Abstract Abstract
Ganoderma is a cosmopolitan genus of mushrooms, which can cause root and butt rot diseases on many tree species. Members of this genus are particularly diverse in tropical regions. Some Ganoderma spp. are medicinally active and therefore are used to treat human diseases or as a dietary supplement. In this study, three Ganoderma strains were collected in tropical southern Thailand. Phylogenetic analyses of combined ITS, LSU, TEF1α and RPB2 sequence data indicated that the three strains grouped in a distinct lineage within laccate Ganoderma. One strain was collected from Surat Thani Province clustered in the G. casuarinicola clade with high statistical support (MLBS = 100% / MPBS = 98% / PP = 0.96), while the other two strains of Ganoderma, collected from Nakhon Si Thammarat Province, formed a distinct well-supported clade (MLBS = 100% / MPBS = 100% / PP = 1.00) and are described here as a new species. Ganoderma casuarinicola is reported here as a new record to Thailand. Morphological differences of the two taxa and their closely related taxa are discussed. Colour photographs of macro and micro morphological characteristics and a phylogenetic tree to show the placement of the new record and new species are provided.
Keywords: Ganodermataceae , medicinal mushroom, molecular phylogeny, morphological characteristics, new species, white rot
Introduction
Ganoderma, a genus of the Ganodermataceae, was established by Karsten (1881) with G. lucidum (Curtis) P. Karst. as the type species. Justo et al. (2017) treated Ganodermataceae as a synonym of Polyporaceae, while Cui et al. (2019) state that Ganoderma was not included in Polyporaceae because their double-walled basidiospores are quite different from Polyporaceae. Relevant characteristics for Ganoderma species delimitation are unique to laccate and non-laccate basidiocarps: truncated double walled basidiospores, an apical germinal pore, a thin and colourless external wall (exosporium) and a dark brown internal wall (endosporium) (Moncalvo and Ryvarden 1997; Zhao 1989; Núñez and Ryvarden 2000; Ryvarden 2004). Ganoderma is a cosmopolitan genus and some of the species are pathogenic, causing white rot diseases on rotting stumps, roots and living trunks (Moncalvo and Ryvarden 1997; Pilotti et al. 2004). Ganoderma are distributed in both tropical and temperate regions, but are particularly diverse in the tropical regions (Cao and Yuan 2013). Index Fungorum records 451 taxa (http://www.indexfungorum.org/; accessed date: 1 June 2019) and MycoBank records 387 taxa (http://www.mycobank.org/; accessed date: 1 June 2019). Ganoderma can be a confusing genus to study due to the highly variable morphological features of the species in this group, including intra-species variations (Ryvarden 2000; Papp et al. 2017; Hapuarachchi et al. 2018a, c; Hapuarachchi et al. 2019a, b).
The genus Ganoderma is economically important, as the members of the genus are regarded as valuable medicinal mushrooms (Dai et al. 2009; Hapuarachchi et al. 2018b). Ganoderma spp. have been used in traditional medicines for hundreds of years in Asian countries. Several Ganoderma species are known to be prolific sources of highly active bioactive compounds such as polysaccharides, proteins, steroids and triterpenoids, such as ganoderic acids (Shim et al. 2004; Qiao et al. 2005; Wang and Liu 2008; Teng et al. 2011; De Silva et al. 2012a, b; De Silva et al. 2013; Li et al. 2018). Those bioactive compounds have a therapeutic potential to treat and remedy many pathological diseases (Sanodiya et al. 2009; Richter et al. 2015; Hapuarachchi et al. 2018b).
Most members of Ganoderma are regarded as plant pathogens for trees, such as G. australe (Jungh.) Bres., which is associated with Castanopsis spp. (Luangharn et al. 2017); G. boninense Pat., which is the causal agent of oil palm basal stem rot (Pilotti 2005); G. dunense, which is associated with Acacia cyclops (Tchoumi et al. 2018); G. leucocontextum T.H. Li, W.Q. Deng, Sheng H. Wu, Dong M. Wang & H.P. Hu, which causes problems to Cyclobalanopsis glauca (Li et al. 2015); G. philippii (Bres. & Henn. ex Sacc.) Bres., which causes problems to tea and rubber (Zakaria et al. 2009); G. tropicum, which grows in a solitary manner on living Dipterocarpus spp. (Luangharn et al. 2019); and the holotype of G. casuarinicola, which was found associated with a living Casuarina equisetifolia tree (Xing et al. 2018).
In Thailand, several Ganoderma species have been reported based on both morphological characteristics and molecular data, including G. australe (Luangharn et al. 2017), G. sichuanense (Thawthong et al. 2017) and G. tropicum (Luangharn et al. 2019). The aims of the present study are to report G. casuarinicola as a new record to Thailand and describe G. thailandicum as a new species from Thailand, based on both morphological characteristics and phylogenetic data.
Methods
Mushroom collections and morphological study
Three specimens of Ganoderma were photographed at the collecting sites: one from a tropical climate at Surat Thani Province and the other two from Prachuap Khiri Khan Province in Thailand during the rainy season. The detailed morphological characteristics of the specimens were recorded, based on fresh materials (Luangharn et al. 2017). Specimens were subsequently dried at 40 °C for 24 hours, covered with wax papers, kept in sealed plastic bags with anhydrous silica gel (Luangharn et al. 2017) and deposited in the Mae Fah Luang University herbarium (MFLU herb.), while being duplicated in the Herbarium of Cryptogams, Kunming Institute of Botany Academia Sinica (HKAS).
Morphological characteristics were determined following the methodology described by Lodge et al. (2004). Colour changes on bruising were recorded in the field. Colours were recorded following Ridgeway (Ridgeway 1912). Micro-morphological characteristics were observed using a compound Carl Zeiss™ SteREO Discovery.V8 Microscope, while basidiospores were photographed using a Scanning Electron Microscope (SEM). Microscopic features and measurements were made from glass slide preparations, staining the tissues with 3–5% potassium hydroxide (KOH), 2% Melzer’s reagent and 3% Congo red reagent (Kreisel and Schauer 1987). Measurements were made using the Tarosoft Image Framework programme v. 0.9.0.7. Basidiospore features, hyphal system, colour, sizes and shapes were recorded and photographed. The description of basidiospore measurements was done by using at least 50 basidiospores from each basidiomata (Miettinen and Larsson 2006). The basidiospore quotient was followed [Q = L/W], where Q, the quotient of basidiospore length to width (L/W) of a basidiospore in side view and Qm, the mean of Q-values ± SD, was calculated considering the mean value of the lengths and widths of basidiospores (Tulloss 2005). The basidiospore size was measured with and without the myxosporium and given as (a–)b–c (–d) (Tulloss 2005).
