. 7). The genes encoding this H and L chain variable domains of the scFv were a somatically mutated sequence derived from the IGHV5–51*04 germline gene, and an unmutated sequence derived from the identical IGKV1–39*01 or IGKV1–39D*01 germline genes (https://www.ncbi.nlm.nih.gov/nuccore/MK937271). Importantly, this scFv was blocked from binding to immobilized Der p 2 by pre-incubation of the allergen with Der p 2-specific mAb 7A1 but not by preincubation of the allergen with Der p 2-specific mAb 1D8 (Fig. 7B).
Figure 7. A human IgE antibody fragment, H04_B04, isolated from a mite allergic subject’s repertoire binds the epitope recognized by mAb 7A1.
A) Two antigen concentrations (30 nM -dark grey- and 120 nM -light grey-) were used to show proof of scFv binding to Der p 2 (with an estimated affinity of 35 nM) as demonstrated by SPR-based real time analysis. B) The scFv was blocked from binding to Der p 2 by preincubation with mAb 7A1 but not with mAb 1D8. The reactivity of the scFv to Der p 2 was tested at different concentrations (undiluted (1x), and diluted 10-fold and 40-fold (10x and 40x, respectively) of scFv, and following pre-incubation of the immobilized allergen with IgG mAbs 1D8 (top plot) and 7A1 (bottom plot), respectively, at two different concentrations. Data represent duplicates from one experiment representative of two performed. Analysis of anti-Der p 2 Fab (C) and scFab binding (D) to wild type Der p 2 and mutants thereof by ELISA, demonstrated that neither of these molecular formats of H04_B04, although they bound Der p 2.0103, recognized the two mutated versions of the allergen, Der p 2.0103 Lys96Glu-Ile97Glu, and Der p 2.0103 Arg31Ala-Lys33Ala. The Fab and scFab constructs were in media (sups) from the cultures that expressed these recombinant proteins (E. coli and P. pastoris, respectively). Data are average ± SD of two experiments from three performed.
This scFv antibody construct was also reformatted into Fab and scFab constructs and produced in E. coli and P. pastoris, respectively, for further characterization, in a failed attempt to find a suitable format for crystallographic purposes. All the constructs tested (scFv, Fab and scFab) tended to form multimers that impair crystallization. Nevertheless, these Fab and scFab were very useful to show binding to Der p 2.0103, but not to the two mutants (Lys96Glu-Ile97Glu, and Arg31Ala-Lys33Ala) (Fig. 7C-D). Interestingly, these results indicate that the reactivity profile of mAb 7A1 and its lack of binding to the Der p 2 mutants resembled that of a member of the IgE repertoire of an allergic subject.
Discussion
Structural analysis of the IgE recognition of allergens is the basis for rational design of recombinant counterparts to be used in the future for immunotherapy. Originally, Der p 2 epitope mapping based on 15 amino acid peptides led to the identification of a sequential IgE antibody binding epitope (11). However, IgE antibody binding to Der p 2 was shown to be susceptible to reduction and alkylation of the allergen, demonstrating the conformational nature of many IgE epitopes on Der p 2 (12). Subsequently, site-directed mutagenesis (13,14) and hydrogen exchange NMR spectroscopy were used to define these conformational epitopes at the residue level (45). Other studies have relied on detailed characterization of reactivity profiles of human monoclonal IgEs derived from combinatorial libraries to allergens, such as Phl p 1, to identify residues of particular importance for IgE-allergen-interaction that defined suitable sites of mutagenesis able to prevent IgE binding (60). However, the most precise approach to define the molecular structure of an epitope is to determine the three-dimensional structure of an allergen with an antibody construct. In the current study, a very detailed analysis (based on X-ray crystallography and NMR data) of a conformational antigenic epitope on the mite allergen Der p 2 is presented. This epitope, also recognized by human IgE, is used to construct Der p 2 variants with reduced capacity to bind IgE.
Previous studies by this group and others reported the structures of allergens such as birch Bet v 1, mite Der p 1 and Der f 1, and cockroach Bla g 2 in complex with Fab or Fab’ fragments of murine IgG mAbs (22-25,30). All of these antibodies were used as surrogates for IgE antibodies because: a) they were reported to inhibit IgE antibody binding to the allergen, which confirmed their epitope overlap with IgE binding site/s, and b) they are homogeneous proteins, which is needed for crystallization purposes (unlike serum IgE that is not possible to crystallize due to its polyclonal nature). This approach allowed the determination of IgE antibody binding sites by detailed site-directed mutagenesis analysis of the IgG epitope. The current study reports the first structure of one of the most important major mite allergens, Der p 2, in complex with a murine IgG mAb (7A1). This antibody was shown to inhibit up to 39% of plasma IgE binding to Der p 2 for 5 mite-allergic patients, in agreement with an up to 30% inhibition previously reported in individual or pooled patients’ sera (45). According to the surface of the Der p 2 molecular structure, a maximum of 3–5 non-overlapping epitopes are estimated to be possibly recognized by monoclonal antibodies at the same time. There is evidence of at least two other epitopes involved in IgE antibody binding on Der p 2, which recognize an area that is opposite to the site where mAb 7A1 binds (45). Therefore, the effect size of the observed inhibition by mAb 7A1 is concordant with the expected contribution of this epitope to serum specific IgE binding to Der p 2. The mAb 7A1 also recognizes Der f 2, the homologous allergen from Dermatophagoides farinae (36). A similar contribution of the corresponding mAb 7A1 epitope to serum IgE was found in a study for Der f 2. The mutation of prolines 34, 95 and 99 (that are located in the proximity of the residues mutated here) to alanine led to reduction of IgE antibody binding, without disruption of the global allergen structure (61). A triple Der f 2 mutant inhibited IgE binding up to ~50% compared to the wild type (up 90%).
