Abstract
Sensory photoreceptor proteins underpin light-dependent adaptations in nature and enable the optogenetic control of organismal behavior and physiology. We identified the bacterial light-oxygen-voltage (LOV) photoreceptor PAL that sequence-specifically binds short RNA stem loops with around 20 nM affinity in blue light and weaker than 1 μM in darkness. A crystal structure rationalizes the unusual receptor architecture of PAL with C-terminal LOV photosensor and N-terminal effector units. The light-activated PAL:RNA interaction can be harnessed to regulate gene expression at the RNA level as a function of light in both bacteria and mammalian cells. The present results elucidate a new signal-transduction paradigm in LOV receptors and conjoin RNA biology with optogenetic regulation, thereby paving the way towards hitherto inaccessible optoribogenetic modalities.
Introduction
Sensory photoreceptor proteins mediate manifold physiological and behavioral adaptations to light across all domains of life1. Light-oxygen-voltage (LOV) receptors, first described in plant phototropins2,3, harness flavin nucleotides as chromophores to achieve sensitivity to blue light. Light-induced formation of a thioether bond between the flavin and a conserved cysteine residue4 triggers hydrogen-bonding rearrangements throughout the LOV protein that culminate in modulation of receptor activity5. Although LOV receptors markedly vary in domain architecture1,6, with but few exceptions7 they transduce signals from N-terminal photosensor to C-terminal effector modules. An α helix, denoted Jα, immediately C-terminal of the LOV photosensor module usually governs this process, e.g., via light-modulated unfolding in plant phototropins8. Numerous, disparate output activities are subject to light control in both natural and engineered LOV receptors9, and thus greatly enrich optogenetics10, i.e. the spatiotemporally precise, non-invasive and reversible control by light of cellular properties and physiology. Although manifold and ingenious strategies for optogenetic intervention are now in place, there is a notable lack of photoreceptors, LOV or otherwise, directly acting on RNA in light-dependent manner.
Against this backdrop, we here report the discovery and mechanistic characterization of the bacterial LOV receptor PAL. We identified short RNA aptamers of fewer than 20 nucleotides in size that strongly bind to PAL under blue light but much less so in darkness. The structure of PAL in its dark-adapted state and functional assays characterize the signaling mechanism and rationalize the uncommon situation of the LOV photosensor at the C terminus of the receptor. As we exemplify for light-dependent regulation of translation in bacteria and mammalian cells, the specific and strongly light-enhanced PAL:RNA interaction unlocks hitherto inaccessible optoribogenetic modalities.
Results
Discovery of the PAL receptor
Striving to identify LOV receptors with hitherto uncharacterized architectures, we searched the sequence databases and identified a novel protein in the gram-positive actinobacterium Nakamurella multipartita11,12 (Uniprot C8XJT7). Based on its architecture comprising Per-ARNT-Sim (PAS)13, ANTAR14 and LOV domains, we dubbed the protein PAL (Fig. 1a). Since, two PAL homologs have been identified in the closely related organism Nakamurella sp. 12Sc4-1 (Supplementary Fig. 1). As ANTAR domains have been characterized as prokaryotic RNA-binding modules regulating gene expression via a transcriptional anti-termination mechanism14–17, we hypothesized that the PAL receptor mediates light-dependent RNA binding, an activity to date not observed in nature. Unusually, the LOV photosensor is situated at the C terminus of the receptor which contrasts with the prevalent N-terminal arrangement and raises the question of how light signals are transduced in PAL. To address these aspects, we isolated the PAL gene by PCR amplification from N. multipartita genomic DNA and confirmed its sequence. We produced the PAL protein by heterologous expression in Escherichia coli and used absorption spectroscopy to confirm flavin chromophore incorporation and canonical, fully reversible LOV photochemistry with a time constant for dark recovery of (2200 ± 50) s at 22°C (Supplementary Fig. 2). Circular dichroism measurements indicated a mixed α/β fold and a melting temperature of (50.2 ± 0.5) °C. Size-exclusion chromatography coupled with multi-angle light scattering revealed that PAL adopts a homodimeric state in solution which is retained upon blue-light illumination. Likewise, the isolated LOV domain of PAL adopts a homodimeric state independent of blue light (Supplementary Fig. 2).
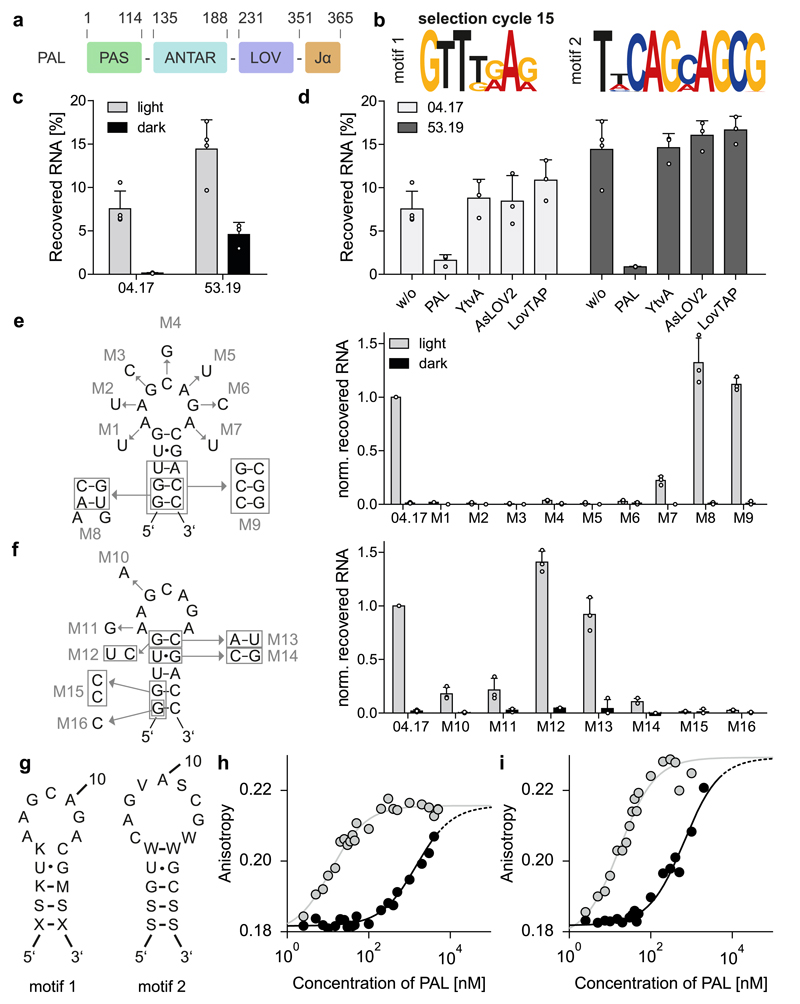
Figure 1. Photoactivated RNA-binding by the light-oxygen-voltage receptor PAL (Uniprot C8XJT7).
a, Domain organization of the light-oxygen-voltage receptor PAL from Nakamurella multipartita with residue numbers on top. b, Next-generation sequencing analysis identifies two PAL-binding motifs dominating in SELEX cycle 15. c-f, 32P-labeled aptamers were analyzed for light-dependent binding to PAL immobilized on streptavidin coated wells. All measurements represent mean ± SD of n ≥ 3 biologically independent replicates. c, Binding of the aptamers 04.17 and 53.19 to PAL under blue light or in darkness. d, Immobilized PAL was incubated with aptamer in the absence or presence of 400 nM of different non-immobilized LOV modules. Only PAL itself could displace the aptamer from the immobilized PAL:RNA complex. e, f, Impact of residue exchanges within the unpaired loop and the base-paired stem of the 04.17 aptamer. g, Consensus sequences and predicted secondary structures for motifs 1 and 2 based on the mutational analyses (degenerate nucleotides are S = G/C, K = G/U, M = A/C, W = A/U, V = A/C/G). h, Titration of the TAMRA-labeled RNA aptamer 04.17 with PAL in the dark (black lines) and following blue-light exposure (grey lines) monitored by fluorescence anisotropy. Lines denote fits to single-site binding isotherms. i, As panel h but for aptamer 53.19. Experiments in panels h and i were repeated three times with similar results.
Aptamer selection and characterization
In initial experiments, PAL bound weakly and unspecifically single-stranded (ss) and double-stranded DNA and RNA, with a slight preference for ssRNA, albeit independently of light (Supplementary Fig. 3). We reasoned that the overall modest affinity for nucleic acids and the lack of light regulation are due to the unspecific nature of the substrates and hence tested binding of PAL to RNA target sequences reported for other, light-inert ANTAR proteins (Supplementary Fig. 4). As none of these sequences exhibited light-regulated affinity to PAL either, we resorted to SELEX18,19 to identify sequences that do. An RNA library comprising a stretch of 40 random nucleotides was incubated with immobilized PAL under blue light (465 nm) (Supplementary Fig. 5). Following removal of non-binding RNA by washing and incubation in darkness, we recovered bound RNA to enrich the library for aptamer variants preferentially binding to PAL under light. We iterated the selection over 15 cycles under increasingly stringent conditions (Supplementary Table 1) and analyzed RNA libraries from individual steps for binding to PAL via RiboGreen-based fluorescence. Relative to the initial cycle 1, the libraries from cycles 9 and 15 displayed enhanced binding to the light-adapted state of PAL (from hereon denoted PALL) (Supplementary Fig. 6a). Analyses by Sanger and next-generation sequencing (Supplementary Fig. 6b) revealed a successive enrichment of specific sequences that bear one of two RNA motifs, named 1 and 2, of 7 and 10 nucleotides length (Fig. 1b and Supplementary Figs. 6c, d and 7a, b). These motifs located to the 3’ and 5’ ends of the RNA library, respectively, as evidenced by an altered nucleotide distribution near these ends in the libraries from cycles 9 and 15 relative to the starting library (Supplementary Fig. 6e-g). Moreover, the enrichment manifested itself as a reduction of the fraction of unique RNA sequences from 100% in cycle 3 to 10% in cycle 15 (Supplementary Fig. 6h-i). In cycles 1-8, the majority of sequencing reads yielded sequences with copy numbers of ten or less, but by cycle 15 more than 80% of all reads corresponded to sequences with copy numbers larger than 10 (Supplementary Fig. 6j). Whereas motif 1 dominated in cycles 3-12, motif 2 only accumulated in later cycles under increased selection pressure (Supplementary Fig. 6k), which was also observed for individual sequences bearing these motifs (Supplementary Fig. 6l-n, Supplementary Dataset 1). We chose the most abundant sequences from cycles 9 and 15 that bear motifs 1 (aptamers 04, 46, 56 and 57) or 2 (aptamers 51, 53, 54, and 55) for further characterization. Secondary structure prediction suggested that both motifs are part of stem loops, which all feature the conserved sequence AGCAG in the loop regions (Supplementary Fig. 8a-c). Interaction measurements confirmed light-enhanced binding by PAL for all tested aptamers (Supplementary Fig. 9a, b). All aptamers showed similar degrees of binding to PALL, but those comprising motif 2 had more pronounced residual binding to the dark-adapted state (PALD). Disruption of motif 1 in the aptamer 46 abolished binding, thus underlining the importance of this motif.
