The available antifungal therapeutic arsenal is limited. The search for alternative drugs with fewer side effects and new targets remains a major challenge. Decyl gallate (G14) is a derivative of gallic acid with a range of biological activities and broad-spectrum antifungal activity. Previously, our group demonstrated the promising anti-Paracoccidioides activity of G14.
Keywords: Paracoccidioides lutzii, decyl gallate, chemical-genetic interaction, N-glycosylation, mechanisms of action
ABSTRACT
The available antifungal therapeutic arsenal is limited. The search for alternative drugs with fewer side effects and new targets remains a major challenge. Decyl gallate (G14) is a derivative of gallic acid with a range of biological activities and broad-spectrum antifungal activity. Previously, our group demonstrated the promising anti-Paracoccidioides activity of G14. In this work, to evaluate the antifungal characteristics of G14 for Paracoccidioides lutzii, a chemical-genetic interaction analysis was conducted on a Saccharomyces cerevisiae model. N-glycosylation and/or the unfolded protein response pathway was identified as a high-confidence process for drug target prediction. The overactivation of unfolded protein response (UPR) signaling was confirmed using this model with IRE1/ATF6/PERK genes tagged with green fluorescent protein (GFP). In P. lutzii, this prediction was confirmed by the low activity of glycosylated enzymes [α-(1,3)-glucanase, N-acetyl-β-d-glucosaminidase (NAGase), and α-(1,4)-amylase], by hyperexpression of genes involved with the UPR and glycosylated enzymes, and by the reduction in the amounts of glycosylated proteins and chitin. All of these components are involved in fungal cell wall integrity and are dependent on the N-glycosylation process. This loss of integrity was confirmed by the reduction in mitochondrial activity, impaired budding, enhancement of wall permeability, and a decrease in viability. These events led to a reduction of the ability of fungi to adhere on human lung epithelial cells (A549) in vitro. Therefore, G14 may have an important role in balancing the inflammatory reaction caused by fungal infection, without interfering with the microbicidal activity of nitric oxide. This work provides new information on the activity of G14, a potential anti-Paracoccidioides compound.
INTRODUCTION
Paracoccidioidomycosis (PCM) is a systemic infection caused by the species complex Paracoccidioides spp., including Paracoccidioides lutzii. This disease occurs in areas with poor socioeconomic conditions, located mainly in Latin America and in active rural areas in Brazil. Despite the availability of antifungal agents for the treatment of PCM, the selection of the therapy depends on the severity of the disease and the type of antifungal agent available (amphotericin B, azoles, and sulfonamides) (1). In addition, this therapy requires a long treatment period, has many cases of relapse, and frequently comes with side effects and high toxicity, resulting in low quality of life for the patient (2). Therefore, none of the currently available therapies ensures the complete elimination of the fungus from the patient (1).
The investment in the development of new antifungal agents with higher efficacy and fewer side effects is of extreme importance in order to increase the treatment options for this disease. However, this development is a challenge since the pathogenic fungi and their host share the same characteristics of eukaryotic cells.
In this respect, the demand for new antifungal compounds, especially those obtained from natural origin or their synthetic derivatives, is of extreme importance, and PCM experts have been working to find new agents from natural sources (3–9).
Chemical studies of ethanolic extract obtained from the leaves of Alchornea glandulosa identified the presence of phenolic compounds, such as gallic acid, which has a range of biological activities, including anti-inflammatory and antifungal activity (10–13). Structurally, modifications were made in this molecule by the addition of variably sized carbon chains, resulting in the formation of alkyl gallates. They have been described as having great antifungal activity against a representative panel of opportunistic pathogenic fungi (14).
Our group demonstrated that decyl gallate (G14) had great antifungal activity in vitro against 18 pathogenic fungi species (including eight different Paracoccidioides isolates). For Paracoccidioides isolates, G14 presented a low MIC and a good selectivity index value (>10) against infection in human lung epithelial cells (A549) and normal fibroblast pulmonary cells (MRC-5) (8). These results showed that G14 is an excellent candidate for a broad-spectrum antifungal prototype and encouraged us to try to discover its mechanism of action.