A pair of IgE antibodies of different and non-overlapping epitope specificities are sufficient to induce histamine release, even with low affinity binding (61,62). Accordingly, the allergen mutated in only one epitope is expected to still be able to activate basophils and induce mediator release, due to the presence of at least two additional epitopes. In fact, no reduction in mediator release was observed in the past for a Der p 1 mutated in only one epitope -data not shown- and for the larger cockroach allergen Bla g 2 mutated in three IgE antibody binding sites (32). Additional epitopes are currently under study, and multiple epitope mutants will be tested in the future for reduction of mediator release versus the wild type allergens. In summary, the epitope defined here might be an important IgE antibody binding site for Der p 2 that contributes to FcεRI receptor cross-linking on effector cells.
In a previous study, hydrogen exchange NMR spectroscopy mapping described the approximate location of the mAb 7A1 epitope as being defined by residues 18–20, 27–37, 56–63, 56–63, 92–106 and 123–129 (45). Subsequent alanine-scanning mutations further identified the epitope as encompassing residues 30, 31, 33, 96, 97, while other candidate amino acids for the epitope (Ser57, Asn93, Glu102), were confirmed to be far from the epitope (45). In the present study, the high-resolution (2.45 Å) structural data provided by X-ray crystallography, coupled with 1H-13C NMR, allowed for a more targeted approach. IgE binding was reduced (plasma IgE) or abolished (phage display-derived IgE antibody construct) through substitution of only two amino acids representing a small proportion of the Der p 2 surface area (170 Å2 for Arg31-Lys33 and 280 Å2 for Lys96-Ile97 are the areas that contribute towards the epitope area, calculated using 6OY4, versus 7,430 Å2 Der p 2 surface area calculated from 1KTJ using PDBePISA), and with minimal alteration to allergen structure. While Der p 2 is stable and maintains the capacity to bind human IgE and murine IgG mAbs under acidic (pH 2), basic (pH 12) and denaturing (6 M guanidine) conditions (12), it is susceptible to structural perturbations by amino acid substitution within its hydrophobic pocket that could drastically increase or decrease protein stability (44). In this study, the residues were substituted on the molecular surface, without causing a change in the overall folding, as proven by immunoassays and NMR. In previous mutagenesis analysis performed by the same authors on other allergens, the substitution of residues on the molecular surface did not necessarily always have such a strong effect on antibody binding (24,31,33). The Der p 2 mutant Lys96Ala was reported to be misfolded in a previous study (14). Here, the substitution of this Lys96 by Glu in combination with the Ile97Glu mutation resulted in a molecule that was still able to bind polyclonal IgE from patients’ plasma, indicating that some IgE conformational epitopes had been preserved regardless of the mutations, in agreement with the proper folding of the mutant. The ability to engineer hypoallergenic variants using the minimum number of mutations will prove invaluable, particularly when applied to other allergens such as Der p 1 which do not share the same resilience to structural perturbations as Der p 2 (12).
Importantly, phage display library technology was able to identify one human IgE antibody construct that recognized an epitope overlapping that of mAb 7A1. Combinatorial antibody libraries represent one of the most reliable ways of isolating human allergen-specific binders that resemble those of the IgE repertoire (63). The clone identified was also unable to bind both double mutants of Der p 2, which were designed based on the epitope recognized by mAb 7A1. Therefore, the binding of the IgE antibody construct derived from an allergic subject’s repertoire depends also on the presence of the residues Lys96-Ile97 and Arg31-Lys33. Indeed, a set of highly similar clones carrying related heavy chain sequences with identical CDR3, and highly similar light chains differing only in few amino acids were isolated. Such dependence also on the light chain to construct a functional binding site through use of combinatorial library technology has been observed for many other allergen-specific IgEs identified in the past (64). This dependence suggests the existence of a very similar heavy-light chain pair also in the allergic subject from which the library was constructed. These findings suggest that human IgE with similar specificity to that of mAb 7A1 exist in at least some allergic subjects.