DNA extraction, PCR amplification and sequencing
Dried internal tissues of the fruiting bodies were used to extract DNA by using the Biospin Fungus Genomic DNA Extraction Kit (BioFlux), following the manufacturer’s instructions. Total reaction mixtures (25 μl) contained 9.5 μl ddH2O, 12.5 μl of PCR master mix, 1 μl of DNA template and 1 μl of each primer (10 μM). The primers used in PCR amplification were: ITS4/ITS5 for internal transcribed spacer gene region (ITS); LROR/LR5 for partial large subunit rDNA gene region (LSU) (Vilgalys and Hester 1990; White et al. 1990); 983F/2218R for partial translation elongation factor 1-alpha gene region (TEF1α) (Sung et al. 2007); and fRPB2-5f/fRPB2-7cR for partial RNA polymerase II second largest subunit gene (RPB2) (Liu et al. 1999). PCR amplification conditions were 3 min at 94 °C, followed by 35 cycles of 95 °C for 30 s, 55 °C for 1 min, 72 °C for 1 min, followed by a final extension at 72 °C for 10 min for ITS and LSU. The amplification condition for TEF1α consisted of initial denaturation at 5.30 min at 95 °C, followed by 35 cycles of 94 °C for 1 min, 57 °C for 30 s and 72 °C for 1.30 min, followed by a final extension at 72 °C for 10 min and 3 min at 94 °C followed by 35 cycles of 95 °C for 1 min, 52 °C for 2 min and 72 °C for 1 min, followed by a final extension at 72 °C for 10 min for RPB2. PCR products were sequenced by Sangon Biotech (Shanghai) Co., Ltd., Shanghai, China.
Phylogenetic analyses
Sequence data, retrieved from GenBank based on previous studies, are listed in Table 1. The sequences were subjected to standard BLAST searches in GenBank to determine the primary identity of the fungal isolates. Amauroderma rugosum Cui 9011 (Li and Yuan 2015) and Tomophagus colossus (Zhou et al. 2015) were selected as the outgroup taxa. All the newly generated sequences were aligned with the combined datasets of ITS, LSU and TEF1α with MAFFT v. 7.309 (Katoh and Standley 2013) and manually adjusted using Bioedit v. 7.2.5 (Hall 1999). Gaps were treated as missing data. Maximum parsimony (MP) analysis was performed with PAUP v. 4.0b10 (Swofford 2002). Maximum likelihood analyses (ML) were estimated by using the software on the CIPRES Gateway platform (Miller et al. 2010) and performed using RAxML-HPC2 on XSEDE (v. 8.2.8) (Stamatakis 2014), then carried out using the raxmlGUI version v. 1.3.1 (Silvestro and Michalak 2011).
Table 1.
Details of the taxa used in the phylogenetic analysis of this study. The newly generated sequences are in bold.
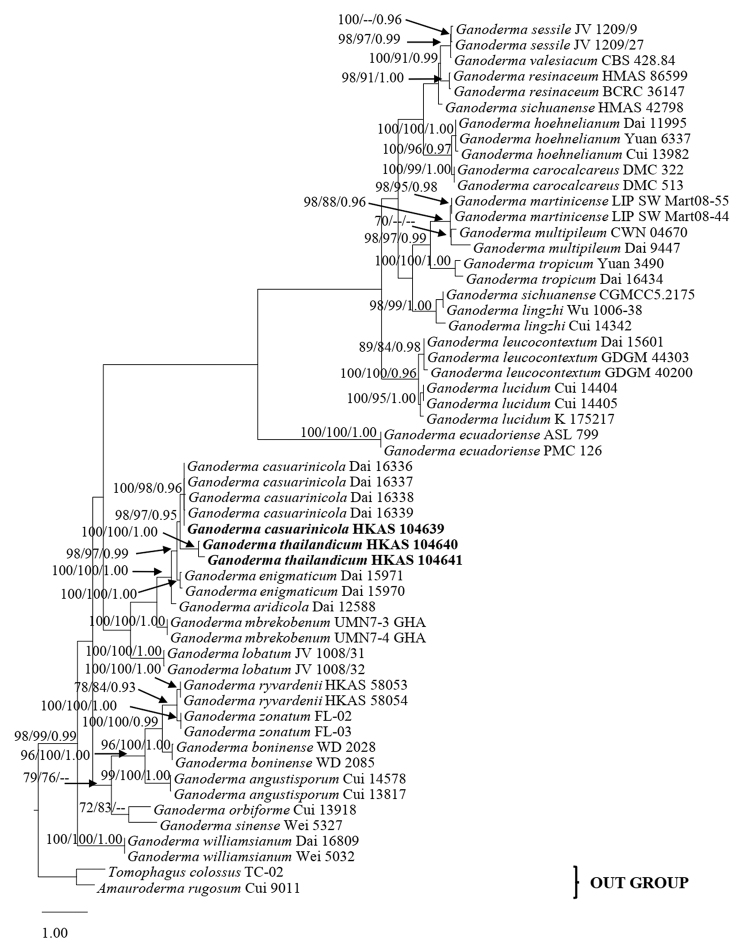
MrModeltest v. 2.3 was used to determine the best-fitting substitution model for each single gene partition and the concatenated dataset for Bayesian analyses (Nylander 2004). Bayesian inference posterior probabilities (PP) with a GTR+I+G model was used for each partition. MrBayes v. 3.2.2 (Huelsenbeck and Ronquist 2001) was used to evaluate PP by Markov Chain Monte Carlo sampling (BMCMC) (Rannala and Yang 1996; Zhaxybayeva and Gogarten 2002). The number of generations was set at 4,000,000, with trees being sampled every 100 generations and a total of 40,000 trees obtained, resulting in an average standard deviation of split frequencies below 0.01. Based on the tracer analysis (Rambaut et al. 2014), the first 20% of trees (8,000 trees) were discarded as the burn-in phase of the analyses represented. The remaining 32,000 trees were used for calculating PP in the majority rule consensus tree (Larget and Simon 1999). ML and MP bootstrap values, equal to or greater than 70% and Bayesian Posterior Probabilities (BP) equal to or greater than 0.95 are presented above each node (Fig. 1). Trees were figured in the FigTree v. 1.4.0 programme (Rambaut 2012), edited using Microsoft Office PowerPoint 2010 and exported to Adobe Illustrator CS v. 3 (Adobe Systems, USA). Sequences derived in this study were deposited in GenBank (http://www.ncbi.nlm.nih.gov).
Figure 1.
Phylogram of Ganoderma thailandicum, obtained from maximum likelihood (RAxML) of combined ITS, LSU, TEF1α and RPB2 datasets. Bootstrap values (BS) from maximum likelihood (ML, left) and Maximum parsimony (MP, middle) greater than 70% and Bayesian posterior probabilities (PP), greater than 0.95, are indicated above the nodes as MLBS/MPBS/PP. The tree is rooted with Amauroderma calcitum Cui 9011 and Tomophagus colossus TC-02. New species and new records are indicated in black bold.
