Abstract
Background The development and adoption of health care common data models (CDMs) has addressed some of the logistical challenges of performing research on data generated from disparate health care systems by standardizing data representations and leveraging standardized terminology to express clinical information consistently. However, transforming a data system into a CDM is not a trivial task, and maintaining an operational, enterprise capable CDM that is incrementally updated within a data warehouse is challenging.
Objectives To develop a quality assurance (QA) process and code base to accompany our incremental transformation of the Department of Veterans Affairs Corporate Data Warehouse health care database into the Observational Medical Outcomes Partnership (OMOP) CDM to prevent incremental load errors.
Methods We designed and implemented a multistage QA) approach centered on completeness, value conformance, and relational conformance data-quality elements. For each element we describe key incremental load challenges, our extract, transform, and load (ETL) solution of data to overcome those challenges, and potential impacts of incremental load failure.
Results Completeness and value conformance data-quality elements are most affected by incremental changes to the CDW, while updates to source identifiers impact relational conformance. ETL failures surrounding these elements lead to incomplete and inaccurate capture of clinical concepts as well as data fragmentation across patients, providers, and locations.
Conclusion Development of robust QA processes supporting accurate transformation of OMOP and other CDMs from source data is still in evolution, and opportunities exist to extend the existing QA framework and tools used for incremental ETL QA processes.
Keywords: electronic medical records, data quality, common data models
Background and Significance
The ability to reproduce or synthesize research across health care systems can be stymied by differences in the underlying structures of the data. The development and implementation of common data models (CDMs) addresses some of the logistical challenges of performing research on data generated from disparate health care systems by using standardized terminology to express clinical information consistently. 1 However, transforming a data system into a CDM is not a trivial task. Despite the prospective usefulness of CDMs for health services and health outcomes research, characteristics of the source data and deficiencies in the data transformation process itself (i.e., the extract, transform, and load [ETL] process) can impact the practical utility and reliability of the CDM within a health care system.
The two main approaches to ETL are incremental and batch. 2 3 Batch ETL consists of loading and transforming all source records each time the ETL process is run. It is straightforward in its internal logic and processes but is resource intensive and may be impractical for large data systems when regularly scheduled releases are expected by its data consumers and stakeholders. Further, model identifiers (i.e., table primary keys) are regenerated with each load, forcing system users to rerun queries to stay consistent with the newly loaded data. Conversely, an incremental load approach is less resource intensive, as it includes only new and modified records in the transformation process, and maintains identifiers across processing instances. However, an incremental load process is more complex with more opportunities for ETL errors to occur requiring additional considerations for quality assurance (QA). Guiding principles of reproducibility and transparency have resulted in general QA documentation related to utilizing electronic medical record (EMR) data for secondary research purposes 4 5 6 and some have been extended for data-quality purposes in CDMs. For example, the framework proposed by Kahn et al was implemented for quality control of the National Patient-Centered Clinical Research Network (PCORnet) 7 but did not address how imperfect incremental ETL into a CDM can affect data quality. Similarly, Post et al described the importance of the timing of load (e.g., nightly, monthly, or quarterly) for incrementally updating local data into the Informatics for Integrating Biology and the Bedside (i2b2) CDM, but did address data-quality issues specific to incremental ETL. 3 The concept of a CDM is not unique to the health care arena. However, health care data have a higher volume of updates than most other settings, and have numerous limitations in patient identifiers and linkages.
The Observational Medical Outcomes Partnership (OMOP) CDM is an open, community-supported CDM that was initiated in 2009. 8 Since then numerous health databases worldwide 9 have converted data into the OMOP CDM and some have published on their approaches to QA. The majority of published QA processes for the OMOP CDM evaluated the degree of information loss through the transformation process and the ability of the transformed data to replicate findings produced by their source data. 10 11 12 13 Others have utilized the open source software application ACHILLES (Automated Characterization of Health Information at Large-Scale Longitudinal Evidence Systems) 8 14 15 to conduct QA and report their transformation's fidelity to the underlying source data. 15 16 The ACHILLES tool enables the assessment of mapping completeness across domains, generates data visualization to characterize data, and indirectly reports of general transformation errors in the destination OMOP domain tables. Development of robust QA processes supporting accurate transformation to OMOP and other similar CDMs (e.g., PCORnet) from source data is still in evolution, and opportunities exist to extend the existing QA framework and tools used for incremental ETL processes.
Objectives
Our objective was to develop a QA process and a code base to accompany our incremental transformation of the Department of Veterans Affairs (VA) EMR data into the OMOP CDM to prevent incremental load errors. Our library of QA scripts and procedures builds upon the open-source tools available for OMOP CDM and extends an existing framework of EMR data-quality research 5 by incorporating some key characteristics and requirements for incremental ETL processes. In this work we describe our QA process, the types of potential errors, and proposed solutions, and highlight areas for specific consideration and future development.