Additional evidence that an IgE antibody binding site had been mutated was provided by IgE inhibition experiments using plasma from five different patients. The data confirmed a consistent decrease in IgE binding to the mutants versus the wild type. Given that the human IgE antibody response is polyclonal, the mutations performed are not expected to completely inhibit plasma IgE antibody binding, but only affect the binding of those IgE that recognize a site overlapping with the mAb 7A1 epitope. This decrease of IgE binding to the mutants could reflect partial or complete abolition of binding of those particular IgE antibodies that overlap with mAb 7A1. In addition, the effect of the mutations differed by patient. For example, the fact that both mutants similarly reached the 100% inhibition at high concentration, with curves displaced to the right versus the wild type, could indicate that all IgEs recognized their epitopes, but an IgE bound to the mutated site with lower affinity. On the other hand, a part of the IgE repertoire of some subjects had lost its ability to fully recognize the Der p 2 mutants (indicated by the fact that 100% inhibition of plasma IgE was not reached -for plasma 35 and 49, and the RK mutant only for plasma 6-). Similarly, the above-mentioned study of the homologous allergen Der f 2 reported that substitution of 1–3 amino acids located close to the equivalent residues mutated here, also affected allergenicity, but not in the same way and for all patients (61). The plasma IgE binding results reported in the current study would be in agreement with the finding that the monoclonal scFv derived from the IgE repertoire of an allergic subject did not recognize the mutated variants of the allergen. The different responses of IgE from different subjects illustrate that a personalized approach to hypoallergen design may be required.
The structural analysis demonstrated a flexibility of Der p 2. Superposition of the X-ray crystal structures of Der p 2 alone and in complex with the Fab of the mAb 7A1 revealed conformational changes in the allergen structure. Antibody binding to one part of the molecule was translated into conformational changes, even in distal parts of the molecule opposite to the mAb 7A1 epitope, which could potentially affect the binding of other antibodies. Nevertheless, mAb 7A1 and either mAb 1D8 or alpha-DpX can simultaneously bind Der p 2. The flexible immunoglobulin-like structure of Der p 2, formed by anti-parallel β-sheets, might contribute to conformational changes in areas that are far from the antibody binding site. It was considered if these changes could be in part attributed to differences in pH of the crystallization conditions of the allergen alone or in complex with the antibody, which include a 2-pH unit difference. The crystal used for determination of Der p 2 structure (PDB code 1KTJ) (35) was grown in conditions that were significantly more acidic than the ones used to obtain the crystal of the Der p 2–7A1Fab complex. Nevertheless, this possibility is unlikely given that the NMR analysis performed here at pH 7.4 (which confirmed the flexibility of Der p 2 upon mAb binding) affected the same residues identified by X-ray crystallography analysis (i.e. Val63, Val116).
The epitope structure and subsequent mutagenesis analysis shows that residues 31, 33, 96 and 97 are important for IgE binding. These residues are conserved across 15 Der p 2 and 16 Der f 2 variants currently listed in the Allergen Nomenclature database (www.allergen.org). Accordingly, the mAb 7A1 recognized all six variants tested: Der p 2.0101, Der p 2.0102, Der p 2.0103, Der f 2.0101, Der f 2.0102 and Der f 2.0103 (65). Therefore, the relevance of these finding in terms of design of hypoallergens could be extended to the cross-reactive allergen Der f 2 that is also recognized by mAb 7A1. As a consequence, a large influence of the variants on effector cell degranulation due to this epitope is not expected (66). As such, mAb 7A1 is a good candidate for “universal” Der p 2 detection. This is in contrast with previous reports of five mAbs that were unable to bind a certain Der p 2 variant (i.e. 1D8 does not bind Der p 2.0101, an Asp114Asn variant) (65,67). In addition, Eur m 2, Lep d 2 and Tyr p 2 variants have at least one amino acid substitution at one of the two positions mutated for each mutant, and these allergens are not recognized by mAb 7A1 (65). These differences are the basis for cross-reactivity or lack thereof among group 2 allergens from different species (65). Overall, these findings indicate that mAb 7A1 would be a good candidate for measuring Der p 2 in complex extracts used for diagnosis and immunotherapy, in contrast to other mAbs that do not recognize all the isoforms (68).
This study presents a detailed analysis of an IgE antibody binding site on Der p 2, one of the most important dust mite allergens. First, the epitope recognized by the murine IgG mAb 7A1, that had been reported to overlap with an IgE antibody binding site on the allergen, was identified, showing the flexibility of the molecule upon antibody binding. This was achieved by determining the X-ray crystal structure of Der p 2 in complex with Fab from the mAb 7A1, combined with NMR studies. Second, a detailed analysis of the epitope defined critical residues for allergen-antibody interactions based on a rational design of site-directed mutagenesis. Finally, the identification of a phage-display antibody derived from human genes that recognizes the same epitope as mAb 7A1, further confirms the relevance of the mAb 7A1 epitope to human health. The identification of amino acid substitutions involved in IgE binding will allow for a rational design of hypoallergens for better therapy, aiming to mitigate adverse reactions that are usually observed in conventional immunotherapy.