Results
Phylogenetic analyses
The phylogenetic analyses included 56 taxa (including the three new sequence data) and the tree was inferred from the combined ITS, LSU, TEF1α and RPB2 sequences, which comprise 3,360 characters with gaps; 623 characters for ITS, 930 characters for LSU, 859 characters for TEF1α and 948 characters for RPB2. The best scoring ML tree is shown in Fig. 1. Tree topologies of the ML and MP were similar to the Bayesian analysis. The dataset represents 26 Ganoderma species, with Amauroderma rugosum Cui 9011 and Tomophagus colossus TC-02 as the outgroup taxa. The dataset comprised 3361 total characters, of which 2378 were constant, 782 variable characters were parsimony-informative and 201 characters were parsimony-uninformative. Phylogenetic analyses indicated the placement of three isolates (HKAS 104639, HKAS 104640 and HKAS 104641) within the laccate Ganoderma clade. Phylogenetic results showed that the tree has two main distinct clades. The phylogenetic tree gave considerably high support for the G. casuarinicola strain HKAS 104639 and is closely related to the laccate G. casuarinicola, as well as the isolates of Guangdong, China, with good support (MLBS = 100% / MPBS = 98% / PP = 0.96), while the two newly isolated strains from this study (HKAS 104640 and HKAS 104641) formed a distinct clade (MLBS = 100% / MPBS = 100% / PP = 1.00) with a sister clade with G. casuarinicola clade (MLBS = 98% / MPBS = 97% / PP = 0.95).
Taxonomy
Ganoderma casuarinicola
J.H. Xing, B.K. Cui & Y.C. Dai., MycoKeys 34: 93–108 (2018)
635D987A-1FC9-5AC5-8A2F-EE5B8DF149C4
Figure 2.
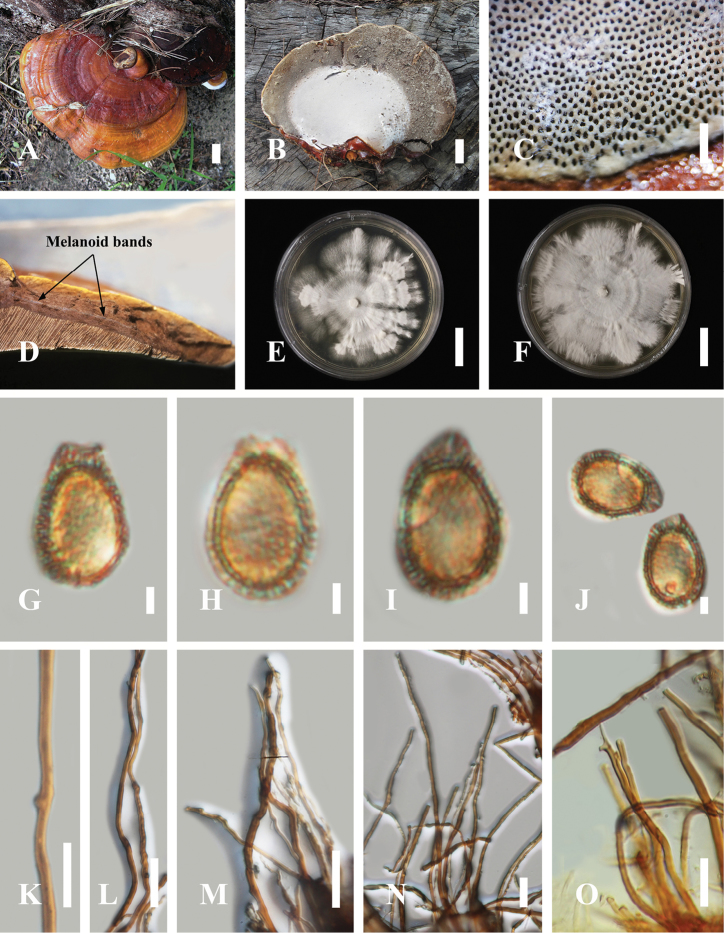
Morphology of Ganoderma casuarinicola (HKAS 104639) A The upper surface of mature basidiocarp B the lower surface of mature basidiocarp C pore characteristics D melanoid bands in the context tissue E, F culture after incubation at 25 °C for 10–14 days on Potato Dextrose Agar (PDA) G–J basidiospores in KOHK clamp connections L thick walled unbranched generative hyphae of context in KOHM, N thin-thick-walled unbranched generative and flexuous skeletal hyphae O thick-walled generative and skeletal hyphae of the tube layers. Scale bars: 2 cm (A, B); 500 µm (C); 2 cm (E, F); 2 µm (G–J); 5 µm (K); 3 µm (L–O).
Description.
Basidiocarps: Substipitate to stipitate. Pileus shape. Annual, applanate and dimidiate when becoming mature, up to 10–16 cm in length, 4– 9 cm in width, up to 0.7–1.2 cm thick. Pileus surface. Distinctively zonate from the base to the margin where the new hyphae are in active development, orange, golden yellow at the base, slightly to reddish-orange, orange red, brownish-red, extended to reddish-brown, red at centre, orange to deep orange extending to the upper margin surface, with yellowish-white to pale yellow under margin surface, strongly laccate, glabrous, glossy, shiny, smooth, spathulate, shallow sulcate when fresh, thin crust overlies the pellis, thicker at the base than the margin, light in weight when dried, non-woody when dried. Context. Mostly yellow to light orange, orange close to crust, reddish-golden, light brown, brown near the tube layers, dense context layer but not fully homogeneous, thick near the base, tough to break when dried; generative hyphae up to 2.10–4.92 μm (x̄ = 3.34, n = 50) in diam., thin walled, almost colourless, some expanded at the apex, unbranched, with clamp connections; binding hyphae 3.67–5.93 µm (x̄ = 4.85, n = 50), almost colourless, thin to thick-walled, branched, with clamp connections; skeletal hyphae abundant, up to 3.49–7.34 μm (x̄ = 5.34, n = 50), almost colourless, thick-walled, unbranched or with very few branches in the distal end, without clamp connections. Hymenophore. Trimitic, heterogeneous, up to 1.4 cm thick, generally yellow slightly to light orange, up to 4 mm thick, the lower layer (close to the tubes) on the upper layers, light brown to brown close to the tubes, presented dark brown, melanoid band. Basidiospores. Ellipsoid to broadly ellipsoid with double wall (ganodermoid) at maturity, yellowish brown, (8.7)10.8–13.5 (14.4) × (6.6)7.6–8.9 (9.8) μm (x̄ = 12.05 × 7.8 μm, n = 50), with Q = 1.38 – 1.45, L = 11.68 µm, W = 8.25 µm (including myxosporium), (7.1)9.9–11.2 (12.1) × (5.2)6.7–7.3 (8.9) μm (x̄ = 10.2 × 6.4 μm, n = 50) μm, with Q = 1.48–1.52, L = 10.65 µm, W = 7.10 µm (excluding outer myxosporium). Tubes. Up to 6–14 mm long, dark brown, hard, woody when dried; generative hyphae 1.0–3.7 µm in diam., occasionally with simple septa, almost colourless, thin-walled with occasionally thick walls, with clamp connections, occasionally branched; skeletal hyphae 2.7–5.1 µm in diam., thick-walled frequently branched at apex; binding hyphae 1.1–3.0 µm in diam., thin to thick-walled, frequently branched at apex. Stipe. Lateral, golden yellow, orange red, up to 8 cm long, 1.8 cm in diam. Margin. Obtuse from the substrate, soft, slippery to the touch when young, tough to break. Pores. Angular to round, 4–6 per mm, up to 128–195 × 148–266 µm (x̄ = 162 × 220 μm, n = 50). Pore surface. White when fresh, turning yellowish-white to pale yellow when dry, reddish-grey when touched, greyish-brown, brownish-grey when wet. Hyphal system. Trimitic, generative hyphae, 2–5 µm in diam., almost colourless, thin-walled or occasionally thick-walled, with clamp connections, occasionally with irregular cuticle cells, light brown to brown in KOH; binding hyphae 3–5 µm, almost colourless, thin to thick-walled, branched, with clamp connections; skeletal hyphae abundant, up to 3–7 μm, almost colourless, thick-walled, unbranched, without clamp connections.