Methods
Data Source
In 2015, VA Informatics and Computing Infrastructure (VINCI) began transforming the VA Corporate Data Warehouse (CDW) health care database into the OMOP CDM for use by its research and operations communities. 17 The CDW contains data dating back to fiscal year 2000 and includes data from inpatient and outpatient encounters, diagnoses, procedures, pharmacies, laboratory tests, vital signs, provider information, and inpatient and outpatient external fee for service payments. Data are sourced from >130 hospitals, >1,000 outpatient and skilled nursing facilities, as well as VA external fee for service claims with over 700,000 non-VA facility inpatient admissions annually. 18 The VA EMR is especially complex, with data being sourced from 130 distinct medical systems that operate on different instances of the same EMR. 19 Historically, local sites were allowed a wide latitude in customization, although in recent years, national standardization has been strongly pursued. Thus, the same clinical concept (e.g., blood glucose laboratory test) may be represented in many different ways, with a unique representation from each distinct VA medical facility.
The initial transformation of the CDW in 2015 adhered to OMOP Version 4.0 specifications, with multiple ETL updates, and the most recent transformation process follows OMOP Version 5.1 conventions. Since the initial transformation, we have had three data releases to incorporate more current data in the OMOP instance, and because of the computational resources required for batch load, the VA's current OMOP ETL processes are run incrementally. In the following sections, we define our QA process as it relates specifically to two data releases of OMOP version 5.1—September 2017 and June 2018 (hereinafter referred to as Release A and Release B, respectively).
Incremental ETL Architecture
We used Transact-SQL stored procedures as our ETL incremental processing tool. A high-level architecture of our incremental ETL process is presented in Fig. 1 . The extract layer is initiated by (1) taking a snapshot of the current source dimension and source fact tables as well as downloading the most current OMOP meta-data. We then apply basic exclusion criteria such as row duplications and filtered date ranges as a preliminary source data cleaning process. Next, begins the transformation layer (2). All source records that have been created, updated, or deleted since the last snapshot are identified through a combination of primary key, CRUD (create, read, update, and delete), and ETL auto-incrementing processing ID. Similarly, at this stage we account for additions and updates to the OMOP CDM (e.g., new or changed concepts or concept relationships). Lastly (3), the transformed data records are mapped to OMOP meta-data and populated into the appropriate OMOP domain tables.
Fig. 1.
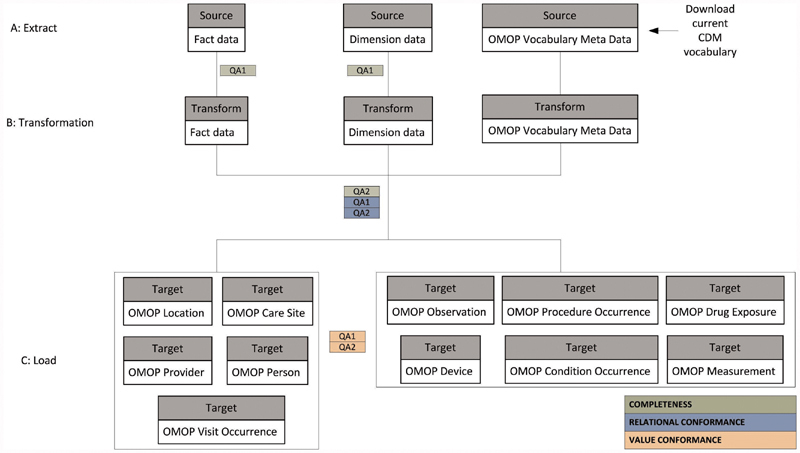
A summary of the data flow from the extract of the source data warehouse, through transformation, and loading into the OMOP data tables. The figure is generic to represent any fact or dimension table appropriate at that stage of ETL. The colors represent the QA element assessed at each stage. ETL, extract, transform, load; OMOP, Observational Medical Outcomes Partnership; QA, quality assurance.
Data-Quality Framework
The harmonized data-quality terminology discussed by Kahn and colleagues in 2016 5 forms the basis of our incremental data-quality framework. The authors amassed the diverse terminology used throughout data-quality literature and developed an ontology with three elements: completeness, conformance, and plausibility. While these three elements are interrelated, the goal of each data-quality check is distinct. Completeness for example is focused on the frequency of missing observations but is not concerned with the data values of the missing observations. However, conformance assures that data representation is in accordance with the constraints of a prespecified referent standard in terms of its structure and derived and independent data values. Similarly, plausibility focuses on whether or not the data values align with truth or are at least clinically possible. Each of these three elements can be affected by a multitude of underlying mechanisms but the focus of our present framework evaluates only the impact of the incremental ETL load process on the completeness and conformance QA elements. Plausibility is not included in the presented QA process because it less likely to be impacted by incremental load, but it is included in later stages of our QA process (not discussed).
The data-quality checks are interspersed across all steps of the transformation process with circular feedback workflow between the VINCI QA and ETL teams: testing, implementing fixes, and retesting until acceptable results are attained. The color scheme in Fig. 1 indicates which QA element— completeness (green), relational conformance (blue), or value conformance (orange) —each step addresses. Below we define each QA element and its purpose, extend the framework by describing key incremental load challenges, describe our ETL solution to overcome those challenges, and lastly describe examples within our process for how the incremental QA framework was employed to ensure that the ETL solution functioned as intended.