Next, we iteratively truncated the aptamers 04 and 53 bearing motifs 1 and 2, respectively, to single RNA hairpins of 17 and 19 nucleotides length (Supplementary Fig. 9c, d). The resultant aptamers, denoted 04.17 and 53.19, fully retained light-dependent interaction with PAL, with 04.17 showing less background binding to PALD than 53.19 (Fig. 1c and Supplementary Fig. 7b). We assessed the specificity of both aptamers in competition assays by incubating immobilized PAL:aptamer complex with several non-immobilized LOV receptors (Fig. 1d). Irrespective of illumination, none of the LOV receptors other than PAL itself could displace 04.17 and 53.19 from the immobilized PAL:RNA complex. Thus, both aptamers possess specificity towards PAL over other photoreceptors. To pinpoint sequence determinants in 04.17 and 53.19 governing PAL binding, we investigated the impact of nucleotide substitutions (Fig. 1e, f and Supplementary Fig. 7c, d). Replacement of any of the seven nucleotides within the loop of 04.17 for the sequence-complementary nucleotides completely abolished PAL binding for positions 1-6 (variants M1-M6) and severely impaired it for the last position (M7) (Fig. 1e). More conservative exchanges of one purine for another purine at positions 1 and 3 retained light-dependent binding, albeit at reduced efficiency (Fig. 1f, M10 and M11). Variations in the stem region (M8, M9, M13) were mostly tolerated provided Watson-Crick base pairing was maintained, whereas disruption of base pairing abolished PAL binding (M15, M16) (Fig. 1f). Replacing a G:U wobble base pair within the stem for G:C in variant M9 entailed strongly reduced binding. Likewise, we probed the 53.19 aptamer and found that single nucleotide replacements in the loop impaired binding albeit less severely than for 04.17 (Supplementary Fig. 7c, d, variants M17-M27). Variations in the stem that maintained Watson-Crick pairing were tolerated but less well than in 04.17 (Supplementary Fig. 7d, M28, M29, M31, M34). Vice versa, substitutions disrupting base pairing (M32, M33) were better accommodated than in 04.17. Alteration of the G:U wobble pair in 53.19 (M30) strongly impaired binding, as it did for 04.17. Taken together, the mutagenesis data allow the determination of consensus motifs for the two aptamers, as shown in Fig. 1g. To obtain quantitative interaction data, we immobilized the aptamers 04.21 and 53.19 and studied their binding to PALD or PALL by surface plasmon resonance (Supplementary Fig. 10a, Table 1). At 25°C, the apparent dissociation constants (KD) for the interaction with PALL amounted to (102 ± 9) nM and (120 ± 9) nM for 04.21 and 53.19, respectively, whereas no binding to PALD could be detected under these conditions (Supplementary Fig. 10b, Table 1). Despite similar KD values, the association and dissociation kinetics for 04.21 were ~ 2.8-fold and 2.4-fold faster than for 53.19. At a temperature of 37°C, suitable for cell-based assays (vide infra), we obtained KD values of (253 ± 12) nM and (153 ± 14) nM (Supplementary Fig. 10c, Table 1), showing that the interaction with 04.21 is more strongly affected by temperature. Omission of Mg2+ ions resulted in weaker binding with KD values at 25°C of (169 ± 40) nM for 04.21 and (651 ± 32) nM for 53.19, indicating that the interaction with 53.19 depends more strongly on magnesium (Supplementary Fig. 10d, Table 1). This loss of affinity of 53.19 relates to a 4.2-fold increase of its dissociation rate, whereas its association rate is similar that at 25°C. We also resorted to fluorescence anisotropy as a complementary technique to monitor in solution the interaction of PAL with the RNA aptamers 04.17 and 53.19 labelled at their 5’ termini with tetramethyl-rhodamine (TAMRA). Under blue light, PALL bound the 04.17 and 53.19 aptamers with dissociation constants KD of (12 ± 1) and (17 ± 2) nM, respectively, but in darkness binding was around 100-fold weaker with affinities larger than 1 μM (Fig. 1h, i). We recorded association kinetics by following fluorescence anisotropy over time after illuminating dark-adapted samples with a brief blue-light pulse (Supplementary Fig. 11a, b). For the 04.17 and 53.19 aptamers, bimolecular association rate constants of (2.0 ± 0.1)·104 M-1 s-1 and (2.2 ± 0.2)·104 M-1 s-1, respectively, resulted from which we calculated dissociation rate constants of (2.4 ± 0.2)·10-4 s-1 and (3.8 ± 0.6)·10-4 s-1. Neither the TAMRA-labelled DNA variants of the aptamers 04.17 and 53.19 showed any binding up to protein concentrations of 10 μM, thus confirming the specificity of PAL for RNA and ruling out unspecific interactions with the TAMRA fluorophore (Supplementary Fig. 11c, d). We next recorded binding isotherms in the regime of strong binding, i.e. at aptamer concentrations well above the KD, and observed a stoichiometry of one aptamer per one PAL dimer (Supplementary Fig. 11e). In competition experiments (Supplementary Fig. 11f, g), the 04.17 and 53.19 aptamers mutually displaced each other from PAL, indicating that they occupy the same binding site. Notably, RNA target sequences for the few other known ANTAR systems14–17 were reported to comprise two degenerate direct repeats of a short stem-loop structure. In at least one of these systems, an isolated single stem loop no longer showed detectable affinity for the ANTAR protein16. That notwithstanding, the presently identified aptamers 04.17 and 53.19 share with a previously reported ANTAR consensus motif16 similar lengths of the base-paired stem and the unpaired loop, and a prevalence for purine nucleotides within the loop.
Table 1. KD determination by surface plasmon resonance.
| condition | k1 [104 M-1 s-1] | k-1 [10-3 s-1] | KD [nM] | |
|---|---|---|---|---|
| 04.21 | 25°C, light | 5.6 ± 1.6 | 5.8 ± 1.8 | 102 ± 9 |
| 25°C, dark | n.d. | n.d. | n.d. | |
| 37°C, light | 1.1 ± 0.7 | 2.7 ± 1.7 | 253 ± 12 | |
| 25°C, light, w/o MgCl2 | 5.2 ± 0.9 | 8.5 ± 0.6 | 169 ± 4 | |
| 53.19 | 25°C, light | 2.0 ± 0.2 | 2.4 ± 0.2 | 120 ± 9 |
| 25°C, dark | n.d. | n.d. | n.d. | |
| 37°C, light | 3.7 ± 0.9 | 5.5 ± 0.9 | 153 ± 14 | |
| 25°C, light, w/o MgCl2 | 1.6 ± 0.2 | 10.0 ± 1.3 | 651 ± 32 | |
Structure-function analysis of PAL
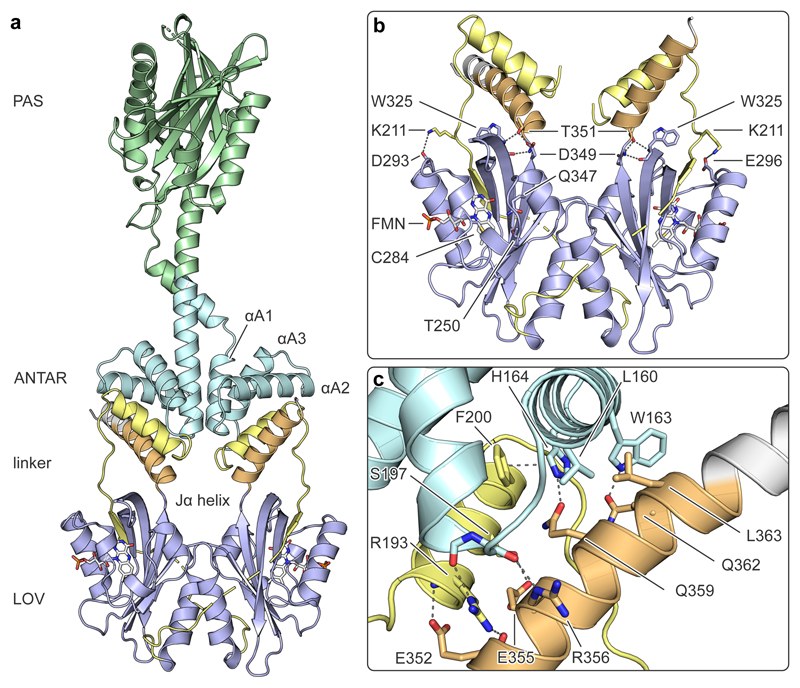
To resolve how the PAL receptor transduces light signals from its C-terminal LOV photosensor to its more N-terminal ANTAR effector, we determined the crystal structure of the receptor in its dark-adapted state PALD at 2.5 Å resolution (Supplementary Table 2). Consistent with its solution behavior, PAL crystallized as a homodimer in parallel orientation (Fig. 2a, Supplementary Fig. 12a-c). Within the dimer, two PAS domains connect through a parallel two-helix bundle to the dimeric, all-helical ANTAR module (helices αA1-αA3). Notably, the PAL structure is bent within this region, which we ascribe to crystal packing (Supplementary Fig. 12a), but which might also be of functional relevance. A long adapter sequence emanates from the ANTAR C terminus and connects to the LOV photosensor dimer. The adapter forms a helix that associates with the helices αA1-αA3 and is succeeded by a long, proline-rich linker. Unexpectedly, this linker wraps around the LOV domains and extends the antiparallel β sheet of the LOV domain by a sixth strand (Fig. 2b). The LOV photosensors can thus point with their C-terminal Jα helices directly towards the ANTAR effector and form contact with the adapter helix. Access to the ANTAR domain is thereby blocked by the LOV module which provides the structural rationale for the observed autoinhibition of RNA binding in darkness (Fig. 2c). Moreover, this novel structural arrangement directly implicates the Jα helices in signal transduction, consistent with their role in diverse receptors with the much more common architecture featuring N-terminal LOV photosensors. Intriguingly, aureochromes, which mediate photomorphological responses in certain algae and diatoms7, share with PAL not only the C-terminal situation of the LOV module but also a long, proline-rich linker immediately preceding this domain. We hence propose that a structural arrangement akin to that of PAL also exists in aureochromes which to date have eluded structural elucidation at full length20,21.