Habitat.
Solitary on Pinus kesiya stumps in pine forests.
Specimen examined.
THAILAND, Surat Thani Province, Phanom District, Khao Sok national park, 8°54'32"N, 98°31'09"E, 427 m elev., 25 June, 2018, LT2018-103 (HKAS 104639).
Ganoderma thailandicum
T. Luangharn, P.E. Mortimer, S.C. Karunarathna & J.C. Xu sp. nov.
B2FF56D7-2287-5F91-834F-E01C886D11B6
831323
Figure 3.
Morphological characteristics of Ganoderma thailandicum (HKAS 104640, HKAS 104641). A, B Mature basidiocarps (HKAS 104640) C lower surface of mature basidiocarp (HKAS 104640) D, E development of young to mature fruiting bodies (HKAS 104641) F lower surface (HKAS 104641) G clamp connections H thick-walled unbranched generative hyphae with clamp connections of context in KOHI thick-walled, skeletal hyphae in KOH without septa J thick-walled sparingly branched skeletal hyphae in Melzer’s reagent K hyphae of tube layers L–Q basidiospores in 3% Congo red reagent. Scale bars: 2 cm (A–F); 10 µm (G); 15 µm (H–K); 3 µm (L–P); 5 µm (Q).
Diagnosis.
Ganoderma thailandicum is characterised by its laccate deep magenta close to stipe, brownish-red at centre and light yellow of active development towards the margin on pileal surface, white pore surface, brownish-red context and absence of melanoid band.
Holotype.
THAILAND, Nakhon Si Thammarat Province, Khanom District, solitary on stump of Pinus merkusii, 10 December 2018, LT2018-105 (HKAS 104640).
Etymology.
The species epithet “thailandicum” refers to the country where the holotype was collected.
Description.
Basidiocarps. Dimidiate, laccate, substipitate to stipitate. Pileus shape. Annual and dimidiate when mature, up to 3–9 cm in length, 3–6 cm in width, up to 0.4–1.8 cm thick at centre of pileus close to the stipe, obtuse from the substrate. Pileus surface. Laccate, glabrous, glossy, smooth, soft, umbonate, distinctly concentrically zonate, greyish-magenta to deep magenta at stipe, greyish-ruby, greyish-red to brownish-red at centre, extended to reddish-orange to slightly pale red with light yellow to vivid yellow of active development towards the margin, thin crust overlaying the pileus, sometimes convex sulcate extending at centre, with distinct concentric zones, with fine furrows at centre extended to the margin, thicker at the base than the margin, consistency hard when young to mature, some cracked when old, non-woody, light in weight when dried. Hymenophore. Trimitic, up to 0.4–2.4 cm thick, heterogeneous with greyish-red close to the upper layers slightly to brownish-red to reddish-brown close to the tubes. Context. Mostly brownish-red to reddish-brown in Melzer’s reagent, absent of melanoid band, with dense context layer. Basidiospores. Ellipsoid to broadly ellipsoid with some globose with double wall (ganodermoid) at maturity, light brown to reddish-brown in Congo red reagent, (6.8)8.4–9.7 (10.2) × (5.8)6.5–7.3 (7.7) μm (x̄ = 9.1 × 6.9 μm, n = 50), with Q = 1.29–1.35, L = 9.13 µm, W = 6.96 µm (including myxosporium), (5.4)7.6–9.6 (10.0) × (4.7)5.8–6.9 (7.4) μm (x̄ = 7.6 × 6.0 μm, n = 50) μm, with Q = 1.32–1.38, L = 8.64 µm, W = 6.42 µm (excluding outer myxosporium). Tubes. Up to 0.5 mm close to margin to 7 mm at centre in length, brown to dark brown, hard, woody when dried; generative hyphae 2.73–4.74 µm in diam., almost colourless, thin-walled with occasionally thick walls, with clamp connections, occasionally branched; skeletal hyphae 3.76–5.81 µm in diam., thick-walled frequently branched at apex; binding hyphae 3.24–5.84 µm in diam., thin to thick-walled, frequently branched at apex. Stipe. Lateral, pale red to vivid red, greyish-red to red when present, with violet brown when mature, different from and darker than pileus, up to 3–5 cm long, 2.5–3.0 cm in diam., 1.8–2.7 cm thick. Margin. Up to 0.4–0.8 cm thick when becoming mature, active growing margin white on the upper and under margin surface when fresh, with a yellow line under the pileus, round, soft, smooth, slippery when touched when young to mature stage, without any zonation, tough when broken. Pores. Angular to round, 4–8 per mm, up to 121–176 × 174–247 µm (x̄ = 155 × 209 μm, n = 50). Pore surface. White when fresh, grey at centre, slightly orange grey at margin, brownish-grey when touched, turning brownish-orange when dry, grey when wet. Hyphal system. Trimitic, light orange to deep orange, reddish-brown in Melzer’s reagent; generative hyphae, 2.65–4.58 µm (x̄ = 3.82, n = 50) in diam., almost colourless, mostly thick-walled, occasionally thin-walled, bearing clamp connections, occasionally with irregular cuticle cells; binding hyphae 3.32–6.28 µm (x̄ = 5.53, n = 50), almost colourless, thin-walled, occasionally branched in the distal end, with clamp connections; skeletal hyphae abundant, up to 3.40–6.78 μm (x̄ = 5.73, n = 50), almost colourless, thick-walled and unbranched. Context. Mostly brownish-red in Melzer’s reagent, reddish-brown, with greyish-red close to crust, dense context layer, agglutinate mass, usually solid in basal part, thick near the base, tough to break when dried; generative hyphae up to 2.80–5.75 μm (x̄ = 4.36, n = 50) in diam., mostly colourless, thick-walled, with clamp connections, occasionally with simple septa; binding hyphae 1.23–4.75 µm (x̄ = 2.49, n = 50), colourless, thin-walled or with a very few branches in the distal end, with clamp connections; abundant skeletal hyphae up to 3.30–7.51 μm (x̄ = 5.75, n = 50), almost colourless, thick-walled, unbranched, with clamp connections and occasionally with simple septa. Cuticle cells. Clavate to narrowly clavate, tuberculate, occasionally with irregular cuticle cells, mostly thick-walled, occasionally thin-walled with simple septa. Basidia. Clavate, with 4 sterigmata, 12.2–19.6 × 8.3–10.9 µm, light brown (5D6) to yellowish in Melzer’s reagent.
Material examined.
THAILAND, Nakhon Si Thammarat Province, Khanom District, solitary on stump of Pinus merkusii, 11°45'58"N, 99°47'43"E, 499 m elev., 10 December 2018, LT2018-105 and LT2018-106, specimens no. HKAS 104640 and HKAS 104641.