Therefore, our study using a chemical-genetic interaction assay (15–19) provided comprehensive information about the effect of specific gene perturbations on the cell’s response to G14 treatment, which, in addition to other assays, had demonstrated that G14 has a strong influence on the N-glycosylation process, highlighting G14 as a potential anti-Paracoccidioides agent.
RESULTS
N-glycosylation/UPR as a target of G14 treatment.
To elucidate the G14 mode of action and cellular target, a chemical-genetic interaction analysis was conducted. A Saccharomyces cerevisiae gene deletion mutant pool was treated with G14 and different controls. The relative fitness of each yeast mutant was quantified by sequencing strain-specific DNA barcodes treated with G14 and comparing fitness levels to that of the dimethyl sulfoxide (DMSO) control. The top 10 deletion mutants susceptible to G14 (Table 1) were subjected to enrichment analysis, and enrichment was found for genes involved in the gene ontology (GO) category cellular/endoplasmic reticulum (ER) unfolded protein response (UPR) (P < 0.01). The deletion mutants of HAC1 and IRE1 were the most hypersensitive to G14. This profile was similar to the chemical-genetic interaction profile of the control agent tunicamycin, which inhibits the N-glycosylation pathway. The Pearson correlation coefficient between G14 and tunicamycin profiles was calculated (0.1906) and showed a moderate level of profile similarity between the two compounds (Fig. 1). Assuming that the chemical inhibitor of a gene product tends to mimic the loss-of-function phenotype of a mutant that inactivates the gene, the chemical-genetic interaction profile for a bioactive compound can resemble the genetic interaction profile for the target. The comparison of the chemical-genetic interaction profile of G14 with the genetic interaction network of S. cerevisiae allowed us to predict the target for the compound as ALG12, which has a genetic interaction profile most significantly correlated to the chemical-genetic interaction profile of G14. ALG12 is a nonessential gene responsible for the addition of alpha-1,6-mannose to dolichol-linked proteins, acting in the dolichol pathway for N-glycosylation.
TABLE 1.
Top 10 deletion mutants sensitive to G14
| Mutant | z-score | Name and/or description |
|---|---|---|
| VPS13 | −10.1938 | Vacuolar protein sorting |
| IRE1 | −14.2456 | Inositol requiring |
| HAC1 | −15.172 | Homologous to Atf/Creb1 |
| UBR2 | −8.99919 | Cytoplasmic ubiquitin-protein ligase |
| CTF3 | −9.53256 | Chromosome transmission fidelity |
| HST3 | −9.29459 | Homolog of SIR2 |
| CTI6 | −8.44196 | Cyc8-Tup1-interacting protein |
| PEX14 | −7.77147 | Peroxisome related |
| YHR003C | −9.02499 | tRNA threonyl-carbamoyl-adenosine dehydratase |
| YKR041W | −8.24314 |
FIG 1.
G14 and tunicamycin had similar profiles. The chemical genetic interaction score (y axis) of each mutant (x axis) was plotted for G14 (A) and tunicamycin (B), highlighting the hypersensitive mutants. The profile similarity was demonstrated by the Pearson correlation coefficient (0.1906) (C).
UPR hyperactivation after G14 challenge.
The gene IRE1 is a mediator of the yeast unfolded protein response (UPR) and coordinates a series of responses by a signal transduction pathway that maintains endoplasmic reticulum homeostasis. Activation of IRE1 is closely linked to the activation of HAC1. Both are essential for the proper functioning of this response (20). The susceptibility to G14 of mutants of IRE1 and HAC1 means that this pathway was essential to yeast survival after G14 exposure. For this reason, a UPR activation assay was performed against G14 in S. cerevisiae yeast strains containing green fluorescent protein (GFP) tags on the IRE1, ATF6, and PERK genes. We investigated if this pathway would be activated after treatment. By fluorescence microscopy observation (Fig. 2A) and with the analysis of the mean fluorescence intensity (MFI) (Fig. 2B), an increase in fluorescence intensity of the cells was verified after 3 h of G14 treatment, and this phenotype persisted after 24 h. Under normal conditions, this pathway has low activity, similar to that of the control DMSO at a nontoxic concentration; however, upon G14 treatment, this pathway was activated, and the transcription products containing the GFP sequence increased. The hyperactivation of the UPR upon treatment revealed that G14 led to stress in the endoplasmic reticulum and affected the correct protein folding.