Supplementary Material
Single-sentence key points that summarize the manuscript’s findings.
The structure of mite Der p 2 in complex with murine IgG mAb7A1 was determined.
Human IgE construct identified from phage display library overlaps with mAb 7A1.
Epitope mutagenesis analysis contributes to designing hypoallergens for therapy.
Acknowledgements
We thank Drs. Alexander C.Y. Foo and Lars C. Pedersen for helpful advice.
Research reported in this publication was supported by the National Institute of Allergy And Infectious Diseases of the National Institutes of Health under Award Number R01AI077653 (to principal investigators A.P., M.D.C., and M.C.; and to J.A.W. and M.O.). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. This research was also supported in part by the Intramural Research Program of the NIH, National Institute of Environmental Health Sciences, Z01-ES102906 (REL), and by the Swedish Research Council (grant numbers 521–2011-3282 and 2016–01720) (to MO). Structural results shown in this report are derived from data collected at Southeast Regional Collaborative Access Team (SER-CAT; 22ID) beamline at the Advanced Photon Source, Argonne National Laboratory. Supporting institutions may be found at https://www.ser.aps.anl.gov/www.ser-cat.org/images/SER-CAT18_Member_list.jpg. Use of the Advanced Photon Source was supported by the U.S. Department of Energy, Office of Science, Office of Basic Energy Sciences, under Contract Nos. DE-AC02–06CH11357 and W-31–109-Eng-38. This work was partially supported by an ASPIRE III grant from the Office of the Vice President of Research at the University of South Carolina.
Abbreviations
- HMQC
Heteronuclear multiple quantum coherence
- H CDRx
Complementarity Determining Region x (1, 2 or 3) from antibody heavy chain
- IgE-scFv
human anti-Der p 2 IgE scFv antibody construct
- L CDRx
Complementarity Determining Region x (1, 2 or 3) from antibody light chain
- NMR
nuclear magnetic resonance
- PDB
Protein Data Bank
- SEC
size exclusion chromatography
- scFab
single chain Fab
- scFv
single-chain variable fragment
- 7A1scFv
mAb 7A1 scFv
- V
variable
References
- 1.Platts-Mills TA, Vervloet D, Thomas WR, Aalberse RC, and Chapman MD. 1997. Indoor allergens and asthma: report of the Third International Workshop. J. Allergy Clin. Immunol 100: S2–24. [DOI] [PubMed] [Google Scholar]
- 2.Portnoy J, Chew GL, Phipatanakul W, Williams PB, Grimes C, Kennedy K, Matsui EC, Miller JD, Bernstein D, Blessing-Moore J, Cox L, Khan D, Lang D, Nicklas R, Oppenheimer J, Randolph C, Schuller D, Spector S, Tilles SA, Wallace D, Seltzer J, and Sublett J. 2013. Environmental assessment and exposure reduction of cockroaches: a practice parameter. J Allergy Clin Immunol 132: 802–808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gold DR, Adamkiewicz G, Arshad SH, Celedon JC, Chapman MD, Chew GL, Cook DN, Custovic A, Gehring U, Gern JE, Johnson CC, Kennedy S, Koutrakis P, Leaderer B, Mitchell H, Litonjua AA, Mueller GA, O’Connor GT, Ownby D, Phipatanakul W, Persky V, Perzanowski MS, Ramsey CD, Salo PM, Schwaninger JM, Sordillo JE, Spira A, Suglia SF, Togias A, Zeldin DC, and Matsui EC. 2017. NIAID, NIEHS, NHLBI, and MCAN Workshop Report: The indoor environment and childhood asthma-implications for home environmental intervention in asthma prevention and management. J Allergy Clin Immunol 140: 933–949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Platts-Mills T, and de Weck A. 1989. Dust mite allergens and asthma A worldwide problem., 83 ed. 416–427. [DOI] [PubMed] [Google Scholar]
- 5.Sánchez-Borges M, Fernandez-Caldas E, Thomas WR, Chapman MD, Lee BW, Caraballo L, Acevedo N, Chew FT, Ansotegui IJ, Behrooz L, Phipatanakul W, Gerth van WR, Pascal D, Rosario N, Ebisawa M, Geller M, Quirce S, Vrtala S, Valenta R, Ollert M, Canonica GW, Calderon MA, Barnes CS, Custovic A, Benjaponpitak S, and Capriles-Hulett A. 2017. International consensus (ICON) on: clinical consequences of mite hypersensitivity, a global problem. World Allergy Organ J 10: 14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Heymann PW, Chapman MD, Aalberse RC, Fox JW, and Platts-Mills TA. 1989. Antigenic and structural analysis of group II allergens (Der f II and Der p II) from house dust mites (Dermatophagoides spp). J. Allergy Clin. Immunol 83: 1055–1067. [DOI] [PubMed] [Google Scholar]