Completeness
Definition and Purpose
Completeness is defined as the degree to which source data, based on rows, are available in their target form. This does not mean that the data values themselves have to be identical; in fact, in line with Kahn's framework, the data values are not yet considered. Rather, we ensure that the amount of data in source form equals the amount of data expected in their target form. The purpose of the completeness data-quality element as it relates to incremental ETL is to evaluate whether all source updates have been accounted for at the transformation level and that updates in source to target mappings have occurred uniformly across all instances of each clinical concept.
Key Incremental Process Challenges
The number of records in the target OMOP tables can differ greatly from that of the originating source data. At both the transformation and load levels, some record count disparities are intended because of deduplication or expansions due to controlled vocabulary mappings, but others may be unwittingly introduced through incremental ETL. The transformation level is focused on tracking changes of individual source records using a trigger-based change data capture approach. This is an effective technique to detect source rows that are new or have been deleted/updated since the last ETL cycle, but is less reliable for administrative deletions (i.e., data still existed in the source but were deleted by the ETL team to meet data inclusion/exclusion criteria). For instance, filtered start and end dates are not flagged as source deletions and need to be handled differently. If these rows are not removed from the transformation level, the record will persist in target data and the volume of data will be above what is expected/intended.
The load level is focused on CDM vocabulary expansion/merge issues. Mappings between source concepts and OMOP standard concepts may be in one-to-one or one-to-many relationships and these relationships can change overtime, even within the same CDM version, to improve data quality and keep pace with evolving vocabularies. If the incremental load process does not trigger a recompute of historical records to account for updated intentional mapping expansions (e.g., from a 1:1 to a 1:2 relationship), the volume of OMOP data will be below what is expected as old records will be represented by fewer rows than are new records. Similarly, if the incremental load process does not trigger a recompute of historical records to account for intentional mapping consolidations (e.g., from a 1:2 to a 1:1 relationship), the volume of OMOP data will be greater than what is expected as old records will be represented by more rows than new records.
Incremental ETL Design Solution
At the transformation layer of ETL, records no longer in the source will trigger a delete from the transformation level tables. For records in the source layer that were intended to be excluded, we automated the removal at the transformation level. For example, current business logic for VA OMOP excludes any data prior to October 01, 1999. At the load level we introduced ETL auto-incrementing processing to update concept identifiers (e.g., concept IDs, concept type IDs) in OMOP tables for historical records. When a concept identifier is updated, we check the OMOP fact table against each transformation table (step B in Fig. 1 ). If the concept ETL identifiers match, there have been no updates to the concept identifiers and the historical records are left unchanged. If the concept ETL identifiers do not match, the fact table is updated with the most current concept identifiers to mirror the transformation table. We iterate through this process: checking for updates to each type of concept identifier (e.g., condition concept ID, condition type concept ID).
Quality Assurance Implementation
We use row-to-row comparisons to assess the completeness element of the ETL process. We first ensure that all inclusion and exclusion criteria were applied consistently across all source dimensions and fact tables. We compare what we observe (transformation level row count) to what we expect (source rows – exclusions = expected transformation rows) ( Fig. 1 , green QA1) and work with the ETL team to perform error analysis to identify the cause of any discrepancies and then implement code to resolve them. See Supplementary Appendix A (available in the online version) for examples of our error analysis process. Next, we ensure accurate mapping expansions and consolidations using row count comparisons between the transformation level and the OMOP level ( Fig. 1 , green QA2). We compare what we observe (OMOP level row count) to what we expect (transformation rows ± additional/fewer rows due to multiple mappings) and resolve any differences.
Relational Conformance
Definition and Purpose
Relational conformance is defined as the degree to which foreign keys throughout the model agree with the OMOP primary keys in the same way they would in source data. The purpose of relational conformance QA as it relates to incremental ETL is to ensure cascading updates and deletions have occurred across all relevant tables in the OMOP model. In other words, if an identifier, such as a provider identifier, has been updated, we check whether or not the update is reflected both in its OMOP parent table (the Provider table) and in all linked tables (e.g., Condition Occurrence, Procedure Occurrence, Visit Occurrence).
Key Incremental Process Challenges
Relational conformance errors can be compounded with dynamic EMRs when changes are made to patient records (mergers, splits, key data value substitutions) and health care personnel (mergers, splits, addition or subtraction of identifying or classifying information) are common between data loads. Similarly, physical addresses of sites of care delivery are vulnerable to frequent updates. These are the most computationally intensive source data changes to accommodate within an incrementally updated CDM representation. For example, there are numerous reasons patients can be assigned multiple source identifiers within a large health care system, and management of unique patient identification and patient record mergers are handled in multiple ways in different organizations. Each patient or provider merger can potentially affect thousands of records across all OMOP tables. Similarly, a slight change to a location such as an addition of a specified county will trigger a new location and care-site identifier to be generated and have downstream effects on millions of records across multiple OMOP tables. If the incremental load does not fully account for identifier changes, new records will align with updated identifiers while historical rows will retain the outdated identifier. Incremental changes to the OMOP meta-data can also cause similar relational conformance errors but are less computationally intensive to integrate.