Figure 2. Structure of PAL in its dark-adapted state.
a, The homodimeric receptor comprises consecutive Per-ARNT-Sim (PAS, green), ANTAR (cyan) and light-oxygen-voltage (LOV, blue) domains. The ANTAR and LOV moieties are joined by a proline-rich adapter segment (yellow), and the LOV domain features a C-terminal helix denoted Jα (orange). b, The LOV monomers dimerize via their N-terminal A’α helices and bind flavin-mononucleotide chromophores. The five-stranded antiparallel β sheets of the PAL LOV domains are extended by sixth strands originating from the adapter segments. Several hydrogen and salt bridges mediate contacts between the LOV core domains and their Jα helices. c, The Jα helices contribute to an interface with the ANTAR domains and an additional helix deriving from the adapter segment.
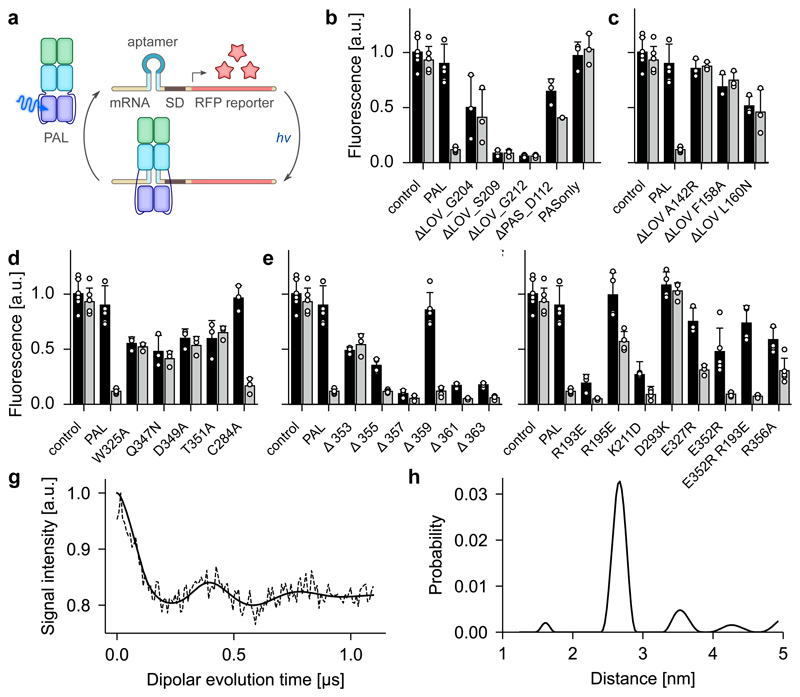
To efficiently probe PAL function, we harnessed the light-dependent RNA binding of PAL for the regulation of gene expression in E. coli. We reasoned that PAL binding to an mRNA might interfere with the cellular transcription/translation machinery and embedded the 04.17 aptamer directly upstream of the Shine-Dalgarno sequence of a red-fluorescent reporter (Fig. 3a). When combined with PAL, reporter fluorescence was unaffected in darkness but around tenfold repressed under blue light. Beyond providing an efficient test bed for PAL activity and regulation, the setup establishes a new optogenetic modality for the regulation of bacterial gene expression9. Using this assay, we varied specific residues and structural regions of PAL to interrogate their role for function and regulation by light (Fig. 3, Supplementary Fig. 13a-c). Western blotting verified the expression of the resulting receptor variants in the assay context (Supplementary Fig. 14). The isolated PAS domain was incapable of down-regulating reporter fluorescence (Fig. 3b). By contrast, a PAL variant that omitted the N-terminal PAS domain retained light-dependent regulation albeit at reduced efficiency. Deletion of the C-terminal LOV module entirely abolished light responsiveness and resulted in low reporter fluorescence, indicative of constitutive RNA binding. Replacements of several conserved amino acids (A142R, F158A, L160N; cf. Supplementary Fig. 1) within the ANTAR domain of this C-terminally truncated PAL variant abolished or severely impaired RNA binding (Fig. 3c). Interestingly, down-regulation of reporter fluorescence was also lost entirely when the adapter helix was additionally removed, suggesting that this segment contributes to the RNA interaction. These results indicate that RNA binding localizes to the ANTAR module of PAL and that the autoinhibitory effect on PAL function observed in darkness is exerted by the LOV module. We next probed individual residue positions throughout the PAL receptor (Fig. 3d-f, Supplementary Fig. 13). Replacement of the conserved Q347 in hydrogen-bonding contact to the FMN chromophore by asparagine abrogated light regulation of PAL and led to constitutive intermediate reporter fluorescence as did the alanine substitutions of W325, D349 and T351, all at the junction of the LOV core to the C-terminal Jα helix, a region generally implicated in signal transduction of LOV proteins22,23 (Fig. 3d). Serial truncation of this helix resulted in impaired light regulation for most variants, with constitutively low reporter fluorescence, indicative of RNA binding, for short deletions, and intermediate reporter fluorescence for more extended deletions (Fig. 3e). Combined with the structural data, we hence ascribe to the Jα helix an important role in relaying light signals from the LOV to the ANTAR module of PAL. To corroborate this view, we substituted numerous residues at the interface between Jα, the adapter helix and helix αA2 of the ANTAR domain (cf. Fig. 2c and Supplementary Fig. 13d). Overall, substitutions near the Jα C terminus, e.g., Q359A, Q362A and L363A, had little effect on reporter fluorescence. By contrast, perturbation of a set of hydrogen bonds formed between the Jα N terminus, the adapter helix and the ANTAR helix αA2 disrupted light regulation, e.g., in the variants R193E, R195E, E352R and R356A (Fig. 3f). Consistent with these findings, the N-terminal segment of the Jα helix is somewhat more conserved across PAL and its N. sp. 12Sc4-1 homologs than the C-terminal part (cf. Supplementary Fig. 1). We next addressed by mutagenesis contact sites between the adapter segment and the LOV photosensor. Several residue exchanges in the region of the LOV strand Gβ had minute effects, but disruption of a salt bridge between D293 in the LOV domain and K211 within the adapter greatly impaired light regulation, with K211D displaying constitutively low and D293K constitutively high fluorescence. Interestingly, neither K211 nor D293 are conserved between PAL and the N. sp. 12Sc4-1 homologs (cf. Supplementary Fig. 1). Taken together, the mutagenesis data crucially implicate the interface between the LOV domain, its Jα helix, the adapter and the ANTAR domain in signal transduction. In the dark-adapted state, an intricate network of interactions keeps PAL in an autoinhibited conformation; free energy perturbations, introduced by mutagenesis or by light during signal transduction, relieve this inhibition.
Figure 3. Functional analysis of PAL.
a, The 04.17 aptamer was embedded near the Shine-Dalgarno (SD) sequence of a red-fluorescent reporter gene. Blue light induces binding of PAL to the aptamer, thereby interfering with expression in E. coli and leading to lower reporter fluorescence. b-f, Using this assay, different regions of PAL were functionally probed in darkness (black bars) and under blue light (grey bars). Data represent mean ± SD of n = 3 biologically independent replicates. b, Truncation variants of PAL in comparison to wild-type PAL positive and empty-vector negative controls. c, Residue exchanges within the ANTAR domain of truncated PAL. d, Residue exchanges within the LOV photosensor module. e, Serial truncations of the C-terminal Jα helix. f, Residue exchanges at the interface between LOV and ANTAR domains. g, Pulsed electron-electron double resonance measurements on the light-induced flavin neutral semiquinone radical state of the PAL variant C284A. h, Evaluation of data from panel g by Tikhonov regularization identifies an inter-flavin distance of (2.7 ± 0.1) nm.
