Exacerbations of chronic Pseudomonas aeruginosa infections are a major treatment challenge in cystic fibrosis due to biofilm formation and hypermutation. We aimed to evaluate different dosage regimens of meropenem and tobramycin as monotherapies and in combination against hypermutable carbapenem-resistant P. aeruginosa. A hypermutable P. aeruginosa isolate (meropenem and tobramycin MICs, 8 mg/liter) was investigated in the dynamic CDC biofilm reactor over 120 h.
KEYWORDS: combination therapy, hypermutators, biofilm infections, antibiotic resistance, dosage regimens
ABSTRACT
Exacerbations of chronic Pseudomonas aeruginosa infections are a major treatment challenge in cystic fibrosis due to biofilm formation and hypermutation. We aimed to evaluate different dosage regimens of meropenem and tobramycin as monotherapies and in combination against hypermutable carbapenem-resistant P. aeruginosa. A hypermutable P. aeruginosa isolate (meropenem and tobramycin MICs, 8 mg/liter) was investigated in the dynamic CDC biofilm reactor over 120 h. Regimens were meropenem as the standard (2 g every 8 h, 30% epithelial lining fluid [ELF] penetration) and as a continuous infusion (CI; 6 g/day, 30% and 60% ELF penetration) and tobramycin at 10 mg/kg of body weight every 24 h (50% ELF penetration). The time courses of totally susceptible and less-susceptible bacteria and MICs were determined, and antibiotic concentrations were quantified by liquid chromatography-tandem mass spectrometry. All monotherapies failed, with the substantial regrowth of planktonic (>6 log10 CFU/ml) and biofilm (≥6 log10 CFU/cm2) bacteria occurring. Except for the meropenem CI (60% ELF penetration), all monotherapies amplified less-susceptible planktonic and biofilm bacteria by 120 h. The meropenem standard regimen with tobramycin caused initial killing followed by considerable regrowth with resistance (meropenem MIC, 64 mg/liter; tobramycin MIC, 32 mg/liter) for planktonic and biofilm bacteria. The combination containing the meropenem CI at both levels of ELF penetration synergistically suppressed the regrowth of total planktonic bacteria and the resistance of planktonic and biofilm bacteria. The combination with the meropenem CI at 60% ELF penetration, in addition, synergistically suppressed the regrowth of total biofilm bacteria. Standard regimens of meropenem and tobramycin were ineffective against planktonic and biofilm bacteria. The combination with meropenem CI exhibited enhanced bacterial killing and resistance suppression of carbapenem-resistant hypermutable P. aeruginosa.
TEXT
Carbapenem-resistant Pseudomonas aeruginosa has been classified by the World Health Organization as one of the top three critical pathogens requiring new antibiotic treatments (1). P. aeruginosa has a particularly large armamentarium of resistance mechanisms and can develop resistance against virtually all antibiotics in monotherapy. Such resistance development often results in treatment failure (2, 3). Respiratory infections caused by P. aeruginosa are a serious clinical problem for patients with cystic fibrosis (CF). Acute infective exacerbations (AIE) of chronic P. aeruginosa infections cause progressive lung function decline followed by respiratory failure (4, 5). Thus, they are a main driver of early death for patients with CF. Indeed, the rates of multidrug-resistance (MDR) for these infections in CF patients are substantially higher than those in patients in an intensive care unit (6).
The ability of P. aeruginosa to become hypermutable and to form a biofilm renders AIE especially difficult to treat (7, 8). Hypermutable isolates (i.e., those with an up to ∼1,000-fold increased mutation rate due to defects in DNA repair or error avoidance systems) account for up to ∼54% of P. aeruginosa strains in CF respiratory infections and are associated with reduced lung function (8–10). Despite an increased mutation rate, hypermutable strains generally do not show reduced fitness in the nutrient-rich environment of the CF lung. Their carriage is highly correlated with MDR, and hypermutation is important for biofilm development (8, 11). The formation of a biofilm hampers antibiotic effectiveness, e.g., via extracellular matrix formation, which reduces antibiotic penetration. Biofilm growth also leads to greater phenotypic diversity and, thus, a greater persistence of infections (12, 13). Therefore, biofilm-associated infections by hypermutable P. aeruginosa are extremely difficult to treat, especially when the hypermutable strains are MDR.