Incremental ETL Design Solution
ETL batch tracking IDs are generated for each record of data across all tables in the transformation layer. At the OMOP load layer, where the load architecture is 1:1 (i.e., one row in the transform table equals one row in the loaded OMOP table), additional custom columns are added to the OMOP table to track the ETL batch ID of the transform. For many-to-one or one-to-many (row expansion or row contraction) capable transforms, a parallel mapping table is maintained to track the transform ETL batch ID and a pointer to the relevant record in the OMOP load table. The ETL process compares the current batch ID in the transform table to the batch ID at the OMOP layer (direct in-table or mapping table), and queues all rows with nonmatching ETL batch IDs for recomputation.
Quality Assurance Implementation
We carry out two closely related steps to ensure that relational conformance has not been violated. First, we introduce orphan checks to prevent identifiers in fact tables from pointing to nonexistent fields in the primary tables ( Fig. 1 , blue QA1). For example, if a person identifier is deprecated and correspondingly removed from the Person table, any lingering Procedure Occurrence records linked to the old person identifier are identified as orphan records, flagged as an error and reconfigured. Second, we introduce source to target referential integrity checks ( Fig. 1 , blue QA2). These checks trace each OMOP record back to its source record to confirm values of each foreign key (e.g., person, provider, and visit identifiers) correspond to the originating source value for those foreign keys. Any deviations are flagged as errors and recomputed as needed.
Value Conformance
Definition and Purpose
Value conformance is defined as the degree to which the values of the transformed data conform to the constraints of the OMOP CDM. The purpose of the value conformance QA element in an incremental ETL environment is to ensure that any updates to source standard concept identifiers (e.g., concepts deprecated from standard to nonstandard status or standard concepts that transfer domains) are consistently applied across all instances of the clinical concepts. In addition, this QA domain includes data checks for quantitative and qualitative value changes to the source data.
Key Incremental Process Challenges
A clinical concept as represented in the source data can have one and only one source concept identifier (e.g., ICD9CM code 250.1 for diabetes with ketoacidosis is represented by the OMOP concept ID 44828793) and be mapped or transformed to its corresponding OMOP standard vocabulary and concept ID(s). For example, ICD9CM code 250.1 is represented in the Systematized Nomenclature of Medicine (SNOMED) vocabulary by the standard OMOP concept ID 443727. Moreover, each concept can reside in one and only one allocated domain table (e.g., Condition Occurrence or Drug Exposure).
Factors related to both source data and the CDM can contribute to incomplete mapping of source concepts to standard concepts. Mapping augmentations to existing meta-data, at the source and CDM levels, are a part of all transforms. For example, with each data release we introduce incremental improvements to drug mappings with the aid of manual annotation from clinical experts and the application of multifaceted mapping algorithms. As a result, many concepts that were unmapped in previous OMOP instances can be successfully mapped in later instances. New source data that have not already been loaded into the OMOP model as well as historical data must account for any revised mappings since the previous data transformed. However, if the incremental transformation process does not account for the improved mappings, historical instances will remain unmapped while new instances will be mapped. Not only can the concept identifier change over time to reflect updated mapping between source and target concepts, the domain of the most current target concept(s) can differ from its previous domain. If the incremental logic fails here, historical instances will remain in the former, incorrect, domain, while new instances will be allocated to the new, correct domain.
Incremental ETL Design Solution
First, we design a process to ingest the OMOP meta-data (all the concept tables), analyze and generate ETL batch IDs for all records, compare the current ingested version with the prior download, and increment ETL batch IDs within the OMOP data so that we can accurately track any changes in the OMOP meta-data between releases. As these ETL batch IDs are tracked within each OMOP table during the load process, we are able to detect OMOP meta-data changes and queue them for downstream recomputation. We also created a manual meta-dataset of tables (concept table and the concept relationship tables) to allow for custom mappings. To manage the incremental process, and integration with future OMOP meta-data releases, we included a date/time stamp and versioning information to the manual mapping layer so that the ETL process can distinguish between a mapping that was forced to be different by the ETL team and a mapping that has changed through underlying OMOP meta-data changes. We prioritize OMOP official release data such that a prior custom mapping will be deprecated if an official concept ID (and relationship) is released later.
Quality Assurance Implementation
Evaluation of unmapped and partially mapped activity occurs at the OMOP level of the ETL process ( Fig. 1 , orange QA1). Metrics for this QA element are expressed both as a percentage of all source concepts (completely unmapped) and percentages of instances of individual source concepts (partial mappings). Here we quantify the volume of data mapped and unmapped in terms of both distinct source concepts (i.e., 25 individual medications are completely or partially unmapped) and total source instances (133,000/250,000 inpatient administrations of the 25 medications are unmapped). The completely unmapped source activity arises when all instances of a source concept are mapped to zero. The latter arises when a portion of the source instances is unmapped or mapped to zero and a portion is mapped to a standard concept. Any source concepts with more than one distinct set of target concepts are flagged for recomputation. Lastly ( Fig. 1 , orange QA2), similar to a function available in ACHILLES, we ensure that each table (e.g., Condition Occurrence) only contains concepts designated to its domain (e.g., condition concept identifiers). Any inconsistencies are flagged and appropriately redistributed to other tables.