Discussion
In this study, we describe a new species of Ganoderma growing on Pinus sp. in tropical southern Thailand, in a well-researched genus. This is not surprising as Hyde et al. (2018) found that up to 96% of species discovered in northern Thailand were new to science. Ganoderma casuarinicola was collected on a Pinus kesiya stump in a pine forest at Surat Thani Province in Thailand, while two collections of Ganoderma thailandicum were collected on Pinus merkusii stumps from Kanom District, Nakhon Si Thammarat Province in Thailand. All three collections grouped as sister taxa to the laccate Ganoderma clade, their morphological characteristics and molecular analyses providing insights to resolve species delimitation. In this study, we introduce G. casuarinicola (HKAS 104639) as a new record to Thailand which grouped with the holotype from Guangdong, China (Fig. 1) with high statistical support (MLBS = 100% / MPBS = 98% / PP = 0.96) and G. thailandicum is described as a new species, the two collections of G. thailandicum (HKAS 104640 and HKAS 104641) grouping together as a distinct clade with 100% ML, 100% MP and 1.00 PP support.
Our findings are consistent with Xing et al. (2018), who demonstrated that G. casuarinicola forms a sister clade with G. aridicola J.H. Xing & B.K. Cui, from South Africa and G. enigmaticum M.P.A. Coetzee, Marinc., M.J. Wingf., from Africa (Coetzee et al. 2015). The morphological differences of these three Ganoderma species were detailed in Xing et al. (2018). Moreover, our study allows us to compare the holotypes of G. casuarinicola from Guangdong and our collection from Thailand. The Guangdong’s G. casuarinicola shows its distinctive sectorial to shell-shaped, 10 cm long and 7 cm wide pileus (Xing et al. 2018), while the Thai G. casuarinicola shows its annual, applanate to dimidiate shape, 3–16 cm long and 1.5–3 cm wide pileus, larger than the Guangdong collection. Our G. casuarinicola collections show longer tubes of 6–14 mm, while the tubes of the Guangdong collection are 9 mm long; however, our collections show a thinner margin (0.8–1.2 cm thick) than the Guangdong collection (2 cm thick) (Xing et al. 2018). Macro-morphological characteristics of our G. casuarinicola share similarities with the holotype collection, such as strongly laccate, shallow sulcate, reddish-brown pileus surface, lateral stipe shape, white pore surface and brown context.
Micro-morphological characteristics of the context layers of both Guangdong and Thai G. casuarinicola share similar characteristics, such as the dense light brown to brown context layers; thin to thick-walled generative hyphae; thin-walled binding hyphae; and thick-walled skeletal hyphae. Although both type specimens and our collection of G. casuarinicola collection have mostly distinctive yellowish-brown basidiospores, Thai G. casuarinicola collections have a smaller size range of (8.7)10.8–13.5 (14.4) × (6.6)7.6–8.9 (9.8) μm than the type of G. casuarinicola (8.3–)9.0–10.2 (–11.5) × (4.5–)5.0–6.0 (–7.0) µm (including myxosporium). However, the type of G. casuarinicola does not have the melanoid band (Xing et al. 2018), while our collection has a dark brown, melanoid band. Although both type specimens and our G. casuarinicola collections are grouped in the same clade, macro-morphologically, their pilei are very different, most probably due to geographical and climatic changes. Boddy et al. (2014) also mentioned that climate change and geography affect fungi in many ways, especially regarding phenological changes of fungal fruiting and the spatial and temporal distribution of hosts.
According to our phylogenetic analyses (Fig. 1), collections of G. thailandicum were grouped as a sister to G. aridicola, G. casuarinicola, and G. enigmaticum as a well-supported clade of 100% ML, 100% MP and 1.00 PP statistical supports. Ganoderma aridicola, G. casuarinicola, G. enigmaticum and G. thailandicum share morphological similarities of laccate to strong laccate upper pileus surface and ellipsoid to broadly ellipsoid basidiospores at maturity. Ganoderma aridicola (Xing et al. 2016), G. casuarinicola (Xing et al. 2018) and G. enigmaticum (Coetzee et al. 2015) are considered as members of the G. lucidum complex and our G. thailandicum is also clustered within the G. lucidum complex, according to the results of the phylogenetic analyses. Our phylogenetic tree showed G. thailandicum clustered together with G. casuarinicola. Although G. thailandicum and G. casuarinicola form a distinctive laccate pileus surface, their macro- and micro-morphological characteristics are quite different. Ganoderma thailandicum can be easily distinguished from G. casuarinicola, by its deep magenta colour near the stipe, brownish-red colour at the centre of the pileus surface and vivid yellow colour at the actively-developed margin, while the fruiting bodies of G. casuarinicola are homogenously brownish-red to reddish-brown at maturity. Ganoderma thailandicum also has a smaller sized pileus (3–9 cm long, 3–6 cm width, 0.4–1.8 cm thick), while G. casuarinicola has a larger pileus (up to 10 cm long, 4–9 cm width, up to 2 cm thick). Ganoderma thailandicum has a smaller pore size (4–8 per mm) than G. casuarinicola (4–6 per mm) and G. thailandicum has narrower basidiospores (6.93 × 9.11 μm; including myxosporium) than G. casuarinicola (8.25 × 11.68 μm; including myxosporium). The basidiopore shapes of G. thailandicum are distinctive, with ellipsoid to broadly ellipsoid or some globose, while basidiospores of G. casuarinicola are mostly ellipsoid to broadly ellipsoid at maturity. Both G. thailandicum and G. casuarinicola are quite similar by having angular to round pore shapes. The differences of G. aridicola and G. enigmaticum have been described in Xing et al. (2016). Ganoderma mbrekobenum can be differentiated from G. casuarinicola and G. thailandicum by its woody to corky texture when dried, with dimitic hyphal system, ovoid and bitunicate basidiospores (Crous et al. 2016).
Casuarina has been reported as a host genus for G. casuarinicola (Xing et al. 2018), which is found in coastal areas, while our G. casuarinicola collection was found on dead Pinus kesiya wood, thus this is the first Pinus host recorded for G. casuarinicola. Based on comprehensive morphological characteristics and molecular analyses, we report G. casuarinicola as a new record to Thailand, with G. thailandicum as a new species from Thailand.
Supplementary Material
Acknowledgements
We appreciate the kind support given by the Key Laboratory for Plant Diversity and Biogeography of East Asia, Kunming Institute of Botany, Chinese Academy of Sciences, Kunming 650201, Yunnan, China; Key Laboratory for Economic Plants and Biotechnology, Kunming Institute of Botany, Chinese Academy of Sciences, Kunming 650201, Yunnan, China and Centre for Mountain Futures (CMF), Kunming Institute of Botany, Kunming 650201, Yunnan, China. We thank the Germplasm Bank of Wild Species, Kunming Institute of Botany, Kunming 650201, Yunnan, China for enabling our molecular laboratory work. Kasiphat Limsakul and Wilawan Punyaboon are acknowledged for their invaluable assistance. Samantha C. Karunarathna thanks the CAS President’s International Fellowship Initiative (PIFI) for funding his postdoctoral research (No. 2018PC0006) and the National Science Foundation of China (NSFC) for funding this work under the project code 31750110478. Peter E. Mortimer thanks the National Science Foundation of China (NSFC) project codes 41761144055 and 41771063 and the South East Asia Biodiversity Resources Institute, CAS, project code Y4ZK111B01. The authors also thank William A. Julian for his editing contributions.