Current antibiotic regimens against P. aeruginosa infections in patients with CF are suboptimal; monotherapy is often ineffective, and combination regimens are used empirically (4, 14). We have demonstrated synergistic bacterial killing and the suppression of resistance emergence in hypermutable P. aeruginosa with a modified combination regimen of meropenem and tobramycin in the dynamic hollow-fiber infection model (HFIM) (15). However, treatment regimens against hypermutable P. aeruginosa have never been evaluated in a dynamic biofilm model, such as the Centers for Disease Control and Prevention biofilm reactor (CBR), that allows examination of antibacterial effects against both planktonic and biofilm bacteria. Thus, the aim of the present study was to use the CBR to simulate the concentration-time profiles for different meropenem and tobramycin dosage regimens observed in the epithelial lining fluid (ELF) of patients with CF when given as monotherapy and in combination and characterize the bacterial killing and resistance suppression of carbapenem-resistant hypermutable P. aeruginosa.
RESULTS
Pharmacokinetic validation, bacterial killing, and emergence of resistance.
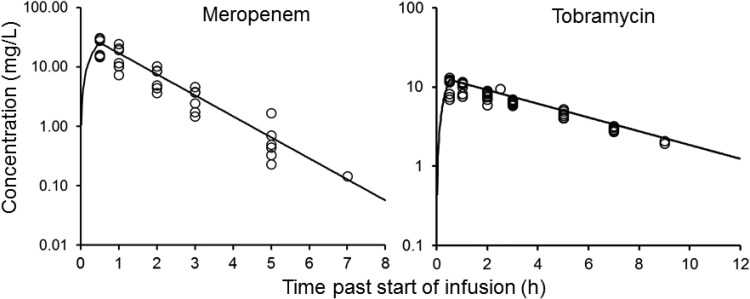
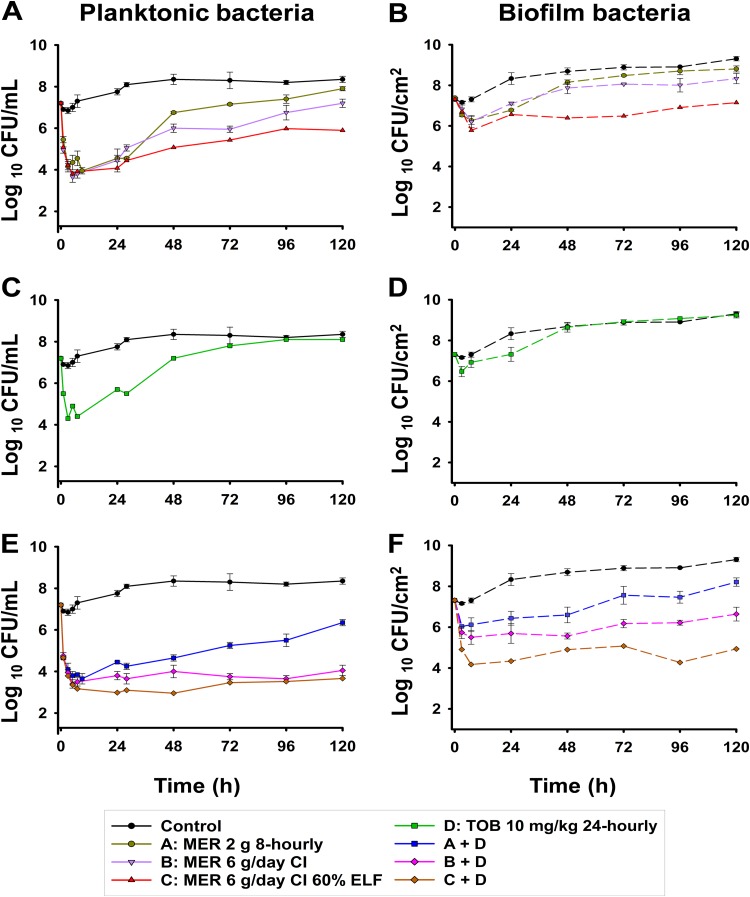
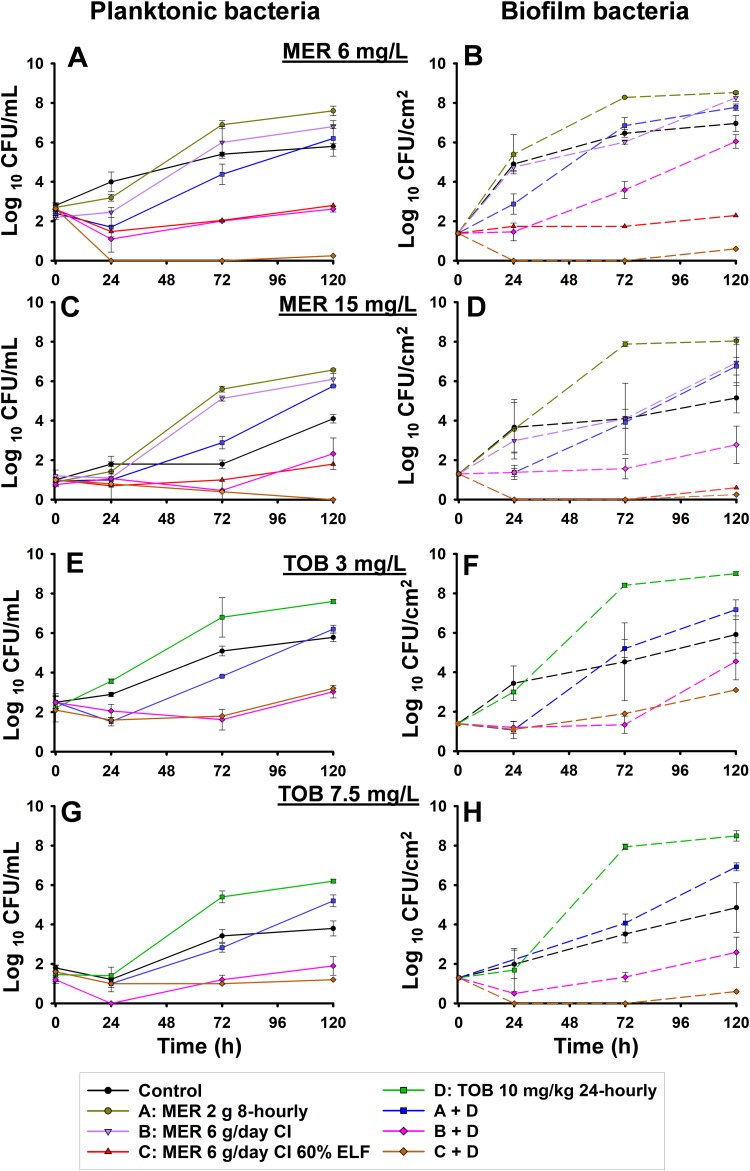
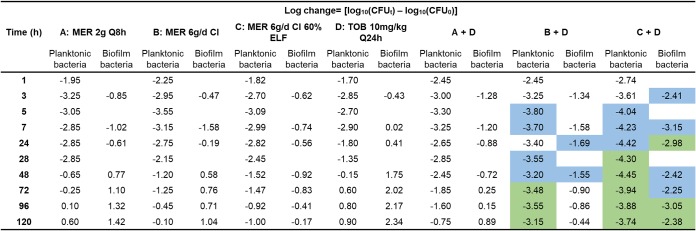
The pharmacokinetic profiles observed in the CBR (Fig. 1) were in good agreement with the targeted exposures (Table 1). The observed meropenem and tobramycin concentrations were, on average, within 10% of the targeted concentrations. The viable count profiles for planktonic and biofilm bacteria are presented in Fig. 2. The counts on antibiotic-containing agar are shown in Fig. 3, log changes in viable counts are shown in Table 2, mutation frequencies are shown in Table 3, and baseline and endpoint MICs are shown in Table 4.
FIG 1.
Pharmacokinetic profiles showing the relationship between the targeted (lines) and measured (symbols) meropenem and tobramycin concentrations in the CBR.
TABLE 1.
Clinically representative ELF concentrations, exposures, and pharmacokinetic/pharmacodynamic indices for meropenem and/or tobramycin in the CBRa
| Treatment | fCmax/fCmin or fCss (mg/liter) | fAUC24 (mg·h/liter) | fCmax/MIC | fT>MIC (%) | fAUC24/MIC |
|---|---|---|---|---|---|
| MER at 2 g every 8 h | 25.4/0.06 | 115 | 3.18 | 22 | |
| MER at 6 g/day as a CI | 4.79 | 115 | 0 | 0 | |
| MER at 6 g/day as a CI (60% ELF penetration) | 9.58 | 230 | 1.20 | 100 | |
| TOB at 10 mg/kg every 24 h | 12.3/0.11 | 64.4 | 1.54 | 8.05 |
MER, meropenem; TOB, tobramycin; CI, continuous infusion; fCmax, unbound maximum concentration; fCmin, unbound minimum concentration before the next dose; fCss, unbound average steady-state concentration; fAUC24, the area under the unbound concentration-time curve over 24 h; fCmax/MIC, the ratio of the fCmax to the MIC; fT>MIC, the cumulative percentage of a 24-h period that the unbound concentrations exceeded 1× MIC; fAUC24/MIC, the ratio of the fAUC24 to the MIC. The simulated half-lives were 0.8 h for meropenem and 3.5 h for tobramycin. No loading dose was administered for intermittent dosing, whereas the modified meropenem dosage regimen (6-g/day CI) was started at the fCss of either 4.79 mg/liter (30% ELF penetration) or 9.58 mg/liter (60% ELF penetration). The simulated ELF penetration was 30% for meropenem, unless it is specified to be 60%, and was 50% for tobramycin.
FIG 2.
Total viable counts for the growth control and treatments with meropenem (MER) and/or tobramycin (TOB) at clinically relevant ELF concentration-time profiles sampled from the medium within the reactor, i.e., planktonic bacteria receiving meropenem monotherapy (A), tobramycin monotherapy (C), or the combination of meropenem and tobramycin (E), and from coupons, i.e., biofilm bacteria receiving meropenem monotherapy (B), tobramycin monotherapy (D), or the combination of meropenem and tobramycin (F). The results are presented as the average ± SE. The y axis starts from the limit of counting.
FIG 3.
Effect of each dosage regimen on the counts of bacteria able to grow on agar plates containing 6 or 15 mg/liter of meropenem or 3 or 7.5 mg/liter of tobramycin. The results are represented as the average ± SE. To differentiate less-susceptible subpopulations from the predominant population, the antibiotic concentrations in agar were based upon Etest MICs, which were 1.5 mg/liter for meropenem and 0.75 mg/liter for tobramycin (9).