FIG 2.
G14 treatment led to UPR hyperactivity. An S. cerevisiae yeast strain with GFP tags on genes belonging to the unfolded protein response (genes IRE1, ATF6, and PERK) was treated with decyl gallate (G14) for 3 and 24 h. (A) GFP-tagged protein expression was observed by fluorescence microscopy. (B) The mean fluorescence intensity (MFI) was measured. Control (W/T), without treatment. *, P < 0.05.
G14 treatment changed N-glycosylated proteins profile of P. lutzii.
Since the mechanism of action of G14 was linked to the N-glycosylation pathway and/or the response of unfolded proteins, the analysis of important enzymes belonging to remodeling and maintenance of the fungal cell wall was indicated. This is based on the idea that these enzymes are dependent on the N-glycosylation process in order to be functional. Supporting this idea, the relative activities of the enzymes α-(1,3)-glucanase, N-acetyl-β-d-glucosaminidase, and α-(1,4)-amylase, after treatment with 0.5 μg/ml of G14 were reduced by almost 50% compared to level of the control without treatment (Fig. 3A).
FIG 3.
G14 changed the normal profiles of N-glycosylated proteins of P. lutzii. The fungal cells were treated with decyl gallate (G14) for 72 h. (A) Percentage of relative activity of the N-glycosylated enzymes was determined for the following: N-acetyl-β-d-glucosaminidase (NAGase), α-(1,3)-glucanase, and α-(1,4)-amylase (α-amylase). Control (W/T), without treatment. (B) Real-time PCR analysis of relative expression was conducted for the genes encoding N-acetyl-β-d-glucosaminidase (nag1), α-(1,4)-amylase (amy), α-(1,3)-glucanase (agn), and the endoplasmic reticulum stress-sensing genes inositol requiring enzyme-1 (ire1) and transcription factor (hac1). (C) Glycosylated proteins present in fungal wall were labeled with Alexa Fluor 488-conjugated concanavalin A, and the mean fluorescence intensity (MFI) was evaluated by flow cytometry. (D) Chitin amount was quantified using calcofluor white label and fluorescence microscopy. Control (W/T), without treatment. *, P < 0.05.
The N-glycosylation process occurs at the posttranslational level and is an essential process for the functioning of these enzymes; we thus tested if the decrease in the activity of the final protein (enzyme) was affected by the decreased expression of the involved genes (Table 2). After treatment with G14, it was observed that all the analyzed genes (agn, nag1, amy, ire1, and hac1) showed an increase in expression (Fig. 3B) compared to the level of gene expression of the sample without treatment.
TABLE 2.
Specific primers for coding genes
| Primer namea | Sequence |
|---|---|
| nag1_F | TCATGCAGAGCTGGAACAAC |
| nag1_R | CATTGTAGCGGTGGTCATTG |
| agn_F | CAACCATGGTCAGCAAACAC |
| agn _R | CTGTAGCCCTGAACCCACAT |
| amy_F | CGGTGACTTGTATGGCCTTT |
| amy _R | CAGTTGGCCATTACATGACG |
| ire1_F | CCCCCAGAGATATCGTGAGA |
| ire1_R | TTAGAACGTGGGGCCTAATG |
| hac1_F | CTTGTCCCCATCCTCTTCAA |
| hac1_R | TTCAAGGATCCCTCCAACTG |
| tub_F | CTGTTACCAGCCTCCCCATA |
| tub_R | TGCTTCAGAAATGGCAGTTG |
F, forward; R, reverse.