Results
Completeness
Table 1 presents a summary of the mapping changes (source to target concept relationships) from Release A to Release B that resulted in either an expansion or consolidation of target rows. The mapping for over 2,000 International Classification of Disease, Ninth Revision, Clinical Modification (ICD9CM) diagnostic codes expanded from one SNOMED code in Release A to two SNOMED codes in Release B. As a result, approximately 38 million historical rows had to be updated. The ICD9CM code that contributed to the majority of this update type was code 296.3 (major depressive disorder, recurrent episode), affecting 9,702,989 historical rows in the Condition Occurrence table. Similarly, 50 ICD9CM codes that were previously mapped to two SNOMEDs in Release A mapped to only one SNOMED in Release B. As a result, more than 2 million rows in the Condition Occurrence table needed to be consolidated, primarily for instances of ICD9CM 796 (other nonspecific abnormal findings). An example of how completeness errors can impact data queries is illustrated in Table 2 .
Table 1. Global impact of intentional expansion and consolidation of source to target mappings between Release A and Release B.
| Source concept frequency | Source vocabulary | Target vocabulary | Previous mapping | New mapping | Historical expansion (rows) |
|---|---|---|---|---|---|
| 2,112 | ICD9CM | SNOMED | 1 | 2 | 37,952,222 |
| 109 | ICD9CM | SNOMED | 1 | 3 | 35,705 |
| 52 | ICD9CM | SNOMED | All other ICD9CM expansions | 17,431 | |
| 50 | ICD9CM | SNOMED | 2 | 1 | 2,180,536 |
| 4 | ICD9CM | SNOMED | 3 | 1 | 140,756 |
| 6 | ICD9CM | SNOMED | 3 | 2 | 11,976 |
Abbreviations: ICD9CM, International Classification of Disease, Ninth Revision, Clinical Modification; SNOMED, Systematized Nomenclature of Medicine.
Table 2. Examples of the impact of failed incremental load on use case queries.
| Data quality element | Release A status | Release B status | Consequence of failed incremental ETL on Release B |
|---|---|---|---|
| Completeness | |||
| Mapping expansion : the ICD9CM 200.51 (primary central nervous system lymphoma) maps to more target concepts than it did previously. | Mapped to one SNOMED code of the Condition domain (malignant lymphoma of lymph nodes); 265 instances of ICD9CM code equated to 265 rows in OMOP Condition Occurrence table. | Maps to the same SNOMED from Release A plus an additional condition SNOMED code 93195001 (primary central nervous system lymphoma); 646 (265 historic and 381 new records) instances of ICD code equate to 1,292 rows in OMOP. | Historical instances of ICD9CM code 200.51 will not be reflected in instances of SNOMED 93195001. We would erroneously report 2,358 instances (90% of truth) of this SNOMED when the true number of instances is 2,358 + 265 = 2,623 records. |
| Relational conformance | |||
| Person identifier merge : person identifiers that were previously used were deprecated and removed from the Person table. Records from the old person identifier were merged with a valid person identifier. | Person ID 1 was associated with 17,119 records across the Condition, Procedure, Drug, Measurement, and Observation OMOP tables. | Person ID 1 merged with another source identifier, replaced with the identifier ID 2, and linked to 17,649 records across the same OMOP tables from Release A. | Only new rows would be attributed to Person ID 2 and historical rows would be erroneously linked to Person ID 1. Only 3% of these patients' records (530) would be attributed to their updated identifier. 17,119 + 530 = 17,649. |
| Value conformance | |||
| Domain change : the Standard concept in the HCPCS vocabulary HCPCS H0004 (behavioral health counseling and therapy, per 15 minutes) previously belonged to the Observation domain and now belongs to the Procedure domain. | There were 2,054,638 instances of HCPCS H0004 found in the Observation table for 310,121 patients. | There were 2,142,664 instances of HCPCS H0004 found in the Procedure Occurrence table for 323,378 patients. | The historical instances would remain in the Observation table and unobserved by conventional standard queries of the Procedure table. We would erroneously report 88,026 instances (4% of truth) when the true number of instances is 2,054,638 + 88,026 = 2,142,664 records. |
Abbreviations: HCPCS, Healthcare Common Procedure Coding System; ICD9CM, International Classification of Disease, Ninth Revision, Clinical Modification; OMOP, Observational Medical Outcomes Partnership; SNOMED, Systematized Nomenclature of Medicine.