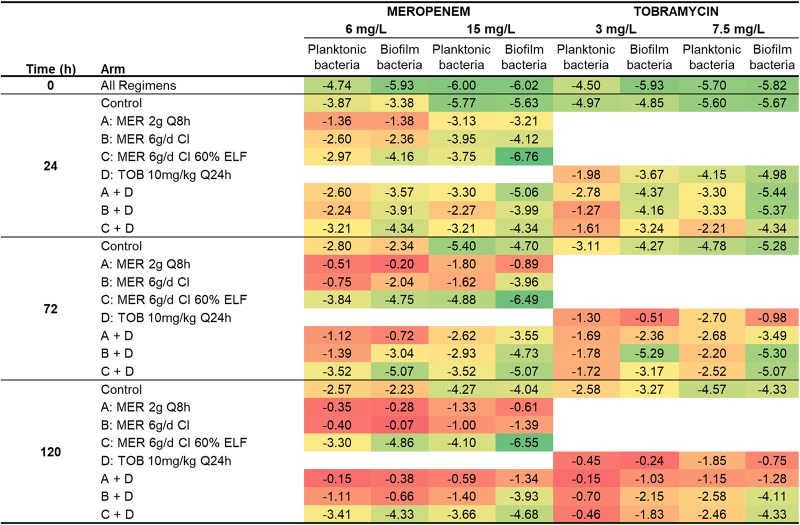
TABLE 2.
Log changes in viable cell counts at various time points with clinically relevant ELF concentrations of meropenem and/or tobramycina
MER, meropenem; TOB, tobramycin; CFUt, number of CFU at time t; CFU0, number of CFU at time zero. The green background indicates synergy (a ≥2-log10 decrease in the number of CFU per milliliter or the number of CFU per square centimeter with the combination compared to the value for its most active component); the blue background indicates a 1.0- to <2-log10 decrease in the number of CFU per milliliter or the number of CFU per square centimeter with the combination compared to the value for its most active component.
TABLE 3.
Log10 mutation frequencies at 6 mg/liter and 15 mg/liter meropenem and 3 mg/liter and 7.5 mg/liter tobramycin for each simulated regimen
MER, meropenem; TOB, tobramycin. The red background indicates a high mutation frequency, i.e., a large proportion of less-susceptible bacteria being present in the total population; the green background indicates a low mutation frequency, i.e., a small proportion of less-susceptible bacteria being present in the total population.
TABLE 4.
MIC values for colonies obtained from antibiotic-containing agar plates at 0 and 120 h for each dosage regimena
| Treatment | Meropenem at 15 mg/liter |
Tobramycin at 7.5 mg/liter |
||||
|---|---|---|---|---|---|---|
| Time (h) | MIC (mg/liter) |
Time (h) | MIC (mg/liter) |
|||
| Planktonic bacteria | Biofilm bacteria | Planktonic bacteria | Biofilm bacteria | |||
| Control | 0 | 16 | 8 | 0 | 16 | 8 |
| 120 | 32 | 16 | 120 | 32 | 16 | |
| MER at 2 g every 8 h | 120 | 128 | 32 | — | — | — |
| MER at 6 g/day as a CI | 120 | 64 | 32 | — | — | — |
| MER at 6 g/day as a CI (60% ELF penetration) | 120 | 16 | NC | — | — | — |
| TOB at 10 mg/kg every 24 h | — | — | — | 120 | 64 | 32 |
| MER at 2 g every 8 h + TOB at 10 mg/kg every 24 h | 120 | 64 | 32 | 120 | 32 | 32 |
| MER at 6 g/day as a CI + TOB at 10 mg/kg every 24 h | 120 | 32 | 16 | 120 | 32 | 16 |
| MER at 6 g/day as a CI (60% ELF penetration) + TOB at 10 mg/kg every 24 h | 120 | NC | NC | 120 | NC | NC |
The agar plates contained meropenem (MER) at 15 mg/liter and tobramycin (TOB) at 7.5 mg/liter. NC, no colonies grew on the antibiotic-containing plates; —, not tested.
Planktonic bacteria.
The starting inoculum (mean ± standard error [SE]) in all arms was 7.19 ± 0.05 log10 CFU/ml (n = 14). P. aeruginosa CW8 in the control chambers grew to 7.75 ± 0.15 log10 CFU/ml by 24 h, and the growth plateaued at ∼8.1 log10 CFU/ml (Fig. 2A). Colonies on drug-containing agar increased approximately in proportion to the growth of the total bacterial population (Fig. 3A, C, E, and G and Table 3).
Meropenem monotherapy simulating the standard regimen (2 g every 8 h) resulted in ∼3-log10-CFU/ml initial killing at 3 h, followed by steady regrowth to ∼7 log10 CFU/ml at 48 h, with slower regrowth toward control values occurring thereafter (Fig. 2A and Table 2). With this regimen, less-susceptible populations increased rapidly, with approximately half of the entire population growing on agar containing meropenem at 6 mg/liter by 120 h. Similar increases were observed on agar containing meropenem at 15 mg/liter, although growth remained ∼1 log lower than the total population at 120 h. Emergence of resistance was observed, with an ∼2.5-log increase of the meropenem-resistant bacteria compared to that for the growth control being seen at 120 h (Fig. 3A and C and Table 3). The MIC of colonies recovered from 15-mg/liter meropenem-containing plates at this time point was 128 mg/liter (Table 4). The bacterial killing achieved with the modified meropenem regimen at 6 g/day as a continuous infusion (CI) simulating 30% and 60% ELF penetration closely matched that achieved with the standard regimen (2 g every 8 h) over the first 24 h, but thereafter growth remained ∼1 log10 CFU/ml (30% ELF penetration) and ∼2 log10 CFU/ml (60% ELF penetration) lower than that for the standard regimen; for the treatment simulating 60% ELF penetration, regrowth at 120 h reached ∼6 log10 CFU/ml (Fig. 2A and Table 2). While the increases in less-susceptible populations with the CI simulating 30% ELF penetration closely matched those for the standard regimen, the less-susceptible populations remained suppressed for 60% ELF penetration (Fig. 3A and C and Table 3). By 72 h with the latter treatment, growth on agar containing meropenem at 6 mg/liter and 15 mg/liter was ∼2 and <1 log10 CFU/ml, respectively. Furthermore, the emergence of resistance was not observed at 120 h, and the MICs of colonies recovered from drug plates were increased by only 1 dilution (MIC, 16 mg/liter) (Table 4). Amplification of resistance with meropenem 30% ELF penetration was more evident, with an ∼2.1-log increase in the meropenem-resistant population at 120 h in comparison to that for the growth control (Fig. 3C and Table 3). The MIC at this time point was 64 mg/liter (Table 4). The tobramycin monotherapy produced rapid (within the first 7 h) initial killing of ∼3 log10 CFU/ml, followed by steady regrowth, such that growth approximated that of the growth control by 72 h (Fig. 2C and Table 2). Amplification of bacteria less susceptible to tobramycin was observed. The proportion of colonies growing on tobramycin-containing agar increased substantially over 120 h, with a large proportion of the entire population growing on plates containing 3 mg/liter tobramycin; on plates containing 7.5 mg/liter tobramycin, growth increased to within ∼2 log10 CFU/ml of the total population (Fig. 3E and G and Table 3). The MIC at 120 h was 64 mg/liter (Table 4).
The combination containing the standard meropenem regimen produced ∼3.3-log10-CFU/ml initial killing at 5 h and regrowth to within ∼2 log10 CFU/ml of the growth of the growth control at 120 h (Fig. 2E and Table 2). The amplification of less-susceptible and resistant bacteria in comparison to that of the growth control was observed. Less-susceptible populations increased dramatically, such that virtually the entire population at 120 h grew on agar containing meropenem at 6 mg/liter and tobramycin at 3 mg/liter (Fig. 3A and E and Table 3). The MIC at this time point was 64 mg/liter for meropenem and 32 mg/liter for tobramycin (Table 4).