Citation
Luangharn T, Karunarathna SC, Mortimer PE, Hyde KD, Xu J (2019) Additions to the knowledge of Ganoderma in Thailand: Ganoderma casuarinicola, a new record; and Ganoderma thailandicum sp. nov. MycoKeys 59: 47–65. https://doi.org/10.3897/mycokeys.59.36823
Contributor Information
Peter E. Mortimer, Email: peter@mail.kib.ac.cn.
Jianchu Xu, Email: jxu@mail.kib.ac.cn.
References
- Boddy L, Büntgen U, Egli S, Gange AC, Heegaard E, Kirk PM, Mohammad A, Kauserud H. (2014) Climate variation effects on fungal fruiting. Fungal ecological 10: 20–33. 10.1016/j.funeco.2013.10.006 [DOI] [Google Scholar]
- Cao Y, Wu SH, Dai YC. (2012) Species clarification of the prize medicinal Ganoderma mushroom “Lingzhi”. Fungal Diversity 56: 49–62. 10.1007/s13225-012-0178-5 [DOI] [Google Scholar]
- Cao Y, Yuan HS. (2013) Ganoderma mutabile sp. nov. from southwestern China based on morphological and molecular data. Mycological Progress 12: 121–126. 10.1007/s11557-012-0819-9 [DOI] [Google Scholar]
- Coetzee MPA, Marincowitz S, Muthelo VG, Wingfield MJ. (2015) Ganoderma species, including new taxa associated with root rot of the iconic Jacaranda mimosifolia in Pretoria, South Africa. IMA Fungus 6(1): 249–256. 10.5598/imafungus.2015.06.01.16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crous PW, Wingfield MJ, Richardson DM, Le Roux JJ, Strasberg D, Edwards J, Roets F, Hubka V, Taylor PWJ, Heykoop M, Martín MP, Moreno G, Sutton DA, Wiederhold NP, Barnes CW, Carlavilla JR, Gené J, Giraldo A, Guarnaccia V, Guarro J, Hernández-Restrepo M, Kolařík M, Manjón JL, Pascoe IG, Popov ES, Sandoval-Denis M, Woudenberg JHC, Acharya K, Alexandrova AV, Alvarado P, Barbosa RN, Baseia IG, Blanchette RA, Boekhout T, Burgess TI, Cano-Lira JF, Čmoková A, Dimitrov RA, Dyakov MY, Dueñas M, Dutta AK, EsteveRaventós F, Fedosova AG, Fournier J, Gamboa P, Gouliamova DE, Grebenc T, Groenewald M, Hanse B, Hardy GEJ, Held BW, Jurjević Z, Kaewgrajang T, Latha KPD, Lombard L, Luangsa-ard JJ, Lysková P, Mallátová N, Manimohan P, Miller AN, Mirabolfathy M, Morozova OV, Obodai M, Oliveira NT, Ordóñez ME, Otto EC, Paloi S, Peterson SW, Phosri C, Roux J, Salazar WA, Sánchez A, Sarria GA, Shin HD, Silva BDB, Silva GA, Smith MT, Souza-Motta CM, Stchige AM, Stoilova-Disheva MM, Sulzbacher MA, Telleria MT, Toapanta C, Traba JM, Valenzuela-Lopez N, Watling R, Groenewald JZ. (2016) Fungal planet description sheets: 400–468. Persoonia 36: 316–458. 10.3767/persoonia.2018.40.10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui BK, Li HJ, Ji X, Zhou JL, Song J, Si J, Yang ZL, Dai YC. (2019) Species diversity, tax- onomy and phylogeny of Polyporaceae (Basidiomycota) in China. Fungal Diversity 97: 137–392. 10.1007/s13225-019-00427-4 [DOI] [Google Scholar]
- Dai YC, Yang ZL, Cui BK, Yu CJ, Zhou LW. (2009) Species diversity and utilization of medicinal mushrooms and fungi in China. International Journal of Medicinal Mushrooms 11: 287–302. 10.1615/IntJMedMushr.v11.i3.80 [DOI] [Google Scholar]
- De Silva DD, Rapior S, Fons F, Bahkali AH, Hyde KD. (2012a) Medicinal mushrooms in supportive cancer therapies: an approach to anti-cancer effects and putative mechanisms of action. Fungal Diversity 55: 1–35. 10.1007/s13225-012-0151-3 [DOI] [Google Scholar]
- De Silva DD, Rapior S, Hyde KD, Bahkali AH. (2012b) Medicinal mushrooms in prevention and control of diabetes mellitus. Fungal Diversity 56: 1–29. 10.1007/s13225-012-0187-4 [DOI] [Google Scholar]
- De Silva DD, Rapior S, Sudarman E, Stadler M, Xu J, Alias SA, Hyde KD. (2013) Bioactive metabolites from macrofungi: ethnopharmacology, biological activities and chemistry. Fungal Diversity 62: 1–40. 10.1007/s13225-013-0265-2 [DOI] [Google Scholar]
- Douanla-Meli C, Langer E. (2009) Ganoderma carocalcareus sp. nov., with crumbly-friable context parasite to saprobe on Anthocleista nobilis and its phylogenetic relationship in G. resinaceum group. Mycological Progress 8: 145–155. 10.1007/s11557-009-0586-4 [DOI] [Google Scholar]
- Hall TA. (1999) BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symposium Series 41: 95–98. [Google Scholar]
- Hapuarachchi KK, Elkhateeb WA, Karunarathna SC, Phengsintham P, Cheng CR, Bandara AR, Kakumyan P, Hyde KD, Daba GM, Wen TC. (2018a) Current status of global Ganoderma cultivation, products, industry and market. Mycosphere 9: 1025–1052. 10.5943/mycosphere/9/5/6 [DOI] [Google Scholar]
- Hapuarachchi KK, Karunarathna SC, Phengsintham P, Kakumyan P, Hyde KD, Wen TC. (2018b) Amauroderma (Ganodermataceae, Polyporales) – bioactive compounds, beneficial properties and two new records from Laos. Asian Journal of Mycology 1: 121–136. 10.5943/ajom/1/1/10 [DOI] [Google Scholar]
- Hapuarachchi KK, Karunarathna SC, Raspé O, De Silva KHWL, Thawthong A, Wu XL, Kakumyan P, Hyde KD, Wen TC. (2018c) High diversity of Ganoderma and Amauroderma (Ganodermataceae, Polyporales) in Hainan Island, China. Mycosphere 9: 931–982. 10.5943/mycosphere/9/5/1 [DOI] [Google Scholar]
- Hapuarachchi KK, Karunarathna SC, McKenzie EHC, Wu XL, Kakumyan P, Hyde KD, Wen TC. (2019a) High phenotypic plasticity of Ganoderma sinense (Ganodermataceae, Polyporales) in China. Asian Journal of Mycology 2: 1–47. 10.5943/ajom/2/1/1 [DOI] [Google Scholar]
- Hapuarachchi KK, Karunarathna SC, Phengsintham P, Yang HD, Kakumyan P, Hyde KD, Wen TC. (2019b) Ganodermataceae (Polyporales): diversity in Greater Mekong Subregion countries (China, Laos, Myanmar, Thailand and Vietnam). Mycosphere 10: 221–309. 10.5943/mycosphere/10/1/6 [DOI] [Google Scholar]
- Huelsenbeck JP, Ronquist F. (2001) MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics 17: 754–755. 10.1093/bioinformatics/17.8.754 [DOI] [PubMed] [Google Scholar]
- Hyde KD, Norphanphoun C, Chen J, Dissanayake AJ, Doilom M, Hongsanan S, Jayawardena RS, Jeewon R, Perera RH, Thongbai B, Wanasinghe DN, Wisitrassameewong K, Tibpromma S, Stadler M. (2018) Thailand’s amazing diversity – up to 96% of fungi in northern Thailand are novel. Fungal Diversity 93: 215–239. 10.1007/s13225-018-0415-7 [DOI] [Google Scholar]