To structurally characterize signal transduction in PAL, we exploited that LOV receptors can retain light sensitivity in the absence of their strictly conserved cysteine moiety. Blue light induces formation of the neutral semiquinone (NSQ) state of the flavin cofactor in cysteine-devoid LOV receptors and thus triggers downstream signaling responses akin to those in the wild-type thioadduct state5. This effect is also present in PAL, as evidenced by retention of wild-type-like activity in the reporter assay upon replacement of the corresponding cysteine 284 by alanine (Fig. 3d). Importantly, the NSQ state is a stable radical and hence amenable to analysis by electron paramagnetic resonance spectroscopy (Fig. 3g). Pulsed electron-electron double resonance (pELDOR) measurements showed a deeply modulated signal indicative of a well-defined distance of (2.7 ± 0.1) nm between the two flavins in PALL which contrasts with a value of 3.2 nm in PALD, as observed in the crystal structure (Fig. 3h). Combined with the results of the reporter gene assay, these data suggest that light prompts an approach of the LOV photosensors which couples to the ANTAR domain via the Jα helices to relieve autoinhibition of RNA binding. Intriguingly, the structure of the PAL LOV dimer closely resembles that in the engineered LOV histidine kinase YF122,23 (Supplementary Fig. 12d), despite this receptor featuring the more prevalent N-terminal situation of the LOV photosensor. However, in YF1, blue light induced a pivoting apart of the LOV dimer and its attached Jα helices, rather than an approach24,25. Remarkably, the flavin-flavin distance in PALL closely resembles that found for a YF1 variant in which the A’α helices are disrupted by mutagenesis24.
PAL-mediated translation control in mammalian cells
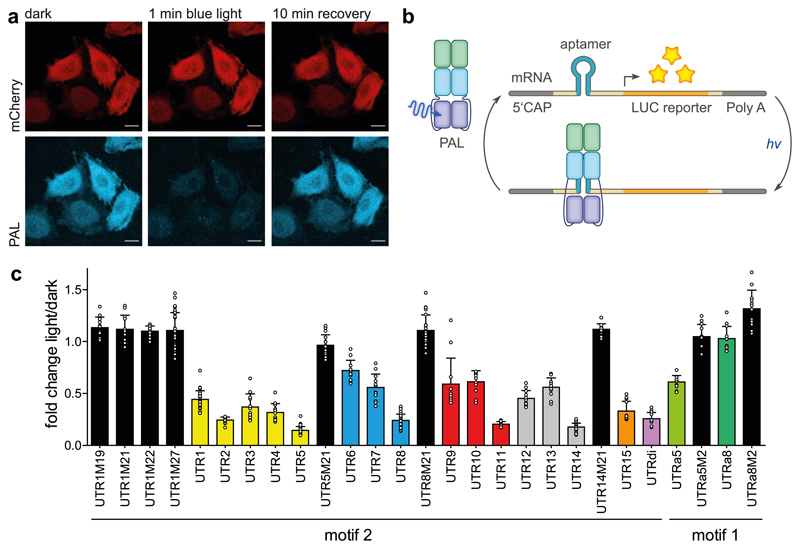
We next investigated whether the light-activated PAL:RNA interaction can be leveraged for the regulation of processes inside mammalian cells26. Functional expression of mCherry-PAL in HeLa cells was assessed by fluorescence of the flavin chromophore (Fig. 4a, Supplementary Fig. 15a-c). The green fluorescence exhibited by PAL was lost upon blue-light application but fully recovered after incubation in darkness, thus indicating intact, reversible photochemistry of PAL inside eukaryotic cells. To establish control by light of mRNA translation, we embedded variants of the aptamer 53 into the 5’-untranslated region (UTR) of a luciferase reporter at different positions relative to the 5’ terminus and the Kozak sequence (Fig. 4b, Supplementary Fig. 16 and Supplementary Table 3). These insertions generally decreased reporter luminescence in the dark although to different extents (Supplementary Fig. 17a-b), depending on their predicted stability27. Reporter luminescence in the light was further reduced to around 40-70% of the levels in darkness for UTR variants 1, 6, 9 and 12, which place the aptamer near the Kozak sequence (Fig. 4c and Supplementary Fig. 16). The light-induced reduction of luminescence was enhanced up to 15-25% of dark levels in the UTR variants 5, 8, 11, 14 and 15 where the aptamer resides closer to the 5’ terminus. The efficiency of repression was modulated by altering the predicted stability of the aptamer stem loop (Supplementary Table 3), with UTR5 exhibiting the best regulation and UTR8 supporting overall higher luciferase expression. Strikingly, the introduction of the single nucleotide exchanges M19-M22 and M27 (cf. Supplementary Fig. 7c, d) abolished any light response. Replacement of the 53.19 aptamer in the UTR5 and UTR8 background for variants of the 04.17 aptamer resulted in an attenuated light-induced reduction of luminescence of 45% for UTRa5 and in no light responsiveness for UTRa8 (Fig. 4c and Supplementary Fig. 17c). Introduction of the mutation M2 (cf. Fig. 1e) abolished light responsiveness in all variants. Evidently, the pronounced RNA sequence specificity of PAL is maintained in eukaryotic cells. As further controls, we introduced four residue substitutions in PAL (K211D, D293E, R193E and W325A), identified in the functional analysis (cf. Fig. 3), and found greatly impaired or no light responsiveness (Supplementary Fig. 17d-e).
Figure 4. Light-dependent regulation of translation in mammalian cells.
a, mCherry-PAL expression in HeLa cells. Green fluorescence of the FMN cofactor of PAL was reduced to background levels upon blue-light exposure but fully recovered during incubation in darkness, thus indicating intact LOV photochemistry. mCherry fluorescence was unaffected by light application. The scale bars denote lengths of 20 μm. b, Light-induced binding of PAL to the 53.19 aptamer embedded in the 5’-untranslated region (5’-UTR) of an mRNA attenuates expression of a Metridia luciferase reporter in HeLa cells. c, Light-dependent translation control for the aptamer variants from panel b. All measurements represent the ratio of reporter luminescence under blue light over darkness (n = 3 biologically independent replicates, mean ± SD). UTR variants M19-M21 harbor single-nucleotide exchanges in the aptamer loop that abolish light responsiveness.
Discussion
In conclusion, we identify the LOV receptor PAL which sequence-specifically binds RNA hairpins upon blue-light absorption, an activity to date neither described in nature nor available by protein engineering. Structural and functional analyses delineate a new paradigm for signal transduction in receptors with C-terminal LOV photosensors. Suspended by a long adapter element, the photosensor module loops back and can thereby regulate the activity of a more N-terminal effector module via interactions with its C-terminal Jα helix. Our findings expand the multi-facetted roles this helix plays and thereby underscore its general importance in diverse LOV receptors, even for specimens with an unusual C-terminal situation of their photosensor units.Aureochrome receptors from algae and diatoms also exhibit C-terminal LOV modules7,20,21 and might hence resemble PAL in their signaling mechanism. Indeed, an autoinhibited dark-adapted state was proposed for the aureochrome receptor from Phaeodactylum tricornutum20. Light absorption is thought to relieve this autoinhibition and enable activation of the receptor, which corresponds to the general sequence of events our data illustrate for PAL. More generally, our findings will inform the rational design of novel LOV receptors for use in optogenetics9.
In a combinatorial approach, we identify short RNA hairpins that tightly and specifically bind PALL. Putatively, these RNA molecules fold into hairpin structures, related to those bound by other ANTAR proteins and to the RNA domains of the iron-responsive element14–17,26. Notably, the identified RNA structures bind to PAL as single hairpins. This aspect sets them apart from the natural RNA molecules that specifically bind to other ANTAR proteins and that comprise two consecutive degenerate hairpins. The identified PAL aptamer sequences stand to benefit the bioinformatic search for natural RNA molecules in Nakamurella multipartita that interact with PAL. Moreover, the small size of these aptamers renders them suitable for the light-dependent regulation of diverse RNA-mediated physiological processes, as we demonstrate at the mRNA level in both prokaryotic and eukaryotic cells. This approach, which we dub optoribogenetics, offers the key advantage of full genetic encodability over a diverse set of optochemical strategies for light control28,29, which generally require the exogenous addition of light-sensitive compounds. Thus, optoribogenetics principally extends to many other RNA species, e.g., micro and long non-coding RNAs, or the guide RNAs in CRISPR/Cas9-related applications, and augurs light-controlled, spatiotemporal access for the versatile investigation of diverse biological processes. As we demonstrated the principal compatibility of PAL with eukaryotic cells, the light-responsive PAL:aptamer pair stands to widely apply to the analysis of RNA-mediated pathways in diverse model organisms.
Online Methods
Molecular biology
The type strain of Nakamurella multipartita was obtained from DSMZ (Deutsche Sammlung von Mikroorganismen und Zellkulturen, no. 44233). The gene encoding PAL was amplified by PCR from genomic N. multipartita DNA and cloned into the pET-28c vector (Novagen) with a C-terminal His6 tag via Gibson assembly30 to yield the expression construct pET-28c-PAL. For the bacterial fluorescence reporter assay (cf. below), the PAL gene was subcloned into a pCDF backbone with a C-terminal myc epitope under control of the arabinose-inducible pBAD promoter to yield the plasmid pCDF-PAL. A PAL gene with Escherichia coli-adapted codon usage, synthesized by GeneArt, served to construct the plasmids pET-28c-PALopt and pCDF-PALopt, respectively. Residue exchanges and truncations in PAL were performed by site-directed mutagenesis and PCR amplification. Oligonucleotide primers were purchased from Integrated DNA Technologies (IDT). The identity of all constructs was confirmed by Sanger DNA sequencing (Eurofins).