The combination treatment with the modified meropenem regimen with 30% ELF penetration produced initial killing of ∼3.8 log10 CFU/ml at 5 h, while that with 60% ELF penetration produced initial killing of ∼4.4 log10 CFU/ml at 24 h. Synergistic bacterial killing (≥2 log10 CFU/ml) was observed from 28 or 72 h onwards with both levels of ELF penetration; regrowth remained suppressed at ∼3.5 to 4 log10 CFU/ml at 120 h (Fig. 2E and Table 2). However, differences in the regrowth of less-susceptible populations were observed. On agar containing meropenem at 15 mg/liter, regrowth of ∼2 log10 CFU/ml was observed at 120 h with the combination simulating 30% ELF penetration of the meropenem CI (MIC, 32 mg/liter). However, virtually no colonies were detected from 24 h onwards at either meropenem plate concentration with the combination simulating 60% ELF penetration of meropenem. Growth on agar containing tobramycin at 7.5 mg/liter was ∼1.5 to 2 log10 CFU/ml at 120 h for both combination treatments involving the meropenem CI, which was ∼2 log10 CFU/ml below the growth control counts (Fig. 3G).
Biofilm-embedded bacteria.
In the growth control, biofilm bacteria grew steadily to ∼9 log10 CFU/cm2 by 72 h and plateaued until 120 h (Fig. 2B); moderate increases in the proportion of less-susceptible populations were observed across 120 h (Fig. 3B, D, F, and H and Table 3). Following 7 h of treatment, bacterial killing from all monotherapy regimens was ∼1 log10 CFU/cm2. After that, regrowth occurred with all regimens except the meropenem CI with 60% ELF penetration, such that growth was within ≤2 log10 CFU/cm2 of that of the control from 48 h onwards; with the CI (60% ELF penetration), growth remained at ∼6.5 to 7.0 log10 CFU/cm2 across 120 h (Fig. 2B and D and Table 2). Substantial increases in less-susceptible populations occurred with the tobramycin and both the standard and CI (30% ELF penetration) meropenem regimens but not with the CI (60% ELF penetration) meropenem regimen (Fig. 3B, D, F, and H and Table 3). With the latter regimen, growth on agar containing meropenem at 6 mg/liter remained steady at ∼2 log10 CFU/cm2 across 120 h, whereas virtually no colonies were detected on agar containing meropenem at 15 mg/liter from 24 h onwards. At 120 h, the MIC was 32 mg/liter for the meropenem standard regimen and the CI at 30% ELF penetration; the MIC for the tobramycin regimen was also 32 mg/liter (Table 4).
The combination containing the standard meropenem regimen simulating 30% ELF penetration produced ∼1-log10-CFU/cm2 initial killing at 3 h, followed by regrowth to within ∼1 log10 CFU/cm2 of the growth of the growth control at 120 h (Fig. 2F and Table 2). Amplification of bacteria less susceptible to meropenem and tobramycin in comparison to that of the growth control was observed (Fig. 3B, D, F, and H and Table 3). The MIC for both meropenem and tobramycin was 32 mg/liter at 120 h (Table 4). In contrast, with the combination regimens simulating the meropenem CI at 30% or 60% ELF penetration, a more substantial antibacterial effect was observed. With the meropenem CI at 30% ELF penetration, biofilm bacteria remained suppressed below ∼6 log10 CFU/cm2 up to 48 h, with only ∼1 log10 CFU/cm2 of growth occurring thereafter (Fig. 2F and Table 2). At 24 and 48 h, bacterial counts were 1 to 2 log10 CFU/cm2 lower for the combination than for the most active monotherapy. Bacteria less susceptible to meropenem and tobramycin grew to within ∼2 log10 CFU/cm2 of the growth of the growth control by 120 h (Fig. 3B, D, F, and H). The MIC for meropenem and tobramycin was 16 mg/liter at this time point (Table 4). The combination simulating the meropenem CI at 60% ELF penetration achieved ∼2.3- to 3.2-log10-CFU/cm2 bacterial killing from 7 h onwards (Fig. 2F and Table 2); synergistic bacterial killing (≥2 log10 CFU/cm2 compared to the killing obtained with the most active monotherapy) was observed at 24, 96, and 120 h (Fig. 2B and F and Table 2). A negligible number (∼0.5 log10 CFU/cm2) of colonies was observed on plates containing tobramycin at 7.5 mg/liter or meropenem at 6 or 15 mg/liter (Fig. 3B, D, F, and H).
DISCUSSION
This study systematically investigated the bacterial killing and resistance suppression of standard versus modified dosage regimens of meropenem and tobramycin against a carbapenem-resistant clinical hypermutable P. aeruginosa isolate in the CBR. The pharmacokinetic profiles simulated were representative of the unbound antibiotic concentrations expected in the ELF of patients with CF.
Pharmacokinetic/pharmacodynamic approaches to optimize the administration of β-lactams, including meropenem, traditionally involve maximizing the cumulative percentage of a 24-h period that the unbound concentrations exceed 1× MIC (fT>MIC) for the infecting pathogen (16, 17). For serious bacterial infections, targets such as 100% fT>4–5×MIC (18–20) have been proposed. The fT>MIC can be modulated by altering the mode of administration. In the present investigations, meropenem was delivered either as a short-term infusion (standard regimen) or as a continuous infusion (CI; modified regimen), representing the extreme modes of administration in clinical practice. Importantly, however, the above-mentioned targets relate to planktonic bacteria; targets for biofilm bacteria are yet to be established and likely to be higher.
In our CBR studies, all meropenem regimens in monotherapy, at both levels of ELF penetration, were unable to suppress regrowth to below ∼6 log10 for both planktonic and biofilm bacteria (Fig. 2). Even the CI with 60% ELF penetration was not successful, despite achieving a 100% fT>MIC. This comprehensive failure of meropenem regimens strongly argues against the use of meropenem as monotherapy against a meropenem-resistant hypermutable P. aeruginosa isolate. For a meropenem-susceptible hypermutable P. aeruginosa strain, we have previously demonstrated in a 10-day HFIM study that the CI of meropenem to achieve a concentration as high as ∼8× MIC was unable to suppress the emergence of less-susceptible planktonic bacteria (15). In the current study, biofilm bacteria were more resilient to meropenem than planktonic bacteria (Fig. 2). The bacterial cells in a biofilm are difficult to kill because of multiple factors. This includes the low metabolic activity of subpopulations located in the inner parts of the biofilm; e.g., low peptidoglycan production affects bacterial killing by meropenem (21). In a recent study of P. aeruginosa in a biofilm, meropenem concentrations substantially higher than those expected in ELF achieved some activity against meropenem-susceptible strains, while there was no activity against a meropenem-resistant strain (22).
For aminoglycosides, such as tobramycin, the ratio of exposure across a 24-h period to the MIC (the area under the unbound concentration-time curve [fAUC]/MIC) and the ratio of the unbound maximum concentration (fCmax) to the MIC (fCmax/MIC) have been correlated with antibacterial activity. An fAUC/MIC of >70 and an fCmax/MIC of 8 to 10 have been proposed as clinical targets (23); in the present study, the corresponding values were ∼8 and ∼1.5 (Table 1). For cationic antimicrobials, such as tobramycin, the presence of extracellular DNA in the biofilm decreases activity via chelation (24, 25). Thus, it is not surprising that the tobramycin regimen was ineffective in suppressing regrowth, resulting in a large increase of less-susceptible planktonic and biofilm bacteria (Fig. 2 and 3). This result was in agreement with that of our previous in vitro study, where tobramycin monotherapy failed to inhibit the regrowth of planktonic hypermutable P. aeruginosa even for fAUC/MIC values of 72 and 168 (26).
In the CBR, the combination containing the standard meropenem regimen (30% ELF penetration) with tobramycin resulted in a significant regrowth of less-susceptible planktonic and biofilm bacteria. Total bacterial counts were within 1 log of those achieved with the most active monotherapy at each time point. Importantly, the combination containing the modified meropenem (CI) regimen achieved enhanced bacterial killing and resistance suppression. This combination suppressed the regrowth and resistance of planktonic bacteria over 5 days at both levels of ELF penetration, demonstrating clear synergy. Although some regrowth of total biofilm bacteria was observed from 48 h onwards when simulating 30% ELF penetration of meropenem, resistant subpopulations remained suppressed compared to the growth control over 5 days. It is not surprising that the combination with standard meropenem dosing was less effective than that with the meropenem CI, as in the former case there were substantial periods with essentially no antibiotic present for activity. The combination simulating 60% ELF penetration synergistically suppressed the regrowth of both total and resistant biofilm bacteria over 5 days. The synergy observed was notable, given that the isolate was meropenem resistant and tobramycin intermediate.