- Index Fungorum (2019) http://www.indexfungorum.org/names/names.asp [Accessed on: 2019-6-1]
- Justo A, Miettinen O, Floudaas D, Ortiz-Santana B, Sjökvist E, Lindner D, Nakasone K, Niemela D, Nakasone K, Niemelö T, Larsson KH, Ryvarden L, Hibbett DS. (2017) A revised family-level classification of the Polyporales (Basidiomycota) 121: 798–894. 10.1016/j.funbio.2017.05.010 [DOI] [PubMed]
- Karsten PA. (1881) Enumeralio boletinearum et polyporearum fennicarum, systemate novo dispositarum. Revue de Mycologie 3: 16–19. [Google Scholar]
- Katoh K, Standley DM. (2013) MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Molecular Biology and Evolution 30: 772–780. 10.1093/molbev/mst010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinge TR, Mih AM. (2011) Ganoderma ryvardense sp. nov. associated with basal stem rot (BSR) disease of oil palm in Cameroon. Mycosphere 2: 179–188. [Google Scholar]
- Kreisel H, Schauer F. (1987) Methoden des mykologischen Laboratoriums. VEB Gustav Fischer Verlag, Jena, 181 pp. [Google Scholar]
- Larget B, Simon DL. (1999) Markov Chain Monte Carlo Algorithms for the Bayesian analysis of phylogenetic trees. Molecular Biology and Evolution 16: 750–759. 10.1093/oxfordjournals.molbev.a026160 [DOI] [Google Scholar]
- Li TH, Hu HP, Deng WQ, Wu SH, Wang DM, Tsering T. (2015) Ganoderma leucocontextum, a new member of the G. lucidum complex from southwestern China. Mycoscience 56: 81–85. 10.1016/j.myc.2014.03.005 [DOI] [Google Scholar]
- Li LF, Liu HB, Zhang QW, Li ZP, Wong TL, Fung HY, Zhang JX, Bai SP, Lu AP, Han QB. (2018) Comprehensive comparison of polysaccharides from Ganoderma lucidum and G. sinense: chemical, antitumor, immunomodulating and gut-microbiota modulatory properties. Scientific Reports 8: 1–12. 10.1038/s41598-018-22885-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li MJ, Yuan HS. (2015) Type studies on Amauroderma species described by J.D. Zhao et al. and the phylogeny of species in China. Mycotaxon 130: 79–89. 10.5248/130.79 [DOI] [Google Scholar]
- Liu YJ, Whelen S, Hall BD. (1999) Phylogenetic relationships among ascomycetes: evidence from an RNA polymerase II subunit. Molecular Biology and Evolution 16: 1799–1808. 10.1093/oxfordjournals.molbev.a026092 [DOI] [PubMed] [Google Scholar]
- Lodge DJ, Ammirati FJ, O’Dell TE, Mueller GM. (2004) Collecting and describing macrofungi. In: Mueller GM, Bills GF, Foster MS (Eds) Biodiversity of Fungi Inventory and Monitoring Methods. Elsevier Academic Press, London, 128–154 pp.
- Luangharn T, Karunarathna SC, Khan S, Xu JC, Mortimer PE, Hyde KD. (2017) Antibacterial activity, optimal culture conditions and cultivation of the medicinal Ganoderma australe, new to Thailand. Mycosphere 8: 1108–1123. 10.5943/mycosphere/8/8/11 [DOI] [Google Scholar]
- Luangharn T, Karunarathna SC, Mortimer PE, Hyde KD, Thongklang N, Xu JC. (2019) A new record of Ganoderma tropicum (Basidiomycota, Polyporales) for Thailand and first assessment of optimum conditions for mycelia production. MycoKeys 51: 65–83. 10.3897/mycokeys.51.33513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miettinen O, Larsson KH. (2006) Trechispora elongata species nova from North Europe. Mycotaxon 96: 193–198. http://hdl.handle.net/10138/42882 [Google Scholar]
- Moncalvo JM, Ryvarden L. (1997) A nomenclatural study of the Ganodermataceae Donk. Synopsis Fungorum 11: 1–114. [Google Scholar]
- MycoBank (2019) http://www.mycobank.org [Accessed on: 2019-6-1]
- Núñez M, Ryvarden L. (2000) East Asian Polypores: Ganoderma taceae and Hymenochaetaceae. Fungiflora, Oslo, Norway, 6–50.
- Nylander JAA. (2004) MrModeltest v.2 program distributed by the author. Evolutionary Biology Centre, Uppsala University.
- Papp V, Dima B, Wasser SP. (2017) What is Ganoderma lucidum in the molecular era? International journal of medicinal mushrooms abbreviation 19(7): 579–593. 10.1615/IntJMedMushrooms.2017021195 [DOI] [PubMed]
- Park YJ, Kwon OC, Son ES, Yoon DE, Han W, Nam JY, Yoo YB, Lee CS. (2012) Genetic diversity analysis of Ganoderma species and development of a specific marker for identification of medicinal mushroom Ganoderma lucidum. African Journal of Microbiology Research 6(25): 5417–5425. 10.5897/AJMR12.846 [DOI] [Google Scholar]
- Pilotti CA. (2005) Stem rots of oil palm caused by Ganoderma boninense: pathogen biology and epidemiology. Mycopathologia 159(1): 129–137. 10.1038/s41598-018-22885-7 [DOI] [PubMed] [Google Scholar]
- Pilotti CA, Sanderson FR, Aitken AB, Armstrong W. (2004) Morphological variation and host range of two Ganoderma species from Papua New Guinea. Mycopathologia 158: 251–265. 10.1023/B:MYCO.0000041833.41085.6f [DOI] [PubMed] [Google Scholar]
- Qiao Y, Yang Y, Dong X, Qiu M. (2005) 13C NMR in the application of new Ganoderma triterpenoids. Journal of Spectroscopy 22(4): 437–456. [Google Scholar]
- Rambaut A. (2012) FigTree version 1.4.0. http://tree.bio.ed.ac.uk/software/soft-ware/figtree/
- Rambaut A, Suchard MA, Xie D, Drummond AJ. (2014) Tracer v16. http://beast.bio.ed.ac.uk/Tracer
- Rannala B, Yang Z. (1996) Probability distribution of molecular evolutionary trees: a new method of phylogenetic inference. Journal of Molecular Evolution 43(3): 304–311. 10.1007/BF02338839 [DOI] [PubMed] [Google Scholar]
- Richter C, Wittstein K, Kirk PM, Stadler M. (2015) An assessment of the taxonomy and chemotaxonomy of Ganoderma. Fungal Divers 71: 1–15. 10.1007/s13225-014-0313-6 [DOI] [Google Scholar]
- Ridgeway R. (1912) Color Standards and Color Nomenclature. Ridgeway, Washington DC, 12–225. 10.5962/bhl.title.144788 [DOI]
- Ryvarden L. (2000) Studies in neotropical polypores 2: a preliminary key to neotropical species of Ganoderma with a laccate pileus. Mycologia 92: 180–191. 10.2307/3761462 [DOI] [Google Scholar]
- Ryvarden L. (2004) Neotropical polypores Part 1. Synopsis Fungorum. Fungiflora, Oslo, 1–227.