Purification of PAL
For protein expression, either the plasmid pET-28c-PAL or pET-28c-PALopt was transformed into CmpX13 E. coli cells31. Bacteria were grown in lysogeny broth (LB) supplemented with 50 μg mL-1 kanamycin and 50 μM riboflavin at 37°C and 225 rpm until an optical density at 600 nm of 0.6 was reached, at which point expression was induced by addition of 1 mM isopropyl β-D-1-thiogalactopyranoside (IPTG). Incubation continued at 16°C and 225 rpm for 16 hours, after which cells were harvested and lysed by ultrasound or fluidizer. The lysate was cleared by centrifugation and applied to an immobilized nickel ion affinity column (Macherey Nagel). Protein was eluted with an imidazole gradient, and elution fractions were analyzed by denaturing polyacrylamide gel electrophoresis (PAGE). Fractions containing pure protein were pooled, dialyzed into buffer A (12 mM HEPES/HCl pH 7.7, 135 mM KCl, 10 mM NaCl, 1 mM MgCl2, 10 % [w/v] glycerol) and concentrated by spin filtration. Protein concentration was determined by absorption spectroscopy using an extinction coefficient of 12,500 M-1 cm-1 at 450 nm. The isolated LOV domain of PAL was purified likewise. For structure determination by X-ray crystallography, selenomethionine (SeMet)-substituted PAL protein was prepared according to published protocols22,32. Purification followed the same steps as for non-substituted protein.
Biochemical characterization of PAL
Spectroscopic analysis of PAL was performed on an Agilent 8435 diode-array spectrophotometer. Absorption spectra were recorded prior to and after saturating illumination with 470-nm light. Following blue-light illumination, the recovery of PAL to its dark-adapted state was monitored over time by absorption at 450 nm.
The secondary structure of PAL was assessed by circular dichroism (CD) spectroscopy on a JASCO J710 spectrophotometer. Before the measurement, PAL was dialyzed into 100 mM sodium phosphate pH 7.5, 300 mM NaCl. Steady-state spectra were recorded for dark-adapted PAL and for protein exposed to saturating 470-nm illumination. The stability of dark-adapted PAL was assessed by thermal denaturation monitored by the CD signal at 210 nm. Experimental data were evaluated by nonlinear least-squares fitting to a two-state unfolding model.
The oligomeric states of PAL and its isolated LOV domain were assessed by size-exclusion chromatography (SEC) on a Superdex 75 column (GE Healthcare) at 4°C in darkness or following 470-nm illumination. Elution profiles were monitored by protein absorption at 280 nm. Both the full-length PAL protein and the isolated LOV module eluted as homogenous peaks at retention times consistent with dimeric species. Blue-light illumination did not alter much the retention times for either protein. To confirm the homodimeric state of the two proteins, they were analyzed by SEC coupled to multi-angle light scattering (MALS, Dawn Heleos, Wyatt) combined with a refractive-index detector (Waters). SEC-MALS analysis for full-length PAL and the isolated LOV domain was performed at 22°C on Superose 6 (GE Healthcare) and Superdex 75 columns, respectively. Molecular mass was calculated with the ASTRA software (Wyatt). All SEC measurements were performed in buffer A, cf. above.
The binding of PAL to nucleic acids was initially characterized by electrophoretic mobility shift assays (cf. Supplementary Fig. 4). To prepare a single-stranded DNA substrate, an oligonucleotide with the arbitrary sequence 5’-GUGAUCCAACCGACGCGACAAGCUAAUGCAAGA-3’ was radio-labelled with 32P at its 5’ end. A double-stranded stranded DNA substrate was obtained by annealing with a non-labeled, reverse complementary oligonucleotide. Single-stranded and double-stranded RNA substrate were prepared likewise, where all thymidine nucleotides were replaced by uridines. 50 pM of each radiolabeled substrate was incubated with increasing amounts of purified PAL protein for 20 minutes in darkness or under blue light (447 nm, 100 mW cm-2). Afterwards, the reaction mix was separated on a 6% (w/v) polyacrylamide gel in Tris borate buffer. The gels were evaluated using a phosphorimager (BioRad Molecular Imager FX).
Selection of aptamers
Library preparation
The library template was purchased from Ella Biotech GmbH (Munich, Germany) as single-stranded DNA, containing a random region of 40 nucleotides. Forward primer (5’- GGGGGAATTCTAATACGACTCACTATAGGGAGGACGATGCGG-3’) and reverse primer (5’-TCTCGGATCCTCAGCGAGTCGTCTG-3’) were used for PCR amplification of the library, and the resulting double-stranded DNA was used as a template for in vitro transcription (5’- GGGAGGACGAUGCGG N40 CAGACGACUCGCUGAGGAUCCGAGA-3’). Transcribed RNA was purified by PAGE and used as starting library for the selection targeting the light-adapted conformation of PAL.
Protein immobilization
For the aptamer selection, PAL protein was biotinylated with a 4-fold excess of EZ-Link Sulfo-NHS-LC-Biotin as per the manufacturer’s instructions (Thermo Fisher Scientific, Darmstadt, Germany) and coupled to streptavidin-coated wells of a plate (Pierce Streptavidin Coated High Capacity Plates, Clear, 8-Well Strip). Wells were washed with 3x 200 μL buffer A. 100 μL of 1.5 μM PAL in buffer A was added, and the coupling was performed in darkness over night at 4°C. Afterwards, wells were washed 3x with 200 μL buffer A.
Aptamer selection
1 nmol purified starting library was incubated with immobilized PAL at 25°C for 30 min under blue light (465 nm, 2.15 mW cm-2) in 100 μL buffer A supplemented with 0.08 mg mL-1 salmon sperm DNA (Thermo Fisher Scientific, Darmstadt, Germany) as competitor. The wells were washed twice under blue light. Fresh buffer was added, and the wells were incubated in the dark for 30 min. The supernatant was collected, and eluted RNA was reverse-transcribed and amplified by PCR. Resulting dsDNA was used as template for in vitro transcription for 20 min at 37°C. Beginning from the second selection cycle, a pre-selection step was performed by incubation of the enriched library with empty streptavidin coated wells. To gradually increase the selection stringency over 15 selection cycles (cf. Table 1), the incubation time was reduced to 1 min, the washing cycles were increased up to one hour, the dark elution time was reduced to 15 min, the amount of competitor was increased to 1 mg mL-1 salmon sperm DNA, 4 mg mL-1 heparin was added, and the amount of biotinylated PAL, immobilized on streptavidin-coated wells, was reduced to 0.03 pmol.
Next-generation sequencing
The starting library and enriched libraries of cycle 3, 5, 7, 8, 9, 10, 11, 12, 13, 14 and 15 were analyzed by next-generation sequencing (NGS) using the Illumina HiSeq1500 platform. Samples were prepared according to33. Briefly, during the first PCR step, twelve different index primers were attached to the different libraries, allowing the analysis of twelve samples in parallel on one lane. PCR products were purified using the NucleoSpin Clean-Up kit (Macherey & Nagel, Düren, Germany), and equal amounts of PCR product of each library were mixed to a final amount of 2 μg DNA. The subsequent adapter ligation step according to manufacturer’s instruction (TruSeq DNA PCR-Free Sample Preparation Kit LT, Illumina) allows the hybridization to the flow cell, and samples were purified by agarose gel purification. Library validation was performed by real-time PCR and quantification using the KAPA library quantification kit (Sigma-Aldrich).
Sequence analysis
Sequencing data were demultiplexed with CASAVA v1.8.2 and analyzed with the COMPAS software. NGS data were analyzed using the in-house AptaNext software and a commercial pattern-analysis software by AptaIT (Planegg, Germany). The 1,000 most abundant patterns of cycle 9 and 15, which include identical sequences with up to five mutations, and sequences obtained from Sanger sequencing of cycle 9 were analyzed by the MEME suite34, resulting in two groups of sequences, which contained motif 1 or motif 2. The most abundant sequences belonging to motifs 1 and 2 were further analyzed by secondary structure prediction with Mfold27. The two most abundant sequences harboring motif 1 were selected from cycle 13, and the four most abundant sequences harboring motif 2 were selected from cycle 15. These sequences and two additional ones identified in cycle 9 by Sanger sequencing were used for further binding studies.
RNA:PAL interaction assays
Radiolabeling and Ribogreen assays
To study the binding of RNA aptamers to light- and dark-adapted PAL via radiography or fluorescence detection, biotinylated PAL was immobilized to streptavidin-coated wells in darkness over night at 4°C, cf. above. Unmodified full-length aptamer or 32P-labeled truncated aptamer were incubated with immobilized PAL in 100 μL buffer A for 30 min at 25°C under light (465 nm, 2,15 mW cm-2) or in darkness, followed by 3 washing steps with 200 μL buffer A for 3 min each. For fluorescence detection, 150 μL RiboGreen (Quant-iT RiboGreen RNA Reagent, Thermo Fisher Scientific, Darmstadt, Germany), diluted 500-fold in 1x TE buffer (10 mM Tris/HCl pH 7.5, 1 mM ethylenediaminetetraacetic acid), was added to each well. After 1 h incubation in the dark, fluorescence intensity was measured on a Tecan Ultra plate reader (Tecan, Crailsheim, Germany) at excitation and emission wavelengths of 500 and 525 nm, respectively. For detection by radiography, wells were washed, and bound RNA was eluted in 100 μL water for 5 min at 95°C. Each fraction was collected, diluted to 1 mL and analyzed by a liquid scintillation counter (Wallac 1414 WIN spectral, PerkinElmer).