The synergistic bacterial killing and suppression of resistance observed in our study may be due to the different mechanisms of action and resistance of each antibiotic. Meropenem inhibits cell wall synthesis via binding to penicillin-binding proteins (27), and the main mechanisms of resistance in P. aeruginosa involve AmpC β-lactamase overexpression, reduced outer membrane porin OprD, and enzymatic inactivation via carbapenemases (28). Tobramycin predominantly acts by protein synthesis inhibition (29) but also by the disruption of the outer bacterial membrane (30–32). Resistance mechanisms against tobramycin include target site modification, enzymatic cleavage, increased expression of MexXY-OprM, and reduced outer membrane permeability (33, 34). For CW8, we previously identified mutations in genes related to both meropenem (oprD, ampC, ampR) and tobramycin (fusA1) resistance (9). Although the effects of each of the antibiotics in the combination were attenuated due to resistance, our results indicate that exposing the bacteria to both antibiotics simultaneously had a beneficial effect. In addition, mechanistic synergy may have been caused by tobramycin enhancing the target site penetration of meropenem (15). We have demonstrated that tobramycin in combination with another carbapenem, imipenem, caused extensive ultrastructural disruption of the outer membrane (32). This mechanistic synergy may apply not only to planktonic bacteria but also to biofilm bacteria.
To the best of our knowledge, this is the first study to examine the activity of the meropenem and tobramycin combination against carbapenem-resistant hypermutable P. aeruginosa in planktonic and biofilm growth by simulating ELF pharmacokinetics. Two studies in the dynamic HFIM (15, 18) and one in vivo infection model study (35) also examined the activity of this combination against P. aeruginosa. It is important to note that these studies involved susceptible isolates and investigated only planktonic growth, and the in vitro studies represented plasma rather than ELF concentrations. Two static concentration time-kill studies previously examined the activity of the same combination at a range of concentrations against susceptible planktonic P. aeruginosa (36, 37). The effect of the meropenem-tobramycin combination on the biofilm biomass of a susceptible P. aeruginosa strain has also been studied using a dynamic flow cell model and microscopy (38). However, that study did not quantify the counts of either biofilm or planktonic bacteria, nor did it examine the emergence of resistance, and the concentrations were higher than those achievable in ELF after intravenous dosing.
The current study has a number of strengths. It is the first study to examine the activity of the meropenem-tobramycin combination against a carbapenem-resistant hypermutable P. aeruginosa isolate. Furthermore, this is the only dynamic in vitro study to utilize concentration-time profiles representative of those in ELF for this combination. Since different levels of ELF penetration of meropenem have been reported (39–41), we examined both 30% and 60% ELF penetration. Importantly, the combination with the modified meropenem regimen achieved enhanced bacterial killing and resistance suppression even at the low ELF penetration. This study was conducted over 5 days of treatment, quantified both biofilm and planktonic bacteria, and evaluated the emergence of resistance. In addition, multiple biological replicates were performed to demonstrate the reproducibility of the total and less-susceptible bacterial counts. It is also important to note some limitations. The comprehensive studies conducted involved one clinical P. aeruginosa isolate. Future studies may be directed at evaluating the effect of the combination against other isolates, investigating its effect on the biofilm structure via confocal microscopy, and developing a mechanism-based mathematical model for antibiotic effects on biofilm bacteria. In addition, as with all other in vitro models, the CBR lacks an immune system. Therefore, future animal studies may be warranted to assess immune system effects on residual populations following the initial bacterial killing by the antibiotics. However, the accurate representation of humanized pharmacokinetic profiles is challenging in animal models due to the differences in clearance and half-life. In addition, given the ethical limitations on study duration inherent in animal studies, suppression of the emergence of resistance, a key component of the present study, is best examined in the CBR, where longer study durations can be employed.
In conclusion, standard regimens of meropenem and tobramycin, both as monotherapy and in combination, were ineffective in suppressing regrowth and the emergence of resistance in both planktonic and biofilm bacteria. Importantly, however, the combination with the meropenem continuous infusion regimen, at both levels of ELF penetration, exhibited enhanced bacterial killing and resistance suppression against carbapenem-resistant hypermutable P. aeruginosa. Thus, this promising combination regimen warrants further evaluation.
MATERIALS AND METHODS
Bacterial isolate, antibiotics, and MICs.
A previously described clinical hypermutable P. aeruginosa isolate (CW8) was employed (9). Hypermutability was defined as a mutation frequency on rifampin-containing agar at least 20-fold higher than that obtained for the control strain, PAO1 (9, 10). Sterile stock solutions of meropenem (lot Maus1025; Kabi, Melbourne, Australia) and tobramycin (lot LC24138; AK Scientific, Union City, MD, USA) were prepared in Milli-Q water immediately prior to each experiment. The MICs determined in duplicate on separate days using agar dilution per Clinical and Laboratory Standards Institute (CLSI) guidelines were 8 mg/liter for each antibiotic (42). Susceptibility and resistance were defined as MICs of ≤2 mg/liter and ≥8 mg/liter, respectively, for meropenem, and ≤4 mg/liter and ≥16 mg/liter, respectively, for tobramycin, per CLSI guidelines (42). The isolate was resistant to meropenem, intermediate to tobramycin, and MDR, based on agar dilution MICs. MDR was defined as nonsusceptibility to at least one antimicrobial agent in three or more antimicrobial categories (43). The biofilm formation capacity of CW8 was confirmed by the crystal violet assay.
In vitro dynamic biofilm model, quantification of bacterial killing, emergence of resistance, and dosage regimens.
The time courses of bacterial killing and the emergence of resistance of planktonic and biofilm-embedded bacteria for the standard and modified regimens of meropenem and tobramycin as monotherapy and in combination were investigated over 120 h using the CBR (Bio Surface Technologies, Bozeman, MT, USA). The CBR model consisted of a 1-liter glass reactor connected to a 10-liter carboy containing sterile drug-free cation-adjusted Mueller-Hinton broth (CAMHB; BD, Sparks, MD, USA) containing 25 mg/liter Ca2+, 12.5 mg/liter Mg2+, and 1% tryptic soy broth (TSB) (CAMHB–1% TSB). Broth was pumped through the model (broth volume in the reactor, 350 ml), along with mixing, and shear was generated by a magnetic stir bar operating at 130 rpm. A hot plate maintained the CAMHB–1% TSB at 35°C. The biofilm formed on removable polycarbonate coupons (diameter, 12.7 mm) located in eight polypropylene coupon holders suspended from the reactor lid (three coupons per holder); the total surface area of each coupon was 2.53 cm2.
The protocol for biofilm growth was similar to that described in our previous study (44). Prior to each experiment, CW8 was subcultured onto cation-adjusted Mueller-Hinton agar (CAMHA) containing 25 mg/liter Ca2+ and 12.5 mg/liter Mg2+ (Media Preparation Unit, University of Melbourne, Melbourne, Australia) and incubated at 35°C for 48 h. Following incubation, 2 or 3 random colonies were selected and grown overnight in 10 ml TSB, from which early-log-phase growth was obtained. A 28-h conditioning phase was then commenced via inoculation of 1 ml of this suspension into the model. Conditioning initially involved 24 h of incubation in drug-free CAMHB–1% TSB. Subsequently, all CAMHB–1% TSB was removed (to expel all planktonic bacteria and allow the amplification of bacteria shedding from the biofilm) and the reactor was refilled with drug-free CAMHB–1% TSB, pumped through the model for 4 h (flow rate, 11.67 ml/min) prior to the commencement of antibiotic treatment (i.e., 0 h) (44). The presence of biofilm on the coupons was confirmed by electron microscopy at 0 h.