- 7.Chapman MD, and Platts-Mills TA. 1980. Purification and characterization of the major allergen from Dermatophagoides pteronyssinus-antigen P1. J Immunol 125: 587–592. [PubMed] [Google Scholar]
- 8.Weghofer M, Grote M, Resch Y, Casset A, Kneidinger M, Kopec J, Thomas WR, Fernandez-Caldas E, Kabesch M, Ferrara R, Mari A, Purohit A, Pauli G, Horak F, Keller W, Valent P, Valenta R, and Vrtala S. 2013. Identification of Der p 23, a peritrophin-like protein, as a new major Dermatophagoides pteronyssinus allergen associated with the peritrophic matrix of mite fecal pellets. J Immunol 190: 3059–3067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mueller GA, Randall TA, Glesner J, Pedersen LC, Perera L, Edwards LL, DeRose EF, Chapman MD, London RE, and Pomés A. 2016. Serological, genomic and structural analyses of the major mite allergen Der p 23. Clin Exp Allergy 46: 365–376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Posa D, Perna S, Resch Y, Lupinek C, Panetta V, Hofmaier S, Rohrbach A, Hatzler L, Grabenhenrich L, Tsilochristou O, Chen KW, Bauer CP, Hoffman U, Forster J, Zepp F, Schuster A, Wahn U, Keil T, Lau S, Vrtala S, Valenta R, and Matricardi PM. 2017. Evolution and predictive value of IgE responses toward a comprehensive panel of house dust mite allergens during the first 2 decades of life. J Allergy Clin Immunol 139: 541–549. [DOI] [PubMed] [Google Scholar]
- 11.van ‘t HW, Driedijk PC, van den Berg M, Beck-Sickinger AG, Jung G, and Aalberse RC. 1991. Epitope mapping of the Dermatophagoides pteronyssinus house dust mite major allergen Der p II using overlapping synthetic peptides. Mol. Immunol. 28: 1225–1232. [DOI] [PubMed] [Google Scholar]
- 12.Lombardero M, Heymann PW, Platts-Mills TA, Fox JW, and Chapman MD. 1990. Conformational stability of B cell epitopes on group I and group II Dermatophagoides spp. allergens. Effect of thermal and chemical denaturation on the binding of murine IgG and human IgE antibodies. J Immunol 144: 1353–1360. [PubMed] [Google Scholar]
- 13.Smith AM, and Chapman MD. 1997. Localization of antigenic sites on Der p 2 using oligonucleotide-directed mutagenesis targeted to predicted surface residues. Clin Exp Allergy 27: 593–599. [DOI] [PubMed] [Google Scholar]
- 14.Reginald K, and Chew FT. 2018. Conformational IgE epitope mapping of Der p 2 and the evaluations of two candidate hypoallergens for immunotherapy. Sci Rep 8: 3391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pomés A 2010. Relevant B cell epitopes in allergic disease. Int Arch Allergy Immunol 152: 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mueller GA, Min J, Foo ACY, Pomés A, and Pedersen LC. 2019. Structural analysis of recent allergen-antibody complexes and future directions. Curr Allergy Asthma Rep 19: 17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Amit AG, Mariuzza RA, Phillips SE, and Poljak RJ. 1986. Three-dimensional structure of an antigen-antibody complex at 2.8 A resolution. Science 233: 747–753. [DOI] [PubMed] [Google Scholar]
- 18.Sheriff S, Silverton EW, Padlan EA, Cohen GH, Smith-Gill SJ, Finzel BC, and Davies DR. 1987. Three-dimensional structure of an antibody-antigen complex. Proc Natl. Acad. Sci. U. S. A 84: 8075–8079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Padlan EA, Silverton EW, Sheriff S, Cohen GH, Smith-Gill SJ, and Davies DR. 1989. Structure of an antibody-antigen complex: crystal structure of the HyHEL-10 Fab-lysozyme complex. Proc Natl. Acad. Sci. U. S. A 86: 5938–5942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mirza O, Henriksen A, Ipsen H, Larsen JN, Wissenbach M, Spangfort MD, and Gajhede M. 2000. Dominant epitopes and allergic cross-reactivity: complex formation between a Fab fragment of a monoclonal murine IgG antibody and the major allergen from birch pollen Bet v 1. J Immunol 165: 331–338. [DOI] [PubMed] [Google Scholar]