To test binding of PAL to the ANTAR target sequences recognized by the Pseudomonas aeruginosa AmiR55, Enterococcus faecalis EutV16, and Klebsiella oxytoca NasR56 proteins, the following sequences were used:
AmiR:
5’- GGGGGAATTCTAATACGACTCACTATAGGGATCAGGTCATGCGCATCAGCGTCGATGTCGCGGGACCGAACCTAACGCATACGCACAGAGCAAATGGGCTCTCCCGGGGTTACCCGGGAGGGCCTTTTTT 3’
EutV:
5’- GGGGGAATTCTAATACGACTCACTATAGGGCAAAGAATCAGAAACACAATGGCGTGTTTTAACAAATCGGCAAAGGAGCCCAAGACTAAGTACGTG 3’
NasR:
5’- GGGGGAATTCTAATACGACTCACTATAGGGAGTGAATAAAAGGTTTTGGGCAGCGCGCCAATGGCGGCGCGTATGTCCAGGGATAAAGGCGTCCAGCGGTGCGTAAGCACCGCCGGGCGCTTTTTT 3’
Surface plasmon resonance measurements
Binding affinities were assessed by surface plasmon resonance on a BIAcore 3000 instrument (GE Healthcare Europe GmbH, Munich, Germany). 50 nM biotinylated aptamers 04.21 (flow cell 2), 53.19 (flow cell 4), and the controls 04.21M2 (flow cell 1) and 53.19M27 (flow cell 3) in 0.5 M NaCl were immobilized on XanTec SAD chips (XanTec bioanalytics) with a flow rate of 10 μL min-1 at 25°C until a response of ~250 response units were reached. PAL in buffer A was incubated in darkness or under blue light and injected at different concentrations and a flow rate of 50 μL min-1 for 240 s at 25°C. The dissociation time was 300 s, followed by a regeneration step (0.3 M NaCl, 0.05% sodium dodecyl sulfate). All buffers were filtered and degassed prior to use. Data were evaluated according to 1:1 binding with drifting baseline with the software BIAevaluation 4.1 (Biacore).
Fluorescence anisotropy
For the analysis of RNA binding by fluorescence anisotropy, variants of the aptamers 04.17 and 53.19 with a 5’-terminal tetramethylrhodamine (TAMRA) fluorophore were synthesized (IDT). Fluorescence measurements were conducted in reaction buffer (buffer A plus 100 μg mL-1 BSA) at a concentration of 4 nM of the TAMRA-labelled RNA. Varying concentrations of dark-adapted PAL were incubated at 22°C with the RNA, and fluorescence anisotropy was recorded with a multi-mode microplate reader (CLARIOstar, BMG labtech) using 540-nm excitation and 590-nm emission filters. The samples were then illuminated with 470-nm light, and fluorescence anisotropy was recorded again. Anisotropy data for dark- and light-adapted PAL were evaluated by nonlinear least-squares fitting to single-site binding isotherms.
| (1) |
To determine the binding stoichiometry of the PAL:aptamer interaction, the concentration of the TAMRA-labelled aptamer was elevated to 200 nM (i.e. well above the KD for the binding interaction under blue light), and binding isotherms were recorded as before. For competition experiments between 04.17 and 53.19, a solution of 100 nM PAL and 50 nM TAMRA-labeled aptamer was titrated with increasing concentrations of unlabeled competitor RNA. Fluorescence anisotropy in the dark and after illumination was determined as before. Data were evaluated according to:
| (2) |
To record association kinetics, 400, 600 and 1000 nM PAL and 4 nM TAMRA-labeled aptamers were pre-incubated in darkness; samples were illuminated and changes in fluorescence anisotropy followed over time. Data were fitted to a pseudo-first-order association model, and resultant unimolecular association rate constants k1 were plotted against PAL concentration to determine a bimolecular association rate constant kbi. Dissociation rate constants were estimated by multiplying kbi with KD.
Structure determination of PAL
Suitable crystallization conditions were determined by sitting-drop vapor diffusion using commercially available sparse-matrix screens (Qiagen). PAL protein in buffer A at concentrations of 4.5 and 9 mg mL-1 was diluted 1:1 with crystallization solution. Crystallization trials were set up under red light with a Phoenix liquid-handling system (Art Robbins Instruments) and incubated in darkness at 4 or 20°C. Crystal growth was monitored by stereomicroscopy under red-light filters. After around 8 weeks, a needle-shaped crystal appeared in one condition (0.1 M bicine pH 9.0, 10% [w/v] PEG 20,000, 2% dioxane). Crystallization was optimized in hanging-drop format by varying pH and precipitant concentration, and subsequently by screening various additives. Needle-shaped crystals of up to several hundred μm length were obtained at 4°C for a PAL protein concentration of 4.5 mg mL-1 and the condition 0.1 M bicine, pH 9.2, 15% (w/v) PEG 20,000, 2% dioxane, 0.8 M imidazole acetate. Single crystals were mounted in loops and rapidly cryo-cooled.
Data collection from crystals of native and SeMet-substituted PAL was carried out on beamline 14.1 at the BESSY II electron storage ring operated by the Helmholtz-Zentrum Berlin35. For the native data set, a total of 1,200 images covering 0.1° each were collected at a wavelength of 0.91814 Å. For the anomalous diffraction data, a total of 14,000 images, comprising two sweeps of 500° and one sweep of 400° at κ angles values of 0, 15 and 30°, respectively, with a rotation per image of 0.1°, were measured using a wavelength of 0.979656 Å. The high-resolution native dataset was processed with xia236 using DIALS37 for indexing, refinement and integration, and POINTLESS38 and AIMLESS39 for merging and scaling. Diffraction was anisotropic with a maximum resolution of 2.51 Å in two dimensions and of 3.20 Å in the third dimension, based on CC1/240. A Wilson plot is shown as Supplementary Fig. 18. The anomalous data sets were processed individually with XDS41 and scaled and merged using XSCALE42. Anomalous pairs were separated in scaling and merging, with the resolution limit set to 3.3 Å. The overall merging statistics of the native and anomalous data are shown in Supplementary Table 2. Structure solution was carried out using the anomalous signal from SeMet incorporated in the protein with the SHELXC/D/E pipeline43. The resolution cutoff for substructure determination was 3.8 Å. SHELXD found fifteen heavy-atom sites with occupancy greater than 35%, with CCall of 0.486 and CCweak of 0.251. SHELXE was able to extend the phase information using the high-resolution native dataset at 2.7 Å resolution and trace part of the backbone of the protein successfully in the original hand, with a CC of 21.14%. Density-modified phases were used for automated model building with BUCCANEER44 and autobuild45. The partial models from the different automatic rebuilding programs were combined, and the structure completed through iterative cycles of manual model building and refinement with phenix.refine46. This process benefitted from the twofold non-crystallographic symmetry within the asymmetric unit between the two PAL dimers. Statistics for the refinement are shown in Supplementary Table 2. The final model comprised two dimers of PAL, with one bound FMN cofactor per monomer, as well as several ligands from the crystallization condition and 158 water molecules. In the Ramachandran plot, 98.93% of residues were in the favored region, 1.07% in the allowed region, and 0.07% in the disallowed region. The structure factors and the final model were deposited in the Protein Data Bank under accession code 6HMJ. Molecular graphics were prepared with PyMOL (Schrödinger).
To ascertain that the high-resolution diffraction data indeed contain meaningful information, we undertook pairwise refinement at resolution cut-offs of 2.51, 2.67 and 2.83 Å. To remove model bias, the coordinates were randomized, and the B-factors were reset according to the Wilson plot (cf. Supplementary Fig. 18). After independent refinement at the three resolution limits, we evaluated the resultant models over a joint resolution range up to 2.83 Å using MOLPROBITY47. As listed in Supplementary Table 4, inclusion of the high-resolution data up to 2.51 Å leads to a lower Rfree over the entire resolution range up to 2.83 Å and within the highest-resolution shell from 2.93 to 2.83 Å. These results indicate that the high-resolution data contain relevant data and hence have to be included in the refinement.
Electron paramagnetic resonance spectroscopy
The C284A variant of PAL was expressed as done for wild-type PAL, concentrated to 45 μM, and transferred into deuterated dialysis buffer supplemented with 50% glycerol. As preliminary spectroscopic measurements indicated that the NSQ yield was poor, 10 mM of the reducing agent TCEP was added. To alleviate light-induced production of detrimental reactive-oxygen species, we employed an oxygen-scavenging system comprising catalase, glucose and glucose-oxidase5. Samples were transferred to capillary tubes, illuminated with blue light (450 nm, 30 mW) for 5 minutes, and rapidly cooled by submerging first in N2-cooled ethanol for 10 s and then in liquid nitrogen. X-band EPR data were recorded at 9.7 GHz on a Bruker BioSpin Elexys E680 X-band spectrometer equipped with a Bruker E580-400U microwave source, Bruker Tera Spec pulsed X-Band microwave bridge and Bruker ER 4118X-MD5 dielectric ring resonator. The necessary amplification of microwaves for the pulsed experiment was achieved with Applied Systems Engineering 117X travelling-wave tube amplifiers. The ELDOR experiment was conducted at 40 K adjusted by an Oxford CF-935 cryostat and controlled with an Oxford ITC503 temperature controller. For the experiments a four-pulse sequence according to48 (probe pulse sequence π/2 - τ1 - π - τ1 - τ2 - π - τ2 - echo with microwave pump pulse (12 ns) on second frequency swept between second and third probe pulse) was utilized with a pulse length for both pulses of 32 ns (the amplitudes were adjusted as required)49. Data were evaluated by Tikhonov regularization50. The obtained electron spin–spin distance is taken as the distance between the flavin C4a atoms carrying the largest spin density in the neutral flavin semiquinone radicals51.
Bacterial reporter gene assay
E. coli Cmpx13 cells were transformed with the arabinose-inducible pCDF-PAL (or, pCDF-PALopt) plasmid and a pET-28c-DsRed-SP reporter plasmid. This reporter plasmid harbors a DsRed expression cassette under control of the T7 promoter and IPTG induction; the 04.17 PAL aptamer was inserted at varying positions near the Shine-Dalgarno (SD) sequence of the DsRed reporter. Nucleotides within the aptamer stem were varied to enable base pairing with the SD sequence. For the assay, starter cultures were grown at 37°C over-night and diluted to an optical density at 600 nm of 0.5. Individual wells of a deep-well microtiter plate containing 700 μL LB medium plus 4 mM arabinose were inoculated with the diluted starter culture and were incubated at 37°C and 600 rpm for 2 h. Cultures were then supplemented with 1mM IPTG and separated into two fractions which were incubated for 16 h at 29°C in darkness or under 40 μW cm-2 470-nm light, respectively. Optical density at 600 nm and DsRed fluorescence of the cultures were measured with a Tecan M200 plate reader according to22. The expression of PAL variants was confirmed by Western blot analysis. To this end, approximately 0.5·109 cells of each culture were lysed, and the lysate was separated by denaturing PAGE. Following semi-dry blotting of the gel (BioRad Transblot Turbo), PAL expression was detected using an anti-myc primary antibody and an alkaline-phosphatase-conjugated secondary antibody.