At 0 h, the flow rate was changed to 4.9 ml/min for all treatments, to simulate a meropenem ELF elimination half-life (t1/2) of ∼0.8 h, reflecting that in patients with CF (45). For tobramycin-containing treatments, tobramycin was supplemented to achieve the required ELF t1/2 of ∼3.5 h (46, 47). For intermittent infusions, the antibiotics were administered using syringe drivers. The meropenem CI was achieved by administering a loading dose at 0 h directly into the reactor to immediately attain the required steady-state ELF concentration and spiking the meropenem stock solution into the carboy to maintain the steady-state concentration. Meropenem and tobramycin have negligible plasma protein binding; therefore, the concentrations used represent unbound (free) concentrations (Table 1). For meropenem, two regimens utilizing the highest FDA-recommended daily dose (6 g/day) were selected: the standard regimen of 2 g three times daily via a 30-min intravenous infusion with an fCmax of 25.3 mg/liter (Table 1) and a modified regimen of 6 g/day as a continuous infusion (CI) with a loading dose to rapidly achieve the unbound average steady-state concentration(fCss; Table 1). The modified regimen was simulated for both 30% and 60% ELF penetration. The pharmacokinetic profiles simulated in the CBR were based on the antibiotic concentrations over time that would be expected in the ELF of CF patients given the respective regimens. These expected unbound antibiotic concentration-time profiles were simulated in silico using the Berkeley Madonna (version 8.3.18) program (48), based on clinical studies and population pharmacokinetic models for CF patients (45, 46). The rate and extent of penetration of meropenem (30%) and tobramycin (50%) into ELF were derived from multiple published studies in patients (39, 41, 47). For meropenem, a higher ELF penetration (60%) based on a healthy volunteer study (40) was also considered. For tobramycin, the highest FDA-recommended daily dose for CF patients (10 mg/kg of body weight) was administered as a 30-min intravenous infusion every 24 h to yield the area under the unbound concentration-time curve over 24 h (fAUC24) of 64.4 mg·h/liter (Table 1). A growth control was also included. With one exception, all control and drug-containing regimens were performed in two replicates. Syringe drivers were tested and flow rates through the CBR were calibrated prior to each study and monitored throughout to ensure that the system was performing optimally.
Samples for viable counting were collected at 0, 1, 2, 3, 5, 7, 24, 28, 48, 72, 96, and 120 h for planktonic bacteria (1 ml) and at 0, 3, 7, 24, 48, 72, 96, and 120 h for biofilm-embedded bacteria. For biofilm bacteria, a coupon holder containing three coupons was aseptically replaced with a blank holder at each time point. The removed coupons were rinsed twice in 10 ml of phosphate-buffered saline (PBS; pH 7.4) to remove planktonic cells and then placed in sterile tubes containing 10 ml PBS. Three alternating 1-min cycles of vortexing and sonication at 43 kHz followed by a final 1 min of vortexing were used to extract the biofilm-embedded cells (44). For the enumeration of the total bacterial population, 100 μl of appropriately diluted sample was manually plated onto drug-free CAMHA and incubated at 35°C for 48 h, due to the slow growth of the hypermutable CW8 isolate. The number of bacteria recovered from the coupons was expressed as the number of log10 CFU per square centimeter. Less-susceptible subpopulations were quantified for planktonic and biofilm bacteria at 0 (pretreatment), 24, 72, and 120 h following the start of treatment by plating 200 μl of appropriately diluted sample onto CAMHA (BD, Sparks, MD, USA) supplemented with meropenem at 6 mg/liter or 15 mg/liter or tobramycin at 3 mg/liter or 7.5 mg/liter. The plates were incubated for 48 h (meropenem) or 72 h (tobramycin) (15). MICs were determined at 0 and 120 h by the agar dilution method for colonies isolated from antibiotic-containing plates.
Pharmacokinetic validation.
For antibiotic-containing regimens, 1-ml samples were collected in duplicate from the CBR at multiple time points across the duration of the study and immediately stored at −80°C. Meropenem and tobramycin in CAMHB–1% TSB were measured using validated liquid chromatography-tandem mass spectrometry assays (15). The protocol for measuring tobramycin and meropenem was similar to that described in our previous study (15), except for slight modifications required by the presence of TSB. Modifications included the gradient of the binary mobile phase, programmed as 0.25% formic acid (A)–acetonitrile (B) at 80:20 that changed over 0.5 min to A-B at 50:50, which was held for 2.51 min, followed by reequilibration to A-B at 80:20 for 4.59 min. The flow rate of the mobile phase was 0.3 ml/min, the column oven temperature was 30°C, and the total run time was 7 min. The lower limit of quantification was 0.10 mg/liter for meropenem and 0.50 mg/liter for tobramycin. The correlation coefficients for the calibration curve of meropenem (range, 0.10 to 50.0 mg/liter) and tobramycin (range, 0.50 to 25.0 mg/liter) were >0.998 and >0.999, respectively. The interday precisions were 1.1 to 5.8% for meropenem and 2.1 to 7.5% for tobramycin; interday accuracies were 96.3 to 106.9% for meropenem and 95.9 to 102.1% for tobramycin.
Pharmacodynamic analysis.
Monotherapy or combination regimens causing a reduction of ≥1 log10 CFU/ml or CFU/cm2 at a specified time relative to the baseline were considered active. Synergy was defined as ≥2 log10-CFU/ml or -CFU/cm2 killing for the combination relative to that for the most active corresponding monotherapy at a specified time. Bacterial counts on antibiotic-containing plates were used to evaluate resistance emergence for different treatment regimens in comparison to the growth control. Mutation frequencies were calculated as the difference between the number of log10 CFU per milliliter (number of log10 CFU per square centimeter) on antibiotic-containing agar and the number of log10 CFU per milliliter (number of log10 CFU per square centimeter) on antibiotic-free agar at the same time point.
ACKNOWLEDGMENTS
We thank Jaime Lora-Tamayo, Instituto de Investigación, Hospital 12 de Octubre, Madrid, Spain, for discussions on the implementation of the CBR. We thank Kate Rogers, Centre for Medicine Use and Safety, Monash University, for experimental assistance.
This work was supported by Australian National Health and Medical Research Council (NHMRC) project grant number APP1101553 to C.B.L., A.O., and R.L.N. and by the Australian Research Council (ARC) Georgina Sweet Award for Women in Quantitative Biomedical Sciences to C.B.L. A.Y.P. was supported by an NHMRC practitioner fellowship (grant number APP1117940). A.O. is supported by the Instituto de Salud Carlos III, Subdirección General de Redes y Centros de Investigación Cooperativa, Ministerio de Economía y Competitividad, Spanish Network for Research in Infectious Diseases (grant number REIPI RD16/0016/0004), cofinanced by the European Development Regional Fund (A Way To Achieve Europe) and operative program Intelligent Growth 2014–2020.
The funders had no role in the study design, data collection and interpretation, or the decision to submit the work for publication. We have no conflicts of interest to declare.