Relational Conformance
The volume of source identifier updates is presented in Table 3 . In the 10 months between Release A and Release B, many new patient, provider, and care-site identifiers were added to source data, and some numbers of site patient records were reassigned to a different patient enterprise identifier. As expected the absolute and relative number of updates to provider and care-site identifiers exceeded those of person identifiers, and collectively, over 200 million records across seven OMOP domain tables needed updating to align the OMOP instances with current source representation. To illustrate the impact of compromised relational conformance, Table 2 shows how incremental ETL errors can lead to the collection of fragmented patient data.
Table 3. Source identifier updates between Release A and Release B.
| Identifier type | Total in Release A table | New in Release B | Deprecated in Release B | Historical rows to be updated | Domains to be updated |
|---|---|---|---|---|---|
| Person | 23,456,405 | 305,434 | 8,090 | 493,039 | 7 |
| Provider | 6,717,950 | 248,825 | 47,582 | 148,042,702 | 7 |
| Care site | 1,174,609 | 118,370 | 71,770 | 59,409,762 | 1 |
Note: Domains evaluated: Condition, Device, Drug, Measurement, Observation, Procedure, and Visit.
Value Conformance
There were updates to concepts from all vocabularies used in the VA between Release A and Release B; most notably there were 138,017 new National Drug Codes (NDCs) added to the model, 1,497 SNOMED codes deprecated from standard to nonstandard status, and 15,413 standard Logical Observation Identifiers Names and Codes (LOINC) concepts switched domains. Table 4 provides a tabular summary of all meta-data changes between Release A and Release B. We only report updates to source vocabularies used in the VA. To illustrate the potential effect of value conformance errors, we focus on standard concepts that change domains over time. Table 2 describes how this type of error can lead to gross underestimation of instances when data incorrectly remained in an old domain.
Table 4. New and updated concepts between Release A and Release B.
| Vocabulary type | New standard and nonstandard concepts | Deprecated standard concepts | Updated to standard concepts | Standard concepts that switched domain |
|---|---|---|---|---|
| CPT4 | 867 | 87 | 1,562 | 0 |
| HCPCS | 463 | 192 | 1,026 | 253 |
| ICD9CM | 0 | NA | NA | NA |
| ICD9Proc | 0 | 0 | 5 | 0 |
| ICD10CM | 418 | NA | NA | NA |
| ICD10PCS | 5,346 | 13 | 0 | 0 |
| LOINC | 8,107 | 461 | 1 | 15,413 |
| NDC | 138,017 | 0 | 7,381 | 0 |
| RxNorm | 2,687 | 428 | 365 | 0 |
| SNOMED | 29,209 | 1,407 | 424 | 6,027 |
| VA product | 256 | NA | NA | NA |
| Total | 185,360 | 2,588 | 10,764 | 21,693 |
Abbreviations: CPT, Current Procedural Terminology, version 4; HCPCS, Healthcare Common Procedure Coding System; ICD9CM, International Classification of Disease, Ninth Revision, Clinical Modification; ICD9Proc, International Classification of Disease, Ninth Revision, Procedural Codes; ICD10CM, International Classification of Disease, Tenth Revision, Clinical Modification; ICD10PCS, International Classification of Disease, Tenth Revision, Procedure Coding System; LOINC, Logical Observation Identifiers Names and Codes; NA, not applicable, nonstandard vocabulary cannot be deprecated or updated to standard; NDC, National Drug Code; SNOMED, Systematized Nomenclature of Medicine.
Discussion
We described a multidimensional QA approach for finding ETL errors while accounting for a changing source data stream and an evolving CDM. We tailored our process to fit with an existing, harmonized data-quality framework 5 for EMR data and extended it to meet the needs of an incremental transformation approach. We found that completeness, value conformance, and relational conformance elements can all be greatly impacted with errors in an incremental ETL process, but a QA process that anticipates specific incremental load issues can expose deficiencies and make it easier to diagnose architectural failure points or gaps in the current logic.
Many industries outside of health care have endorsed the concept of a CDM and have described causes of data-quality issues in ETL. 20 21 However, there are unique challenges in health care that require differences in the way the data are handled and loaded. While data are subject to flux at some level in most industries, this is more common in health care data. We found the most resource-intensive incremental update involved revisions to patient, provider, and location source identifiers. Failure to fully account for updates has potential to greatly compromise relational conformance. We illustrated the impact of compromised relationship conformance as it related to patient identifiers, showing fragmented health records linked to the new identifier, but gaps in data linkage would similarly occur for provider and location identifiers. Referential integrity checks between source and target rows ensure that the most up-to-date identifiers persist throughout the model.