Translational control in mammalian cells
Mammalian expression of PAL
The plasmids pmCherry-C1 and pMetLuc2-Control were purchased from Takara Clontech (Kyoto, Japan). pmCherry-PAL and pMetLuc plasmids with aptamers in the 5’UTR were generated using the In-Fusion HD EcoDry Cloning Kit (Takara Clontech, Kyoto, Japan). Previous PCR amplification used the primer pair:
-
5’- CTCAAGCTTCGAATTCATGAAGGTGAACCGGCCC 3’
5’- TAGATCCGGTGGATCCCTACGACGCCAGCTGCTCCAA 3’
pmCherry-C1 was linearized using EcoRI and BamHI. Primers for the aptamer inserts were:
-
5’- CAGAGCTGGTTTAGTGAACCGTCAGATC 3’
5’- CACCTTGATGTCCATGGTGGCG 3’
Primers for the linearized pMetLuc2-Control plasmid were:
-
5’- CGCCACCATGGACATCAAGGT 3’
5’- GATCTGACGGTTCACTAAACCAGCTCTG 3’
HeLa cells (CLS Cell Lines Service GmbH, Eppelheim, Germany) were cultured in DMEM medium (high glucose, GlutaMAXTM, Thermo Fisher Scientific, Darmstadt, Germany) supplemented with 10% fetal calf serum (FCS, Sigma-Aldrich, St. Louis, Missouri, USA) at 37ºC and 5% CO2, and were passaged every 2 to 3 days.
Photoswitching in HeLa cells
For fluorescence microscopy, 2·105 cells were seeded in black 24-well plates with clear bottom (μ-plate, ibidi, Martinsried, Germany). After 24 h, the cells were transfected with 0.6 μg pmCherry-PAL using FuGENE HD (Promega, Fitchburg, Wisconsin, United States) according to manufacturer’s instructions. After 48 h, cells were analyzed by confocal laser scanning microscopy (LSM 710, Zeiss, Oberkochen, Germany). Fluorescence of mCherry (excitation [ex] / emission [em]: 543 / 596-696 nm) and PAL (ex / em: 405 / 488-529 nm) in its dark-adapted state was monitored, respectively. To assess intact photochemistry of PAL, samples were irradiated for 1 min with 465 nm light and fluorescence was recorded. Afterwards, cells were incubated in darkness for 10 min before fluorescence was recorded again.
For the plate-reader-based assay 2·106 cells were seeded in clear 6-well plates (Sarstedt AG & Co., Rheinbach, Germany). After 24 h, the cells were transfected with 2 μg pmCherry-PAL using FuGENE HD according to manufacturer’s instructions. After 48 h, medium was removed, cells were collected, centrifuged and resuspended in 1x DPBS. The cell suspension was added to a white 96-well plate (lumitrac200, Greiner) and analyzed for fluorescence of mCherry (ex / em: 587 / 610 nm) and PAL (ex / em: 390 / 510 nm) using an Enspire plate reader (PerkinElmer, Waltham, Massachusetts, USA). Samples were irradiated with 465 nm light for 1 min, and fluorescence was recorded. Cells were then incubated in darkness for 30 minutes followed by fluorescence measurement. The illumination, incubation and measurement steps were repeated three times.
Translational control in mammalian cells
5·104 HeLa cells per well were seeded in two separate 24-well plates and incubated in the absence of light for 24 h at 37°C and 5% CO2. After 24 h, medium was replaced by OptiMEM, and cells were co-transfected with 450 ng pmCherry-PAL and 50 ng pMetLuc reporter with 2 μL Lipofectamine 2000 (Thermo Fisher Scientific, Darmstadt, Germany) according to manufacturer’s instructions. Transfected cells were incubated for 4 h at 37ºC and 5% CO2 in the presence of blue light (465 nm, 106 μW cm-2, 30 s pulses) or in darkness. The transfection mix was replaced by DMEM with 10% FCS, and plates were incubated at 18 h under the specified light regime. For the assay, 5 μL luciferase substrate dissolved in buffer according to manufacturer’s instructions (Ready-To-Glow Secreted Luciferase, Takara Clontech, Kyoto, Japan) were added to wells of a white 96-well plate (lumitrac200, Greiner). 50 μL supernatant of the cell culture was added, and the reaction was incubated for 25 min. The luminescence signal was measured using an Enspire plate reader (PerkinElmer, Waltham, Massachusetts, USA) with an integration time of 5 seconds.
Sequence and data analysis
The Genbank database was searched for LOV proteins using custom Python scripts and Biopython. The retrieved sequences were annotated with HMMER52 using the Pfam domain family profiles53. Unless otherwise stated, nonlinear least-squares fitting of experimental data was performed with the program Fit-o-mat54.
Supplementary Material
Acknowledgements
We thank members of the Mayer and Möglich laboratories for discussion; H. Ruwe and C. Schmitz-Linneweber (HU Berlin) for help in the initial analysis of nucleic-acid binding; M. Humenik for assistance with SEC-MALS. Funding through a Sofja-Kovalevskaya Award of the Alexander-von-Humboldt Foundation (to A.M.); a Consolidator grant 615381 by the European Research Council (to G.M.); and Deutsche Forschungsgemeinschaft (MO2192/6-1, MA3442/5-1, SFB-1089/P1 and Sfb-1078/B4) is gratefully acknowledged.
Footnotes
ORCID identifiers: J.K. 0000-0003-3029-3505; G.M. 0000-0003-3010-4049; A.M. 0000-0002-7382-2772
Data and materials availability
Coordinates and structure-factor amplitudes of the PAL crystal structure that support the findings of this study have been deposited in the Protein Data Bank with the accession code 6HMJ (http://www.rcsb.org/structure/6hmj). Additional raw data or materials are available from the corresponding authors upon reasonable request.
Author contributions
A.M.W. established light-dependent SELEX protocol, conducted SELEX cycles 10-15, characterized PAL:RNA interactions via filter retention, Cherenkov measurements, Ribogreen fluorescence and surface plasmon resonance, designed eukaryotic reporter gene constructs, contributed to sequence analysis of enrichment, established and performed switching experiments of PAL in eukaryotes, and wrote the manuscript. J.K. purified PAL, characterized PAL:RNA interaction by fluorescence anisotropy, developed bacterial reporter assay, conducted structure-function analysis, contributed to EPR experiments, and wrote the manuscript. T.Z. isolated and cloned the PAL gene from N. multipartita; purified, biochemically analyzed and crystallized PAL; and contributed to structure solution and refinement. She demonstrated preferential RNA binding by EMSA, adapted the light-dependent SELEX protocol, conducted SELEX cycles 1-9, and characterized PAL:RNA interactions via EMSA and Ribogreen fluorescence. S.P. designed, performed, and analyzed reporter gene constructs for eukaryotic gene expression and Ribogreen assays of ANTAR sequences. C.R. designed, performed, and analyzed reporter gene constructs for eukaryotic gene expression, performed sequence analysis of enrichment, established and performed switching experiments of PAL in eukaryotes. L.S. purified PAL and conducted fluorescence anisotropy experiments. G.P. designed, performed, and analyzed reporter gene constructs for eukaryotic gene expression and contributed to Cherenkov measurements. S.M. solved and refined the PAL crystal structure. A.K. performed and analyzed reporter gene constructs for eukaryotic gene expression. M.J. purified PAL C284A and conducted EPR measurements. S.S. conducted EPR measurements and analyzed EPR data. L.L.B. performed sequence analysis of enrichment. C.S. advised on crystallography. R.B. supervised EPR measurements and analyzed EPR data. G.M. conceived, designed and supervised research and wrote the manuscript. A.M. identified the PAL gene, conceived, designed and supervised research and wrote the manuscript.
Competing interests
The authors declare no competing interest.