REFERENCES
- 1.Tacconelli E, Carrara E, Savoldi A, Harbarth S, Mendelson M, Monnet DL, Pulcini C, Kahlmeter G, Kluytmans J, Carmeli Y, Ouellette M, Outterson K, Patel J, Cavaleri M, Cox EM, Houchens CR, Grayson ML, Hansen P, Singh N, Theuretzbacher U, Magrini N. 2018. Discovery, research, and development of new antibiotics: the WHO priority list of antibiotic-resistant bacteria and tuberculosis. Lancet Infect Dis 18:318–327. doi: 10.1016/S1473-3099(17)30753-3. [DOI] [PubMed] [Google Scholar]
- 2.Carmeli Y, Troillet N, Eliopoulos GM, Samore MH. 1999. Emergence of antibiotic-resistant Pseudomonas aeruginosa: comparison of risks associated with different antipseudomonal agents. Antimicrob Agents Chemother 43:1379–1382. doi: 10.1128/AAC.43.6.1379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Livermore DM. 2002. Multiple mechanisms of antimicrobial resistance in Pseudomonas aeruginosa: our worst nightmare? Clin Infect Dis 34:634–640. doi: 10.1086/338782. [DOI] [PubMed] [Google Scholar]
- 4.Langan KM, Kotsimbos T, Peleg AY. 2015. Managing Pseudomonas aeruginosa respiratory infections in cystic fibrosis. Curr Opin Infect Dis 28:547–556. doi: 10.1097/QCO.0000000000000217. [DOI] [PubMed] [Google Scholar]
- 5.Bhagirath AY, Li Y, Somayajula D, Dadashi M, Badr S, Duan K. 2016. Cystic fibrosis lung environment and Pseudomonas aeruginosa infection. BMC Pulm Med 16:174. doi: 10.1186/s12890-016-0339-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Stefani S, Campana S, Cariani L, Carnovale V, Colombo C, Lleo MM, Iula VD, Minicucci L, Morelli P, Pizzamiglio G, Taccetti G. 2017. Relevance of multidrug-resistant Pseudomonas aeruginosa infections in cystic fibrosis. Int J Med Microbiol 307:353–362. doi: 10.1016/j.ijmm.2017.07.004. [DOI] [PubMed] [Google Scholar]
- 7.Winstanley C, O’Brien S, Brockhurst MA. 2016. Pseudomonas aeruginosa evolutionary adaptation and diversification in cystic fibrosis chronic lung infections. Trends Microbiol 24:327–337. doi: 10.1016/j.tim.2016.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Oliver A. 2010. Mutators in cystic fibrosis chronic lung infection: prevalence, mechanisms, and consequences for antimicrobial therapy. Int J Med Microbiol 300:563–572. doi: 10.1016/j.ijmm.2010.08.009. [DOI] [PubMed] [Google Scholar]
- 9.Rees VE, Deveson Lucas DS, López-Causapé C, Huang Y, Kotsimbos T, Bulitta JB, Rees MC, Barugahare A, Peleg AY, Nation RL, Oliver A, Boyce JD, Landersdorfer CB. 2019. Characterization of hypermutator Pseudomonas aeruginosa isolates from patients with cystic fibrosis in Australia. Antimicrob Agents Chemother 63:e02538-18. doi: 10.1128/AAC.02538-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Oliver A, Canton R, Campo P, Baquero F, Blazquez J. 2000. High frequency of hypermutable Pseudomonas aeruginosa in cystic fibrosis lung infection. Science 288:1251–1254. doi: 10.1126/science.288.5469.1251. [DOI] [PubMed] [Google Scholar]
- 11.Lujan AM, Macia MD, Yang L, Molin S, Oliver A, Smania AM. 2011. Evolution and adaptation in Pseudomonas aeruginosa biofilms driven by mismatch repair system-deficient mutators. PLoS One 6:e27842. doi: 10.1371/journal.pone.0027842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.del Pozo JL, Patel R. 2007. The challenge of treating biofilm-associated bacterial infections. Clin Pharmacol Ther 82:204–209. doi: 10.1038/sj.clpt.6100247. [DOI] [PubMed] [Google Scholar]
- 13.Costerton JW, Stewart PS, Greenberg EP. 1999. Bacterial biofilms: a common cause of persistent infections. Science 284:1318–1322. doi: 10.1126/science.284.5418.1318. [DOI] [PubMed] [Google Scholar]
- 14.Guillot E, Sermet I, Ferroni A, Chhun S, Pons G, Zahar JR, Jullien V. 2010. Suboptimal ciprofloxacin dosing as a potential cause of decreased Pseudomonas aeruginosa susceptibility in children with cystic fibrosis. Pharmacotherapy 30:1252–1258. doi: 10.1592/phco.30.12.1252. [DOI] [PubMed] [Google Scholar]
- 15.Landersdorfer CB, Rees VE, Yadav R, Rogers KE, Kim TH, Bergen PJ, Cheah S-E, Boyce JD, Peleg AY, Oliver A, Shin BS, Nation RL, Bulitta JB. 2018. Optimization of a meropenem-tobramycin combination dosage regimen against hypermutable and nonhypermutable Pseudomonas aeruginosa via mechanism-based modeling and the hollow-fiber infection model. Antimicrob Agents Chemother 62:e02055-17. doi: 10.1128/AAC.02055-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Leggett JE, Fantin B, Ebert S, Totsuka K, Vogelman B, Calame W, Mattie H, Craig WA. 1989. Comparative antibiotic dose-effect relations at several dosing intervals in murine pneumonitis and thigh-infection models. J Infect Dis 159:281–292. doi: 10.1093/infdis/159.2.281. [DOI] [PubMed] [Google Scholar]
- 17.Turnidge JD. 1998. The pharmacodynamics of beta-lactams. Clin Infect Dis 27:10–22. doi: 10.1086/514622. [DOI] [PubMed] [Google Scholar]
- 18.Tam VH, Schilling AN, Neshat S, Poole K, Melnick DA, Coyle EA. 2005. Optimization of meropenem minimum concentration/MIC ratio to suppress in vitro resistance of Pseudomonas aeruginosa. Antimicrob Agents Chemother 49:4920–4927. doi: 10.1128/AAC.49.12.4920-4927.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bergen PJ, Bulitta JB, Kirkpatrick CM, Rogers KE, McGregor MJ, Wallis SC, Paterson DL, Lipman J, Roberts JA, Landersdorfer CB. 2016. Effect of different renal function on antibacterial effects of piperacillin against Pseudomonas aeruginosa evaluated via the hollow-fibre infection model and mechanism-based modelling. J Antimicrob Chemother 71:2509–2520. doi: 10.1093/jac/dkw153. [DOI] [PubMed] [Google Scholar]
- 20.Roberts JA, Ulldemolins M, Roberts MS, McWhinney B, Ungerer J, Paterson DL, Lipman J. 2010. Therapeutic drug monitoring of beta-lactams in critically ill patients: proof of concept. Int J Antimicrob Agents 36:332–339. doi: 10.1016/j.ijantimicag.2010.06.008. [DOI] [PubMed] [Google Scholar]
- 21.Ciofu O, Tolker-Nielsen T. 2019. Tolerance and resistance of Pseudomonas aeruginosa biofilms to antimicrobial agents-how P. aeruginosa can escape antibiotics. Front Microbiol 10:913. doi: 10.3389/fmicb.2019.00913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gomez-Junyent J, Benavent E, Sierra Y, El Haj C, Soldevila L, Torrejon B, Rigo-Bonnin R, Tubau F, Ariza J, Murillo O. 2019. Efficacy of ceftolozane/tazobactam, alone and in combination with colistin, against multidrug-resistant Pseudomonas aeruginosa in an in vitro biofilm pharmacodynamic model. Int J Antimicrob Agents 53:612–619. doi: 10.1016/j.ijantimicag.2019.01.010. [DOI] [PubMed] [Google Scholar]