- Sanodiya BS, Thakur GS, Baghel RK, Prasad GB, Bisen PS. (2009) Ganoderma lucidum: a potent pharmacological macrofungus. Current Pharmaceutical Biotechnology 10: 717–742. 10.2174/138920109789978757 [DOI] [PubMed] [Google Scholar]
- Shim SH, Ryu J, Kim JS, Kang SS, Xu Y, Jung SH, Lee YS, Lee S, Shin KH. (2004) New lanostane-type triterpenoids from Ganoderma applanatum. Journal of Natural Products 67: 1110–1113. 10.1021/np030383p [DOI] [PubMed] [Google Scholar]
- Silvestro D, Michalak I. (2011) raxmlGUI: a graphical front-end for RAxML. Organisms Diversity and Evolution 12(4): 335–337. 10.1007/s13127-011-0056-0 [DOI] [Google Scholar]
- Song J, Xing JH, Decock C, He XL, Cui BK. (2016) Molecular phylogeny and morphology reveal a new species of Amauroderma (Basidiomycota) from China. Phytotaxa 260: 47–56. 10.11646/phytotaxa.260.1.5 [DOI] [Google Scholar]
- Stamatakis A. (2014) RAxML v. 8: A tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30: 1312–1313. 10.1093/bioinformatics/btu033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sung GH, Sung JM, Hywel-Jones NL, Spatafora JW. (2007) A multi-gene phylogeny of Clavicipitaceae (Ascomycota, Fungi): identification of localized incongruence using a combinational bootstrap approach. Molecular Phylogenetics and Evolution 44(3): 1204–1223. 10.1016/j.ympev.2007.03.011 [DOI] [PubMed] [Google Scholar]
- Swofford DL. (2002) PAUP*. Phylogenetic analysis using parsimony (*and Other Methods), Version 4.0 beta version. Sinauer Associates, Sunderland, Massachusetts.
- Tchoumi JMT, Coetzee MPA, Rajchenberg M, Wingfield MJ, Roux J. (2018) Three Ganoderma species, including Ganoderma dunense sp. nov., associated with dying Acacia cyclops trees in South Africa. Australasian Plant Pathology 47: 431–447. 10.1007/s13313-018-0575-7 [DOI] [Google Scholar]
- Teng BS, Wang CD, Yang HJ, Wu JS, Zhang D, Zheng M, Fan ZH, Pan D, Zhou P. (2011) A protein tyrosine phosphatase 1B activity inhibitor from the fruiting bodies of Ganoderma lucidum (Fr.) Karst and its hypoglycemic potency on streptozotocin-induced type 2 diabetic mice. Journal of Agricultural and Food Chemistry 59: 6492–6500. 10.1021/jf200527y [DOI] [PubMed] [Google Scholar]
- Thawthong A, Hapuarachchi KK, Wen TC, Raspé O, Thongklang N, Kang JC, Hyde KD. (2017) Ganoderma sichuanense (Ganodermataceae, Polyporales) new to Thailand. MycoKeys 22: 27–43. 10.3897/mycokeys.22.13083 [DOI] [Google Scholar]
- Tulloss RE. (2005) Amaniteae: Amanita, Limacella, & Torrendia. by Pierre Neville & Serge Poumarat, etc. (Book Review). Mycotaxon 92: 474–484. [Google Scholar]
- Vilgalys R, Hester M. (1990) Rapid genetic identification and mapping of enzymatically amplified ribosomal DNA from several Cryptococcus species. Journal of Bacteriology 172(8): 4238–4246. 10.1128/jb.172.8.4238-4246.1990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang F, Liu JK. (2008) Highly oxygenated lanostane triterpenoids from the fungus Ganoderma applanatum. Chemical and Pharmaceutical Bulletin 56: 1035–1037. 10.1248/cpb.56.1035 [DOI] [PubMed] [Google Scholar]
- Welti S, Courtecuisse R. (2010) The Ganoderma taceae, in the French West Indies (Guadeloupe and Martinique). Fungal Diversity 43: 103–126. 10.1007/s13225-010-0036-2 [DOI] [Google Scholar]
- White TJ, Bruns T, Lee S, Taylor JW. (1990) Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In: Innis MA, Gelfand DH, Sninsky JJ, White TJ. (Eds) PCR Protocols: a Guide to Methods and Applications.Academic Press, New York, 315–322. 10.1016/B978-0-12-372180-8.50042-1 [DOI]
- Xing JH, Song J, Decock C, Cui BK. (2016) Morphological characters and phylogenetic analysis reveal a new species within the Ganoderma lucidum complex from South Africa. Phytotaxa 266: 115–124. 10.11646/phytotaxa.266.2.5 [DOI] [Google Scholar]
- Xing JH, Sun YF, Han YL, Cui BK, Dai YC. (2018) Morphological and molecular identification of two new Ganoderma species on Casuarina equisetifolia from China. MycoKeys 34: 93–108. 10.3897/mycokeys.34.22593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao YJ, Wang XC, Wang B. (2013) Epitypification of Ganoderma sichuanense J.D. Zhao and X.Q. Zhang (Ganodermataceae). Taxon 62(5): 1025–1031. 10.12705/625.10 [DOI] [Google Scholar]
- Zakaria L, Ali NS, Salleh B. (2009) Molecular analysis of Ganoderma species from different hosts in Peninsular Malaysia. Journal of Biological Sciences 9: 12–20. 10.3923/jbs.2009.12.20 [DOI] [Google Scholar]
- Zhao JD. (1989) The Ganodermataceae in China. Bibliotheca Mycologica 132: 1–176. [Google Scholar]
- Zhaxybayeva O, Gogarten JP. (2002) Bootstrap, Bayesian probability and maximum likelihood mapping: exploring new tools for comparative genome analyses. BMC Genomics 3(1): 4 10.1186/1471-2164-3-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou LW, Cao Y, Wu SH, Vlasák J, Li DW, Li MJ, Dai YC. (2015) Global diversity of the Ganoderma lucidum complex (Ganoderma taceae, Polyporales) inferred from morphology and multilocus phylogeny. Phytochemistry 114: 7–15. 10.1016/j.phytochem.2014.09.023 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.