- 21.Padavattan S, Schirmer T, Schmidt M, Akdis C, Valenta R, Mittermann I, Soldatova L, Slater J, Mueller U, and Markovic-Housley Z. 2007. Identification of a B-cell epitope of hyaluronidase, a major bee venom allergen, from its crystal structure in complex with a specific Fab. J Mol Biol 368: 742–752. [DOI] [PubMed] [Google Scholar]
- 22.Li M, Gustchina A, Alexandratos J, Wlodawer A, Wunschmann S, Kepley CL, Chapman MD, and Pomés A 2008. Crystal structure of a dimerized cockroach allergen Bla g 2 complexed with a monoclonal antibody. J Biol Chem 283: 22806–22814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li M, Gustchina A, Glesner J, Wunschmann S, Vailes LD, Chapman MD, Pomés A, and Wlodawer A. 2011. Carbohydrates contribute to the interactions between cockroach allergen Bla g 2 and a monoclonal antibody. J Immunol 186: 333–340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chruszcz M, Pomés A, Glesner J, Vailes LD, Osinski T, Porebski PJ, Majorek KA, Heymann PW, Platts-Mills TA, Minor W, and Chapman MD. 2012. Molecular determinants for antibody binding on group 1 house dust mite allergens. J Biol Chem 287: 7388–7398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Osinski T, Pomés A, Majorek KA, Glesner J, Offermann LR, Vailes LD, Chapman MD, Minor W, and Chruszcz M. 2015. Structural analysis of Der p 1-antibody complexes and comparison with complexes of proteins or peptides with monoclonal antibodies. J Immunol 195: 307–316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Orengo JM, Radin AR, Kamat V, Badithe A, Ben LH, Bennett BL, Zhong S, Birchard D, Limnander A, Rafique A, Bautista J, Kostic A, Newell D, Duan X, Franklin MC, Olson W, Huang T, Gandhi NA, Lipsich L, Stahl N, Papadopoulos NJ, Murphy AJ, and Yancopoulos GD. 2018. Treating cat allergy with monoclonal IgG antibodies that bind allergen and prevent IgE engagement. Nat Commun 9: 1421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mitropoulou AN, Bowen H, Dodev TS, Davies AM, Bax HJ, Beavil RL, Beavil AJ, Gould HJ, James LK, and Sutton BJ. 2018. Structure of a patient-derived antibody in complex with allergen reveals simultaneous conventional and superantigen-like recognition. Proc. Natl. Acad. Sci U. S. A 115: E8707–E8716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Niemi M, Janis J, Jylha S, Kallio JM, Hakulinen N, Laukkanen ML, Takkinen K, and Rouvinen J. 2008. Characterization and crystallization of a recombinant IgE Fab fragment in complex with the bovine beta-lactoglobulin allergen. Acta Crystallogr. Sect. F. Struct. Biol Cryst. Commun 64: 25–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Padavattan S, Flicker S, Schirmer T, Madritsch C, Randow S, Reese G, Vieths S, Lupinek C, Ebner C, Valenta R, and Markovic-Housley Z. 2009. High-affinity IgE recognition of a conformational epitope of the major respiratory allergen Phl p 2 as revealed by X-ray crystallography. J Immunol 182: 2141–2151. [DOI] [PubMed] [Google Scholar]
- 30.Spangfort MD, Mirza O, Ipsen H, Van Neerven RJ, Gajhede M, and Larsen JN. 2003. Dominating IgE-binding epitope of Bet v 1, the major allergen of birch pollen, characterized by X-ray crystallography and site-directed mutagenesis. J Immunol 171: 3084–3090. [DOI] [PubMed] [Google Scholar]
- 31.Glesner J, Wunschmann S, Li M, Gustchina A, Wlodawer A, Himly M, Chapman MD, and Pomés A. 2011. Mechanisms of allergen-antibody interaction of cockroach allergen Bla g 2 with monoclonal antibodies that inhibit IgE antibody binding. PLoS ONE 6: e22223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Woodfolk JA, Glesner J, Wright PW, Kepley CL, Li M, Himly M, Muehling LM, Gustchina A, Wlodawer A, Chapman MD, and Pomés A. 2016. Antigenic determinants of the bilobal cockroach allergen Bla g 2. J Biol Chem 291: 2288–2301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Glesner J, Vailes LD, Schlachter C, Mank N, Minor W, Osinski T, Chruszcz M, Chapman MD, and Pomés A. 2017. Antigenic determinants of Der p 1: specificity and cross-reactivity associated with IgE antibody recognition. J. Immunol. 198: 1334–1344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mueller GA, Smith AM, Williams DC Jr., Hakkaart GA, Aalberse RC, Chapman MD, Rule GS, and Benjamin DC. 1997. Expression and secondary structure determination by NMR methods of the major house dust mite allergen Der p 2. J. Biol. Chem 272: 26893–26898. [DOI] [PubMed] [Google Scholar]