Changes to the CDM were common between data releases, where changes to the mappings had an influence on data completeness and changes to concepts themselves affected value conformance. Specifically, relationship changes between ICD9 codes and SNOMED codes necessitated downstream recalculation of millions of records and concept changes forced redistribution of rows across the entire model. There were numerous changes to the standard status of concepts as well as domain changes across multiple vocabularies used in VA between Release A and Release B. Unlike source data model changes that may not consider the schedule of a transform to a CDM, a team can choose when to implement new OMOP meta-data, although it is encouraged to stay current with Observational Health Data Sciences and Informatics (OHDSI) recommendations. When an ETL design solution fails to recognize updated concept relationships, there will be anomalies to data completeness. The magnitude of error will vary from system to system, but ultimately will result in inadequate capture of clinical concepts; either an underestimation when relationships are expanded or an overestimation when relationships are consolidated. Row counts between transformation and target tables are easy to execute and offer an effective approach to ensure that CDM meta-data updates are consistently applied to current and historical data. We chose to only report changes to vocabularies that were used in the VA, but the notion of mapping expansions and consolidations is applicable to any of the OMOP CDM vocabularies.
Just as source data and the CDM are ever evolving, the ETL architecture is also a living process. Our QA procedure currently uses a combination of primary key, CRUD operations, and ETL auto-incrementing processing IDs, but it must also adjust over time to incorporate new source data domains, adapt to new versions of the CDM, or perhaps respond to a shift in available computing resources at the VA.
Our proposed QA approach acts as a safety net to catch current but also future gaps or missteps that arise as the ETL process matures. We described results of our QA process as it related to two specific data releases that were spaced 10 months apart, September 2017 and June 2018. The magnitude of new and updated source records will decrease as the interval of time between releases narrows, but the transform is still equally as vulnerable to consistency and conformance errors if there are gaps or failures to the incremental ETL architecture.
We presented our QA process in a descriptive manner that may not be directly transferable to other health care systems. However, our findings highlight the necessity to closely consider incremental load errors when transforming source data into a CDM, and to our knowledge there have been no publications that explicitly describe the consequences of fallible incremental ETL. The few published papers in which QA efforts have been described focused efforts on mapping completeness and data visualization or fidelity of source data through study replication. 10 11 12 13 14 15 16 Previous research on the utility of EMR data for research purposes has highlighted the importance of assessing data plausibility 5 and data sufficiency. 22 All of these are important data-quality considerations that should be assessed alongside incremental QA efforts. Supplementary Appendix B (available in the online version) lists key considerations for incremental transforming data into the OMOP CDM. Future steps such as extending current open source QA tools to include elements specific to incremental ETL could be advantageous to the wider OMOP community.
Conclusion
Incrementally transforming EHR into a CDM offers many benefits over batch load but is more complex with more opportunities for ETL errors. A multidimensional QA process that anticipates errors throughout the ETL process—from extract, to transform and through load— can successfully identify errors that would otherwise compromise completeness, relational and value conformance data-quality elements. Development of robust QA supporting accurate transformation of OMOP and other CDMs from source data is still in evolution, and much opportunity exists to extend the existing QA framework and tools used for incremental ETL QA processes.
Clinical Relevance Statement
Incrementally transforming EMR data from their native source form to a target CDM is challenging and can introduce errors that could impact its utility for research. Due to the idiosyncrasies of each unique data source, the one-size-fits-all approach to QA is not realistic. Nonetheless, all approaches to incremental QA control should consider, to some extent, both source data quality and the execution of the ETL process.
Multiple Choice Questions
-
The Department of Veterans Affairs transformed its source electronic health care data into which common data model?
PCORnet.
i2b2.
OMOP.
CDW.
Correct Answer: The correct answer is option c, OMOP. While other health care systems have transformed their native data into PCORnet and i2b2, the VA instantiated an instance of OMOP in 2015 and is working to provide regular data releases to its research and operation community. The VA CDW was the source data model that was transformed into the OMOP CDM.
-
Which data-quality elements can be affected by a fallible incremental ETL process?
Plausibility and consistency
Consistency and completeness
Completeness, value conformance, and relational conformance
Value conformance
Data are not affected by incremental ETL
Correct Answer: The correct answer is option c, completeness, value conformance, and relational conformance. Answers (a)–(d) all contain data-quality elements that should, to some extent, be evaluated in all EHR data-quality processes; however, completeness, value, and relational conformance can be directly affected by an imperfect ETL process. The frequency and magnitude of each error type depend on multiple things including where in the ETL pipeline the failure point occurred.
Funding Statement
Funding This work was supported using resources and facilities at the VA Salt Lake City Health Care System and the VA Informatics and Computing Infrastructure (VINCI), VA HSR RES 13–457.
Conflict of Interest None declared.
Authors' Contributions
Conception and design: K.E.L., S.A.D., M.E.M.; acquisition of data: K.E.L., B.V., A.C., D.P., K.H.; analysis: K.E.L., B.V., A.C., D.P., E.H., K.H.; interpretation: K.E.L., S.A.D., S.L.D., M.E.M.; drafting of manuscript: K.E.L., S.A.D., M.E.M.; and critical revision of the manuscript for important intellectual content: all authors.
Protection of Human and Animal Subjects
The study was performed in compliance with the World Medical Association Declaration of Helsinki on Ethical Principles for Medical Research Involving Human Subjects. All research was conducted with the approval by the University of Utah Institutional Review Board and the VA Salt Lake City Health Care System Research and Development Committee.