It was possible to observe that the G14 treatment reduced the amount of glycosylated proteins present in the fungal cell wall. The MFI of concanavalin A (ConA) (Fig. 3C), which is a lectin able to bind to glycosylated proteins, decreased 58% upon treatment, and calcofluor white (CFW) (Fig. 3D), which binds specifically with chitin, which is present in the cell wall and dependent on N-glycosylated-enzymes, decreased 50%.
G14 impaired P. lutzii development and viability.
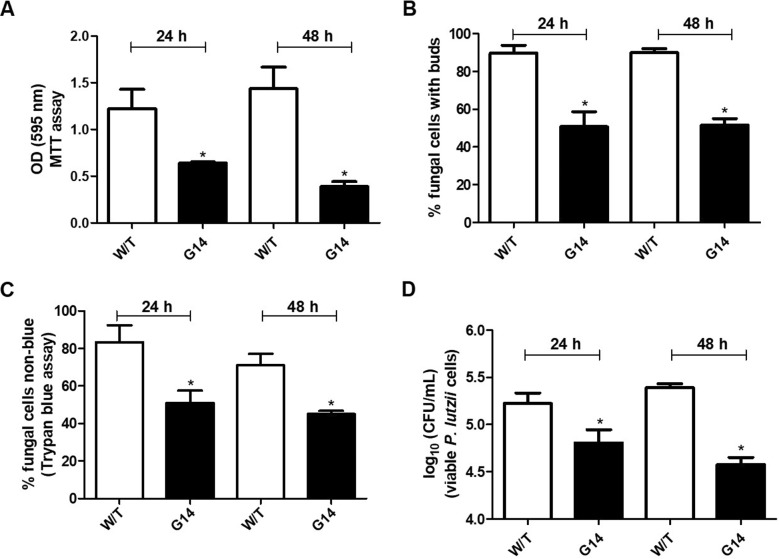
An MTT (3-[4,5-dimethylthiazol-2-yl]-2,5-diphenyl tetrazolium bromide) assay showed that G14 treatment led to 42% and 65% reductions in mitochondrial activity after 24 h and 48 h of treatment, respectively, compared to the levels of the control without treatment (Fig. 4A). The treatment damaged the development of P. lutzii cells since the treated cells showed a budding percentage of 50% at both times, indicating a decrease in new bud formation, while the control without treatment presented budding in about 90% of fungal cells (Fig. 4B). By trypan blue assay, we observed reductions of 39% and 53% in the rate of fungal cells excluding the blue dye after 24 and 48 h of G14 treatment, respectively (Fig. 4C). In the CFU analysis, reduction of 62% and 85% in growth, respectively, were observed after treatment with G14 (Fig. 4D). Together, these data demonstrated that G14 treatment impaired the maintenance of the fungal wall, reducing the cellular viability of the fungus.
FIG 4.
The development of P. lutzii cells was reduced after G14 treatment. Comparative analysis between the untreated control (W/T, without treatment) and of P. lutzii cells treated with decyl gallate (G14) after 24 h and 48 h. (A) MTT assay for mitochondrial activity of P. lutzii cells. OD, optical density. (B) Development assay with analysis of percentage of P. lutzii cells with buds. (C) Trypan blue assay for analysis of fugal wall permeability. (D) CFU assay to analyze the recovery of viable fungal cells. *, P < 0.05.
G14 treatment reduced adhesion of P. lutzii.
Although we observed a tendency toward a reduction of the internalization of P. lutzii in A549 cells after G14 treatment, this reduction was not significant compared to levels in the control without treatment. On the other hand, adhesion, an essential step in Paracoccidioides sp. infection, decreased 75% after G14 treatment (Fig. 5A). In order to know if the treatment affected the internalized fungal cells, CFU enumeration was performed. The results showed that there was not a reduction in CFU counts compared to level of the control without treatment (Fig. 5B). Even though the G14 treatment had not reached the internalized fungi in A549 cells, G14 showed important activity on a fungal virulence factor, adhesion.
FIG 5.