- 35.Derewenda U, Li J, Derewenda Z, Dauter Z, Mueller GA, Rule GS, and Benjamin DC. 2002. The crystal structure of a major dust mite allergen Der p 2, and its biological implications. J Mol Biol 318: 189–197. [DOI] [PubMed] [Google Scholar]
- 36.Ovsyannikova IG, Vailes LD, Li Y, Heymann PW, and Chapman MD. 1994. Monoclonal antibodies to group II Dermatophagoides spp. allergens: murine immune response, epitope analysis, and development of a two-site ELISA. J Allergy Clin Immunol 94: 537–546. [DOI] [PubMed] [Google Scholar]
- 37.Minor W, Cymborowski M, Otwinowski Z, and Chruszcz M. 2006. HKL-3000: the integration of data reduction and structure solution-from diffraction images to an initial model in minutes. Acta Crystallogr. D. Biol Crystallogr 62: 859–866. [DOI] [PubMed] [Google Scholar]
- 38.Vagin A, and Teplyakov A. 2010. Molecular replacement with MOLREP. Acta Crystallogr. D. Biol Crystallogr 66: 22–25. [DOI] [PubMed] [Google Scholar]
- 39.Winn MD, Ballard CC, Cowtan KD, Dodson EJ, Emsley P, Evans PR, Keegan RM, Krissinel EB, Leslie AG, McCoy A, McNicholas SJ, Murshudov GN, Pannu NS, Potterton EA, Powell HR, Read RJ, Vagin A, and Wilson KS. 2011. Overview of the CCP4 suite and current developments. Acta Crystallogr. D. Biol Crystallogr 67: 235–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cowtan K 2006. The Buccaneer software for automated model building. 1. Tracing protein chains. Acta Crystallogr. D. Biol Crystallogr 62: 1002–1011. [DOI] [PubMed] [Google Scholar]
- 41.Emsley P, and Cowtan K. 2004. Coot: model-building tools for molecular graphics. Acta Crystallogr. D. Biol Crystallogr 60: 2126–2132. [DOI] [PubMed] [Google Scholar]
- 42.Murshudov GN, Skubak P, Lebedev AA, Pannu NS, Steiner RA, Nicholls RA, Winn MD, Long F, and Vagin AA. 2011. REFMAC5 for the refinement of macromolecular crystal structures. Acta Crystallogr. D. Biol Crystallogr 67: 355–367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Williams CJ, Headd JJ, Moriarty NW, Prisant MG, Videau LL, Deis LN, Verma V, Keedy DA, Hintze BJ, Chen VB, Jain S, Lewis SM, Arendall WB III, Snoeyink J, Adams PD, Lovell SC, Richardson JS, and Richardson DC. 2018. MolProbity: More and better reference data for improved all-atom structure validation. Protein Sci 27: 293–315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nakamura H, Ohkuri T, So T, and Ueda T. 2016. Relationship between the magnitude of IgE production in mice and conformational stability of the house dust mite allergen, Der p 2. Biochim. Biophys. Acta 1860: 2279–2284. [DOI] [PubMed] [Google Scholar]
- 45.Mueller GA, Smith AM, Chapman MD, Rule GS, and Benjamin DC. 2001. Hydrogen exchange nuclear magnetic resonance spectroscopy mapping of antibody epitopes on the house dust mite allergen Der p 2. J Biol Chem 276: 9359–9365. [DOI] [PubMed] [Google Scholar]
- 46.Goto NK, Gardner KH, Mueller GA, Willis RC, and Kay LE. 1999. A robust and cost-effective method for the production of Val, Leu, Ile (delta 1) methyl-protonated 15N-, 13C-, 2H-labeled proteins. J Biomol. NMR 13: 369–374. [DOI] [PubMed] [Google Scholar]
- 47.Ledsgaard L, Kilstrup M, Karatt-Vellatt A, McCafferty J, and Laustsen AH. 2018. Basics of antibody phage display technology. Toxins. (Basel) 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Reefer AJ, Carneiro RM, Custis NJ, Platts-Mills TA, Sung SS, Hammer J, and Woodfolk JA. 2004. A role for IL-10-mediated HLA-DR7-restricted T cell-dependent events in development of the modified Th2 response to cat allergen. J Immunol 172: 2763–2772. [DOI] [PubMed] [Google Scholar]
- 49.Reijonen H, and Kwok WW. 2003. Use of HLA class II tetramers in tracking antigen-specific T cells and mapping T-cell epitopes. Methods 29: 282–288. [DOI] [PubMed] [Google Scholar]
- 50.Levin M, King JJ, Glanville J, Jackson KJ, Looney TJ, Hoh RA, Mari A, Andersson M, Greiff L, Fire AZ, Boyd SD, and Ohlin M. 2016. Persistence and evolution of allergen-specific IgE repertoires during subcutaneous specific immunotherapy. J Allergy Clin Immunol 137: 1535–1544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Andréasson U, Flicker S, Lindstedt M, Valenta R, Greiff L, Korsgren M, Borrebaeck CA, and Ohlin M. 2006. The human IgE-encoding transcriptome to assess antibody repertoires and repertoire evolution. J Mol Biol 362: 212–227. [DOI] [PubMed] [Google Scholar]