Supplementary Material
References
- 1.Voss E A, Makadia R, Matcho A et al. Feasibility and utility of applications of the common data model to multiple, disparate observational health databases. J Am Med Inform Assoc. 2015;22(03):553–564. doi: 10.1093/jamia/ocu023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jörg T, DeBloch S.Towards generating ETL processes for incremental loadingIn: ACM International Conference Proceeding Series; September 10–12, 2008; Coimbra, Portugal. pp. 101–110
- 3.Post A R, Ai M, Kalsanka Pai A, Overcash M, Stephens D S. Architecting the data loading process for an i2b2 research data warehouse: full reload versus incremental updating. AMIA Annu Symp Proc. 2018;2017:1411–1420. [PMC free article] [PubMed] [Google Scholar]
- 4.Kahn M G, Brown J S, Chun A T et al. Transparent reporting of data quality in distributed data networks. EGEMS (Wash DC) 2015;3(01):1052. doi: 10.13063/2327-9214.1052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kahn M G, Callahan T J, Barnard J et al. A harmonized data quality assessment terminology and framework for the secondary use of electronic health record data. EGEMS (Wash DC) 2016;4(01):1244. doi: 10.13063/2327-9214.1244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hersh W R, Cimino J, Payne P R et al. Recommendations for the use of operational electronic health record data in comparative effectiveness research. EGEMS (Wash DC) 2013;1(01):1018. doi: 10.13063/2327-9214.1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Qualls L G, Phillips T A, Hammill B G et al. Evaluating foundational data quality in the National Patient-Centered Clinical Research Network (PCORnet®) EGEMS (Wash DC) 2018;6(01):3. doi: 10.5334/egems.199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hripcsak G, Duke J D, Shah N H et al. Observational Health Data Sciences and Informatics (OHDSI): opportunities for observational researchers. Stud Health Technol Inform. 2015;216:574–578. [PMC free article] [PubMed] [Google Scholar]
- 9.OHDSI. Available at:https://www.ohdsi.org. Accessed August 30, 2019
- 10.Makadia R, Ryan P B. Transforming the premier perspective hospital database into the Observational Medical Outcomes Partnership (OMOP) common data model. EGEMS (Wash DC) 2014;2(01):1110. doi: 10.13063/2327-9214.1110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Matcho A, Ryan P, Fife D, Reich C. Fidelity assessment of a clinical practice research datalink conversion to the OMOP common data model. Drug Saf. 2014;37(11):945–959. doi: 10.1007/s40264-014-0214-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schwalm M, Raoul T, Chu D et al. PRM59 - Conversion of a French electronic medical record (EMR) database into the Observational Medical Outcomes Partnership common data model. Value Health. 2017;20(09):A741. [Google Scholar]
- 13.Maier C, Lang L, Storf H et al. Towards implementation of OMOP in a German university hospital consortium. Appl Clin Inform. 2018;9(01):54–61. doi: 10.1055/s-0037-1617452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Klann J G, Joss M AH, Embree K, Murphy S N. Data model harmonization for the All Of Us Research Program: transforming i2b2 data into the OMOP common data model. PLoS One. 2019;14(02):e0212463. doi: 10.1371/journal.pone.0212463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yoon D, Ahn E K, Park M Y et al. Conversion and data quality assessment of electronic health record data at a Korean tertiary teaching hospital to a common data model for distributed network research. Healthc Inform Res. 2016;22(01):54–58. doi: 10.4258/hir.2016.22.1.54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.You S C, Lee S, Cho S Y et al. Conversion of national health insurance service-national sample cohort (NHIS-NSC) database into Observational Medical Outcomes Partnership-common data model (OMOP-CDM) Stud Health Technol Inform. 2017;245:467–470. [PubMed] [Google Scholar]
- 17.FitzHenry F, Resnic F S, Robbins S L et al. Creating a common data model for comparative effectiveness with the Observational Medical Outcomes Partnership. Appl Clin Inform. 2015;6(03):536–547. doi: 10.4338/ACI-2014-12-CR-0121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.U.S. Department of Veterans Affairs. National center for veterans analysis and statistics [11/13/2017]. Available at:https://www.va.gov/vetdata/Utilization.asp. Accessed August 30, 2019
- 19.Fihn S D, Francis J, Clancy C et al. Insights from advanced analytics at the Veterans Health Administration. Health Aff (Millwood) 2014;33(07):1203–1211. doi: 10.1377/hlthaff.2014.0054. [DOI] [PubMed] [Google Scholar]
- 20.Singh R, Singh S. A description of classification of causes of data quality problems in data warehousing. International Journal of Computer Science Issues. 2010;7(03):41–49. [Google Scholar]
- 21.Rupali G, Singh J. A review of contemporary data quality issues in data warehouse ETL environment. Journal on Today's Ideas – Tomorrow's Technologies. 2014;2(02):153–160. [Google Scholar]
- 22.Weiskopf N G, Bakken S, Hripcsak G, Weng C. A data quality assessment guideline for electronic health record data reuse. EGEMS (Wash DC) 2017;5(01):14. doi: 10.5334/egems.218. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