In vitro G14 decreased adhesion rate of P. lutzii in human pneumocytes. After 6 h of P. lutzii infection in A549 cells (human pneumocyte type I line), treatment with decyl gallate (G14) was performed for 24 h. (A) A quenching assay was performed to obtain the rate of adhesion and internalization by flow cytometry analysis. (B) The viability of internalized fungal cells was assessed by CFU counts in BHI agar plates. Control (W/T), without treatment. *, P < 0.05.
G14 improved the antifungal activity of macrophages.
When macrophages were treated only with G14, we observed a reduction of 62% in the nitrite concentration compared to that in macrophages stimulated with lipopolysaccharide (LPS). The same was observed during the infection of macrophages by P. lutzii. When the infection was treated with G14, we observed a reduction of 55% in the nitrite concentration compared to the level in the control without treatment (Fig. 6A). These results showed the anti-inflammatory activity of G14. We observed a reduction in CFU numbers when the infection was treated with G14 (Fig. 6B), suggesting that G14 contributed to macrophage activity against P. lutzii.
FIG 6.
G14 decreased nitrite production by macrophages infected in vitro with P. lutzii. A RAW 264.7 macrophage monolayer was stimulated with LPS (positive control) and infected with P. lutzii for 6 h and subsequently treated with decyl gallate (G14) for 24 h. (A) Griess assay was performed to verify the ability of G14 to modulate nitrite production. (B) The infection was plated on BHI agar plates, and CFU counts were determined. Control (W/T), without treatment. *, P < 0.05.
DISCUSSION
In order to know the mechanism of action of G14 in Paracoccidioides spp., we used a chemical-genetic interaction assay using S. cerevisiae as a model to predict the mechanism of action of G14 (decyl gallate). In the chemical-genetic interaction, a sublethal concentration of a growth-inhibitory compound was screened, and the deletion mutants with hypersensitive profiles were highlighted. The next step was the comparison of a chemical-genetic interaction profile to a compendium of genetic interactions (synthetic lethal). A synthetic lethal genetic interaction is characterized by the deletion of a single gene, resulting in viable mutants. However, a lethal phenotype is observed when double deletions are combined in the same mutant strain. Gene deletion alleles that show chemical-genetic interactions with a compound should also be synthetically lethal or synthetically sick as a result of a mutation in the compound target gene. Genetic profile similarities are measured for all gene pairs by computing Pearson correlation coefficients (PCC) from the complete genetic interaction matrix. These profiles predict the pathways and targets inhibited by treatment (15–19).
Based on the chemical-genetic interaction profile, we predicted that G14 affects the N-glycosylation and protein folding pathways. The profile was similar to that of one of our control compounds, tunicamycin. This compound is classified as a nucleoside antibiotic and inhibits N-glycosylation by blocking the first step in the synthesis of the dolichol-linked oligosaccharide, the transfer of UDP-N-acetylglucosamine to dolichol phosphate (21).
This result was confirmed when the chemical-genetic interaction profile of G14 was compared to the genetic interaction network of S. cerevisiae, allowing the prediction of its putative molecular target, alpha-1,6-mannosyltransferase (ALG12). ALG12 is localized in the endoplasmic reticulum (ER) and is responsible for the addition of alpha-1,6-mannose to dolichol-linked Man7GlcNAc2 and acts in the dolichol pathway for N-glycosylation (22).
N-glycosylation is an essential process for posttranslational modification of proteins. Some proteins are synthesized in the ribosomal portion of the rough endoplasmic reticulum (RER), where they assume their correct conformation. After this, there is an addition of a precursor oligosaccharide by transferase. The process of linking the protein with the oligosaccharide is called N-glycosylation (23).
It is known that N-glycosylation is an important process for fungal cells. The fungal cell wall is composed of polysaccharides, such as glucans, chitin, lipids, and proteins, some of which are highly glycosylated (glycoproteins). Since the fungal cell wall is essential for osmotic homeostasis maintenance, morphogenesis, and maintenance of the cellular form and for a role in mechanical resistance, it is considered a promising target for the development of new agents (24). In Paracoccidioides this role is not different; N-glycosylated proteins are crucial for many biological processes (25).