- 52.Säll A, Walle M, Wingren C, Muller S, Nyman T, Vala A, Ohlin M, Borrebaeck CAK, and Persson H. 2016. Generation and analyses of human synthetic antibody libraries and their application for protein microarrays. Protein Eng Des Sel 29: 427–437. [DOI] [PubMed] [Google Scholar]
- 53.Engberg J, Andersen PS, Nielsen LK, Dziegiel M, Johansen LK, and Albrechtsen B. 1995. Phage-display libraries of murine and human antibody Fab fragments. Methods Mol Biol 51: 355–376. [DOI] [PubMed] [Google Scholar]
- 54.Koerber JT, Hornsby MJ, and Wells JA. 2015. An improved single-chain Fab platform for efficient display and recombinant expression. J Mol Biol 427: 576–586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wilson PC, de BO, Liu YJ, Potter K, Banchereau J, Capra JD, and Pascual V. 1998. Somatic hypermutation introduces insertions and deletions into immunoglobulin V genes. J Exp Med 187: 59–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ohlin M, and Borrebaeck CA. 1998. Insertions and deletions in hypervariable loops of antibody heavy chains contribute to molecular diversity. Mol Immunol 35: 233–238. [DOI] [PubMed] [Google Scholar]
- 57.Sprangers R, Velyvis A, and Kay LE. 2007. Solution NMR of supramolecular complexes: providing new insights into function. Nat Methods 4: 697–703. [DOI] [PubMed] [Google Scholar]
- 58.Hamel DJ, and Dahlquist FW. 2005. The contact interface of a 120 kD CheA-CheW complex by methyl TROSY interaction spectroscopy. J Am Chem Soc. 127: 9676–9677. [DOI] [PubMed] [Google Scholar]
- 59.Xu JL, and Davis MM. 2000. Diversity in the CDR3 region of V(H) is sufficient for most antibody specificities. Immunity 13: 37–45. [DOI] [PubMed] [Google Scholar]
- 60.Levin M, Rydnert F, Kallstrom E, Tan LW, Wormald PJ, Lindstedt M, Greiff L, and Ohlin M. 2013. Phl p 1-specific human monoclonal IgE and design of a hypoallergenic group 1 grass pollen allergen fragment. J Immunol 191: 551–560. [DOI] [PubMed] [Google Scholar]
- 61.Nakazawa T, Takai T, Hatanaka H, Mizuuchi E, Nagamune T, Okumura K, and Ogawa H. 2005. Multiple-mutation at a potential ligand-binding region decreased allergenicity of a mite allergen Der f 2 without disrupting global structure. FEBS Lett. 579: 1988–1994. [DOI] [PubMed] [Google Scholar]
- 62.Christensen LH, Holm J, Lund G, Riise E, and Lund K. 2008. Several distinct properties of the IgE repertoire determine effector cell degranulation in response to allergen challenge. J Allergy Clin Immunol 122: 298–304. [DOI] [PubMed] [Google Scholar]
- 63.Gadermaier E, Levin M, Flicker S, and Ohlin M. 2014. The human IgE repertoire. Int. Arch. Allergy Immunol 163: 77–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Persson H, Sadegh MK, Greiff L, and Ohlin M. 2007. Delineating the specificity of an IgE-encoding transcriptome. J Allergy Clin Immunol 120: 1186–1192. [DOI] [PubMed] [Google Scholar]
- 65.Smith AM, Benjamin DC, Hozic N, Derewenda U, Smith WA, Thomas WR, Gafvelin G, van Hage-Hamsten M, and Chapman MD. 2001. The molecular basis of antigenic cross-reactivity between the group 2 mite allergens. J Allergy Clin Immunol 107: 977–984. [DOI] [PubMed] [Google Scholar]
- 66.Christensen LH, Riise E, Bang L, Zhang C, and Lund K. 2010. Isoallergen variations contribute to the overall complexity of effector cell degranulation: effect mediated through differentiated IgE affinity. J Immunol 184: 4966–4972. [DOI] [PubMed] [Google Scholar]
- 67.Hakkaart GA, Chapman MD, Aalberse RC, and van RR. 1998. Immune-reactivity of recombinant isoforms of the major house dust mite allergen Der p 2. Clin. Exp. Allergy 28: 169–174. [DOI] [PubMed] [Google Scholar]
- 68.Park JW, Kim KS, Jin HS, Kim CW, Kang DB, Choi SY, Yong TS, Oh SH, and Hong CS. 2002. Der p 2 isoallergens have different allergenicity, and quantification with 2-site ELISA using monoclonal antibodies is influenced by the isoallergens. Clin Exp Allergy 32: 1042–1047. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.