References
- 1.Möglich A, Yang X, Ayers RA, Moffat K. Structure and function of plant photoreceptors. Annu Rev Plant Biol. 2010;61:21–47. doi: 10.1146/annurev-arplant-042809-112259. [DOI] [PubMed] [Google Scholar]
- 2.Christie JM, et al. Arabidopsis NPH1: a flavoprotein with the properties of a photoreceptor for phototropism. Science. 1998;282:1698–1701. doi: 10.1126/science.282.5394.1698. [DOI] [PubMed] [Google Scholar]
- 3.Crosson S, Moffat K. Photoexcited structure of a plant photoreceptor domain reveals a light-driven molecular switch. Plant Cell. 2002;14:1067–1075. doi: 10.1105/tpc.010475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Salomon M, et al. An optomechanical transducer in the blue light receptor phototropin from Avena sativa. PNAS. 2001;98:12357–12361. doi: 10.1073/pnas.221455298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yee EF, et al. Signal transduction in light-oxygen-voltage receptors lacking the adduct-forming cysteine residue. Nat Commun. 2015;6 doi: 10.1038/ncomms10079. 10079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Glantz ST, et al. Functional and topological diversity of LOV domain photoreceptors. Proc Natl Acad Sci U S A. 2016;113:E1442–1451. doi: 10.1073/pnas.1509428113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Takahashi F, et al. AUREOCHROME, a photoreceptor required for photomorphogenesis in stramenopiles. Proc Natl Acad Sci U S A. 2007;104:19625–19630. doi: 10.1073/pnas.0707692104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Harper SM, Neil LC, Gardner KH. Structural basis of a phototropin light switch. Science. 2003;301:1541–1544. doi: 10.1126/science.1086810. [DOI] [PubMed] [Google Scholar]
- 9.Losi A, Gardner KH, Möglich A. Blue-Light Receptors for Optogenetics. Chem Rev. 2018;118:10659–10709. doi: 10.1021/acs.chemrev.8b00163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Deisseroth K, et al. Next-generation optical technologies for illuminating genetically targeted brain circuits. J Neurosci. 2006;26:10380–10386. doi: 10.1523/JNEUROSCI.3863-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yoshimi Y, Hiraishi A, Nakamura K. Isolation and Characterization of Microsphaera multipartita gen. nov., sp. nov., a Polysaccharide-Accumulating Gram-Positive Bacterium from Activated Sludge. International Journal of Systematic and Evolutionary Microbiology. 1996;46:519–525. [Google Scholar]
- 12.Tice H, et al. Complete genome sequence of Nakamurella multipartita type strain (Y-104) Stand Genomic Sci. 2010;2:168–175. doi: 10.4056/sigs.721316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Möglich A, Ayers RA, Moffat K. Structure and signaling mechanism of Per-ARNT-Sim domains. Structure. 2009;17:1282–1294. doi: 10.1016/j.str.2009.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shu CJ, Zhulin IB. ANTAR: an RNA-binding domain in transcription antitermination regulatory proteins. Trends Biochem Sci. 2002;27:3–5. doi: 10.1016/s0968-0004(01)02036-9. [DOI] [PubMed] [Google Scholar]
- 15.Wilson CA, Kreychman J, Gerstein M. Assessing annotation transfer for genomics: quantifying the relations between protein sequence, structure and function through traditional and probabilistic scores. J Mol Biol. 2000;297:233–49. doi: 10.1006/jmbi.2000.3550. [DOI] [PubMed] [Google Scholar]
- 16.Ramesh A, et al. The mechanism for RNA recognition by ANTAR regulators of gene expression. PLoS Genet. 2012;8:e1002666. doi: 10.1371/journal.pgen.1002666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.DebRoy S, et al. Riboswitches. A riboswitch-containing sRNA controls gene expression by sequestration of a response regulator. Science. 2014;345:937–940. doi: 10.1126/science.1255091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tuerk C, Gold L. Systematic evolution of ligands by exponential enrichment: RNA ligands to bacteriophage T4 DNA polymerase. Science. 1990;249:505–510. doi: 10.1126/science.2200121. [DOI] [PubMed] [Google Scholar]
- 19.Ellington AD, Szostak JW. In vitro selection of RNA molecules that bind specific ligands. Nature. 1990;346:818–822. doi: 10.1038/346818a0. [DOI] [PubMed] [Google Scholar]
- 20.Heintz U, Schlichting I. Blue light-induced LOV domain dimerization enhances the affinity of Aureochrome 1a for its target DNA sequence. Elife. 2016;5:e11860. doi: 10.7554/eLife.11860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Banerjee A, Herman E, Kottke T, Essen L-O. Structure of a Native-like Aureochrome 1a LOV Domain Dimer from Phaeodactylum tricornutum. Structure. 2016;24:171–178. doi: 10.1016/j.str.2015.10.022. [DOI] [PubMed] [Google Scholar]
- 22.Diensthuber RP, Bommer M, Gleichmann T, Möglich A. Full-length structure of a sensor histidine kinase pinpoints coaxial coiled coils as signal transducers and modulators. Structure. 2013;21:1127–1136. doi: 10.1016/j.str.2013.04.024. [DOI] [PubMed] [Google Scholar]
- 23.Möglich A, Ayers RA, Moffat K. Design and signaling mechanism of light-regulated histidine kinases. J Mol Biol. 2009;385:1433–1444. doi: 10.1016/j.jmb.2008.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Engelhard C, Diensthuber RP, Möglich A, Bittl R. Blue-light reception through quaternary transitions. Scientific Reports. 2017;7 doi: 10.1038/s41598-017-01497-7. 1385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Berntsson O, et al. Sequential conformational transitions and α-helical supercoiling regulate a sensor histidine kinase. Nat Commun. 2017;8:284. doi: 10.1038/s41467-017-00300-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hentze MW, et al. Identification of the iron-responsive element for the translational regulation of human ferritin mRNA. Science. 1987;238:1570–1573. doi: 10.1126/science.3685996. [DOI] [PubMed] [Google Scholar]
- 27.Zuker M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003;31:3406–3415. doi: 10.1093/nar/gkg595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ankenbruck N, Courtney T, Naro Y, Deiters A. Optochemical Control of Biological Processes in Cells and Animals. Angewandte Chemie International Edition. 2018;57:2768–2798. doi: 10.1002/anie.201700171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lotz TS, et al. A light-responsive RNA aptamer for an azobenzene derivative. Nucleic Acids Res. 2019;47:2029–2040. doi: 10.1093/nar/gky1225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gibson DG, et al. Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat Methods. 2009;6:343–345. doi: 10.1038/nmeth.1318. [DOI] [PubMed] [Google Scholar]
- 31.Mathes T, Vogl C, Stolz J, Hegemann P. In vivo generation of flavoproteins with modified cofactors. J Mol Biol. 2009;385:1511–1518. doi: 10.1016/j.jmb.2008.11.001. [DOI] [PubMed] [Google Scholar]
- 32.Doublié S. Preparation of selenomethionyl proteins for phase determination. Meth Enzymol. 1997;276:523–530. [PubMed] [Google Scholar]
- 33.Tolle F, Mayer G. Preparation of SELEX Samples for Next-Generation Sequencing. Methods Mol Biol. 2016;1380:77–84. doi: 10.1007/978-1-4939-3197-2_6. [DOI] [PubMed] [Google Scholar]
- 34.Bailey TL, et al. MEME Suite: tools for motif discovery and searching. Nucleic Acids Res. 2009;37:W202–W208. doi: 10.1093/nar/gkp335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mueller U, et al. Facilities for macromolecular crystallography at the Helmholtz-Zentrum Berlin. J Synchrotron Radiat. 2012;19:442–449. doi: 10.1107/S0909049512006395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Winter G. xia2: an expert system for macromolecular crystallography data reduction. J Appl Cryst, J Appl Crystallogr. 2010;43:186–190. [Google Scholar]
- 37.Parkhurst JM, et al. Robust background modelling in DIALS. J Appl Crystallogr. 2016;49:1912–1921. doi: 10.1107/S1600576716013595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Evans P. Scaling and assessment of data quality. Acta Cryst D. 2006;62:72–82. doi: 10.1107/S0907444905036693. [DOI] [PubMed] [Google Scholar]
- 39.Evans PR, Murshudov GN. How good are my data and what is the resolution? Acta Cryst D. 2013;69:1204–1214. doi: 10.1107/S0907444913000061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Karplus PA, Diederichs K. Linking crystallographic model and data quality. Science. 2012;336:1030–1033. doi: 10.1126/science.1218231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kabsch W. XDS. Acta Cryst D. 2010;66:125–132. doi: 10.1107/S0907444909047337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kabsch W. Integration, scaling, space-group assignment and post-refinement. Acta Cryst D. 2010;66:133–144. doi: 10.1107/S0907444909047374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sheldrick GM. A short history of SHELX. Acta Cryst A. 2008;64:112–122. doi: 10.1107/S0108767307043930. [DOI] [PubMed] [Google Scholar]
- 44.Cowtan K. The Buccaneer software for automated model building. 1. Tracing protein chains. Acta Cryst D. 2006;62:1002–1011. doi: 10.1107/S0907444906022116. [DOI] [PubMed] [Google Scholar]
- 45.Terwilliger TC, et al. Iterative model building, structure refinement and density modification with the PHENIX AutoBuild wizard. Acta Cryst D. 2008;64:61–69. doi: 10.1107/S090744490705024X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Afonine PV, et al. Towards automated crystallographic structure refinement with phenix.refine. Acta Cryst D. 68:352–367. doi: 10.1107/S0907444912001308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chen VB, et al. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Cryst D. 2010;66:12–21. doi: 10.1107/S0907444909042073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Martin RE, et al. Determination of End-to-End Distances in a Series of TEMPO Diradicals of up to 2.8 nm Length with a New Four-Pulse Double Electron Electron Resonance Experiment. Angewandte Chemie International Edition. 1998;37:2833–2837. doi: 10.1002/(SICI)1521-3773(19981102)37:20<2833::AID-ANIE2833>3.0.CO;2-7. [DOI] [PubMed] [Google Scholar]
- 49.Engelhard C, et al. A structural model for the full-length blue light-sensing protein YtvA from Bacillus subtilis, based on EPR spectroscopy. Photochem Photobiol Sci. 2013;12:1855–1863. doi: 10.1039/c3pp50128k. [DOI] [PubMed] [Google Scholar]
- 50.Jeschke G. DEER Distance Measurements on Proteins. Annual Review of Physical Chemistry. 2012;63:419–446. doi: 10.1146/annurev-physchem-032511-143716. [DOI] [PubMed] [Google Scholar]
- 51.Weber S, Möbius K, Richter G, Kay CWM. The Electronic Structure of the Flavin Cofactor in DNA Photolyase. J Am Chem Soc. 2001;123:3790–3798. doi: 10.1021/ja003426m. [DOI] [PubMed] [Google Scholar]
- 52.Eddy SR. Profile hidden Markov models. Bioinformatics. 1998;14:755–763. doi: 10.1093/bioinformatics/14.9.755. [DOI] [PubMed] [Google Scholar]
- 53.Finn RD, et al. Pfam: clans, web tools and services. Nucleic Acids Res. 2006;34:D247–D251. doi: 10.1093/nar/gkj149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Möglich A. An Open-Source, Cross-Platform Resource for Nonlinear Least-Squares Curve Fitting. J Chem Educ. 2018;95:2273–2278. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.