G14 affected the same pathway affected by tunicamycin, which had an effect on wall composition and development of Paracoccidioides spp. A study by Dos Reis Almeida et al. (26) showed that upon tunicamycin treatment, the level of N-glycosylation was lower, and a reduction of α-(1,4)-amylase activity, a glycosylated enzyme, was observed.
Thinking about glycosylated proteins, analysis of the relative activity of essential glycosylated enzymes present in the cell wall of P. lutzii, such as α-(1,3)-glucanase, N-acetyl-β-d-glucosaminidase, and α-(1,4)-amylase, after G14 treatment was proposed. These enzymes are responsible for covalently joining components to form a three-dimensional matrix of chitins, glucans, and glycoproteins. It is known that the loss of any of these components may impair the growth, morphology, and viability of the fungal cell (24). Our results showed that G14 treatment of P. lutzii reduced the relative activity of these enzymes, suggesting that G14 impairs the N-glycosylation process. Interestingly, the analysis of the expression of genes encoding these enzymes showed upregulation of these genes upon G14 treatment. We believe that this could possibly be a compensatory mechanism for diminished enzymatic activity due to the lack of N-glycosylation (27).
In order to demonstrate the extent of the treatment effect on the N-glycosylation pathway, we used the markers concanavalin A, which has a high affinity for glycosylated structures (28), and calcofluor white, which binds to chitin, a cell wall component dependent on the glycosylated enzymes that we tested in this study (28). The fluorescence of both markers upon treatment with G14 was lower than that in fungal cells without treatment. This reduction suggests that glycosylated enzymes as well chitin amounts are decreased after G14 treatment, probably due to its action in damaging the N-glycosylation process of these components.
When the N-glycosylation process is compromised, unfolded proteins are formed. The consequence is the generation of toxic signals to cells, leading to a stress situation. In eukaryotes, the endoplasmic reticulum stress response is orchestrated by at least three genes: IRE1, ATF6, and PERK. Together, these sensors coordinate the unfolded protein response (UPR), which is responsible for circumventing this situation and restoring homeostasis. In fungi, neither ATF6 nor PERK has been described, but IRE1 is considered the main mediator of homeostasis in response to unfolded proteins. The activation of IRE1 is closely linked to the activation of HAC1, and both are essential for the proper cellular response to this stress (20). In this regard, first of all, UPR activation upon G14 treatment was analyzed in a S. saccharomyces strain with GFP-tagged IRE1, ATF6, and PERK genes. Increased expression of the protein complex was observed by fluorescence microscopy, supporting the prediction that G14 interferes with the N-glycosylation of proteins, activating the UPR. Corroborating these results, real-time PCR analysis of IRE1 and HAC1 genes after G14 treatment revealed the activation of the UPR by the upregulation of these genes.
If the execution of the UPR is unsuccessful, the apoptosis response is induced. This response is deeply linked to mitochondrion activity since there is a physiological and functional interaction between the mitochondrial and ER organelles. This connection is essential to maintain cellular homeostasis and viability (29). The MTT assay confirmed the chronic activation of UPR signaling in which G14 decreases mitochondrial activity and the viability rate of P. lutzii cells. The low mitochondrial activity impairs fungal metabolism and the composition and formation of the cell wall. As a consequence, there was interference with bud formation (25, 30), as shown after the G14 treatment of P. lutzii cells. The effects of G14 on fungal viability and wall damage were confirmed by CFU enumeration and trypan blue assays. A trypan blue assay confirmed that G14 impairs the fungal cell wall, and CFU enumeration confirmed that a consequence of the weaker fungal cell was a loss of viability.
All of these results confirmed that the fungal wall is important to fungal viability and highlighted how the N-glycosylation process is vital to the integrity of the fungal cell wall. For Paracoccidioides spp. the composition of the cell wall is considered an important virulence factor, and its components are important for some properties, such as adhesion to host cells (31).