- 23.Roberts JA, Abdul-Aziz MH, Lipman J, Mouton JW, Vinks AA, Felton TW, Hope WW, Farkas A, Neely MN, Schentag JJ, Drusano G, Frey OR, Theuretzbacher U, Kuti JL. 2014. Individualised antibiotic dosing for patients who are critically ill: challenges and potential solutions. Lancet Infect Dis 14:498–509. doi: 10.1016/S1473-3099(14)70036-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mulcahy H, Charron-Mazenod L, Lewenza S. 2008. Extracellular DNA chelates cations and induces antibiotic resistance in Pseudomonas aeruginosa biofilms. PLoS Pathog 4:e1000213. doi: 10.1371/journal.ppat.1000213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chiang WC, Nilsson M, Jensen PO, Hoiby N, Nielsen TE, Givskov M, Tolker-Nielsen T. 2013. Extracellular DNA shields against aminoglycosides in Pseudomonas aeruginosa biofilms. Antimicrob Agents Chemother 57:2352–2361. doi: 10.1128/AAC.00001-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rees VE, Bulitta JB, Oliver A, Tsuji BT, Rayner CR, Nation RL, Landersdorfer CB. 2016. Resistance suppression by high-intensity, short-duration aminoglycoside exposure against hypermutable and non-hypermutable Pseudomonas aeruginosa. J Antimicrob Chemother 71:3157–3167. doi: 10.1093/jac/dkw297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Davies TA, Shang W, Bush K, Flamm RK. 2008. Affinity of doripenem and comparators to penicillin-binding proteins in Escherichia coli and Pseudomonas aeruginosa. Antimicrob Agents Chemother 52:1510–1512. doi: 10.1128/AAC.01529-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Poole K. 2011. Pseudomonas aeruginosa: resistance to the max. Front Microbiol 2:65. doi: 10.3389/fmicb.2011.00065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lambert PA. 2002. Mechanisms of antibiotic resistance in Pseudomonas aeruginosa. J R Soc Med 95(Suppl 41):22–26. [PMC free article] [PubMed] [Google Scholar]
- 30.Kadurugamuwa JL, Clarke AJ, Beveridge TJ. 1993. Surface action of gentamicin on Pseudomonas aeruginosa. J Bacteriol 175:5798–5805. doi: 10.1128/jb.175.18.5798-5805.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bulitta JB, Ly NS, Landersdorfer CB, Wanigaratne NA, Velkov T, Yadav R, Oliver A, Martin L, Shin BS, Forrest A, Tsuji BT. 2015. Two mechanisms of killing of Pseudomonas aeruginosa by tobramycin assessed at multiple inocula via mechanism-based modeling. Antimicrob Agents Chemother 59:2315–2327. doi: 10.1128/AAC.04099-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yadav R, Bulitta JB, Schneider EK, Shin BS, Velkov T, Nation RL, Landersdorfer CB. 2017. Aminoglycoside concentrations required for synergy with carbapenems against Pseudomonas aeruginosa determined via mechanistic studies and modeling. Antimicrob Agents Chemother 61:e00722-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Angus BL, Carey AM, Caron DA, Kropinski AM, Hancock RE. 1982. Outer membrane permeability in Pseudomonas aeruginosa: comparison of a wild-type with an antibiotic-supersusceptible mutant. Antimicrob Agents Chemother 21:299–309. doi: 10.1128/aac.21.2.299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Guenard S, Muller C, Monlezun L, Benas P, Broutin I, Jeannot K, Plesiat P. 2014. Multiple mutations lead to MexXY-OprM-dependent aminoglycoside resistance in clinical strains of Pseudomonas aeruginosa. Antimicrob Agents Chemother 58:221–228. doi: 10.1128/AAC.01252-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Louie A, Liu W, Fikes S, Brown D, Drusano GL. 2013. Impact of meropenem in combination with tobramycin in a murine model of Pseudomonas aeruginosa pneumonia. Antimicrob Agents Chemother 57:2788–2792. doi: 10.1128/AAC.02624-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Drusano GL, Liu W, Fregeau C, Kulawy R, Louie A. 2009. Differing effects of combination chemotherapy with meropenem and tobramycin on cell kill and suppression of resistance of wild-type Pseudomonas aeruginosa PAO1 and its isogenic MexAB efflux pump-overexpressed mutant. Antimicrob Agents Chemother 53:2266–2273. doi: 10.1128/AAC.01680-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tam VH, Schilling AN, Lewis RE, Melnick DA, Boucher AN. 2004. Novel approach to characterization of combined pharmacodynamic effects of antimicrobial agents. Antimicrob Agents Chemother 48:4315–4321. doi: 10.1128/AAC.48.11.4315-4321.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Haagensen J, Verotta D, Huang L, Engel J, Spormann AM, Yang K. 2017. Spatiotemporal pharmacodynamics of meropenem- and tobramycin-treated Pseudomonas aeruginosa biofilms. J Antimicrob Chemother 72:3357–3365. doi: 10.1093/jac/dkx288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lodise TP, Sorgel F, Melnick D, Mason B, Kinzig M, Drusano GL. 2011. Penetration of meropenem into epithelial lining fluid of patients with ventilator-associated pneumonia. Antimicrob Agents Chemother 55:1606–1610. doi: 10.1128/AAC.01330-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wenzler E, Gotfried MH, Loutit JS, Durso S, Griffith DC, Dudley MN, Rodvold KA. 2015. Meropenem-RPX7009 concentrations in plasma, epithelial lining fluid, and alveolar macrophages of healthy adult subjects. Antimicrob Agents Chemother 59:7232–7239. doi: 10.1128/AAC.01713-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Frippiat F, Musuamba FT, Seidel L, Albert A, Denooz R, Charlier C, Van Bambeke F, Wallemacq P, Descy J, Lambermont B, Layios N, Damas P, Moutschen M. 2015. Modelled target attainment after meropenem infusion in patients with severe nosocomial pneumonia: the PROMESSE study. J Antimicrob Chemother 70:207–216. doi: 10.1093/jac/dku354. [DOI] [PubMed] [Google Scholar]
- 42.Clinical and Laboratory Standards Institute. 2016. Performance standards for antimicrobial susceptibility testing: twenty-fifth informational supplement. M100-S26 Clinical and Laboratory Standards Institute, Wayne, PA. [Google Scholar]
- 43.Magiorakos A-P, Srinivasan A, Carey RB, Carmeli Y, Falagas ME, Giske CG, Harbarth S, Hindler JF, Kahlmeter G, Olsson-Liljequist B, Paterson DL, Rice LB, Stelling J, Struelens MJ, Vatopoulos A, Weber JT, Monnet DL. 2012. Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: an international expert proposal for interim standard definitions for acquired resistance. Clin Microbiol Infect 18:268–281. doi: 10.1111/j.1469-0691.2011.03570.x. [DOI] [PubMed] [Google Scholar]
- 44.Lora-Tamayo J, Murillo O, Bergen PJ, Nation RL, Poudyal A, Luo X, Yu HY, Ariza J, Li J. 2014. Activity of colistin combined with doripenem at clinically relevant concentrations against multidrug-resistant Pseudomonas aeruginosa in an in vitro dynamic biofilm model. J Antimicrob Chemother 69:2434–2442. doi: 10.1093/jac/dku151. [DOI] [PubMed] [Google Scholar]
- 45.Bui KQ, Ambrose PG, Nicolau DP, Lapin CD, Nightingale CH, Quintiliani R. 2001. Pharmacokinetics of high-dose meropenem in adult cystic fibrosis patients. Chemotherapy 47:153–156. doi: 10.1159/000063216. [DOI] [PubMed] [Google Scholar]
- 46.Hennig S, Standing JF, Staatz CE, Thomson AH. 2013. Population pharmacokinetics of tobramycin in patients with and without cystic fibrosis. Clin Pharmacokinet 52:289–301. doi: 10.1007/s40262-013-0036-y. [DOI] [PubMed] [Google Scholar]
- 47.Carcas AJ, García-Satué JL, Zapater P, Frías-Iniesta J. 1999. Tobramycin penetration into epithelial lining fluid of patients with pneumonia. Clin Pharmacol Ther 65:245–250. doi: 10.1016/S0009-9236(99)70103-7. [DOI] [PubMed] [Google Scholar]
- 48.Macey RI. 1996–2010 Berkeley Madonna, version 8.3.18 441. University of California, Berkeley, CA. [Google Scholar]