The dynamics of the interaction of Paracoccidioides brasiliensis with human lung epithelial cells (A549) was previously studied by our group (32), and we demonstrated that the interaction rate of the fungi with these cells increases progressively after 3 h of contact, and with 5 h the internalization process was observed. Considering this knowledge, it was evaluated if the destabilization of the fungal cell wall by G14 treatment in vitro influences the interaction with the host cell. For this, A549 cells were challenged with P. lutzii for 6 h, ensuring that the cells were adhered and internalized. From this point, the interaction was treated with G14 for 24 h. In comparison with the rate for the control, we observed a significant reduction in the interaction rate, and after applying the quenching test, it was possible to conclude that this reduction was in the adhesion rate since no differences in rates of internalization of fungi were observed. The action of G14 in reducing the adhesion rate could be explained by the destabilization this compound caused to the P. lutzii cell wall, as shown in this study. Although the numbers of CFU of the internalized fungal cells were not different between the untreated control and the treated cells, the prevention of fungal adherence to host cells could be an important tool to avoid fungal colonization at different sites, helping to avoid a systemic infection.
Since G14 was able to decrease the Paracoccidioides sp. infection, hindering proper fungal adhesion to host cells, we verified whether G14 could help the host control Paracoccidioides infection. Paracoccidioides spp. are able to induce the modulation of a chronic inflammatory response via the nitric oxide pathway and to exert a negative modulatory effect on granuloma formation in PCM (33, 34). We evaluated the in vitro effect of G14 on nitric oxide modulation in RAW 264.7 macrophages infected with P. lutzii. G14 treatment significantly reduced nitrite production. We suggest that G14 may play an important role in balancing the inflammatory reaction caused by fungal infection without interfering with the microbicidal activity of nitric oxide. However, when CFU analysis of fungal cells internalized by macrophages was performed, a significant reduction upon treatment was observed, which could mean that G14 treatment had a positive influence on other microbicidal mechanisms of the macrophages.
Antifungal agents acting in the modulation of nitric oxide could be considered excellent candidates for therapy in pathophysiological processes. Lopes et al. (12) demonstrated the anti-inflammatory activity of an alkyl gallate, which has a well-described antioxidant activity and could act as an anti-inflammatory agent and protect human cells against oxidative damage caused by reactive oxygen species. In this way, G14 could also act as an antioxidant compound and be able to reduce the inflammation and damage generated during fungal infection.
In conclusion, through this work we know more about the action of G14, which is involved in the impairment of the N-glycosylation pathway. This effect damages the maintenance of the fungal wall, which results in an important decrease in the infection process in vitro, in addition to the important anti-inflammatory role of G14. This study highlights the need for continued studies to promote this compound as a new antifungal agent.
MATERIALS AND METHODS
Decyl gallate (G14).
G14 was obtained and provided by Instituto de Química, UNESP (Araraquara, Brazil), according to Morais et al. (35) and prepared according to de Paula e Silva et al. (8).
Chemical-genetic interaction assay.
A chemical-genetic interaction assay for G14 was performed as described previously by Piotrowsk et al. (36–38) using a S. cerevisiae deletion collection pool containing 310 mutants that are functionally representative of all major cellular processes (15, 36). The optimal inhibitory concentration of G14 for a chemical-genetic interaction assay was determined using an eight-point dose-response assay, and a concentration of 7 μg/ml of G14 was determined (70 to 80% growth versus solvent control in yeast extract-peptone [YEP]-galactose medium after 24 h of growth). The deletion collection pool was grown with 7 μg/ml of G14 for 48 h at 30°C. The controls were DMSO, tunicamycin (glycosylation inhibitor), methyl methanesulfonate (MMS; DNA damage inducing agent), micafungin (inhibitor of β-1,3-glucan production of), bortezomib (proteasome inhibitor), and benomyl (microtubule depolymerizing agent), which have known chemical-genetic interaction profiles. After the incubation with G14, the genomic DNA of the deletion collection pool was extracted using an Epicentre MasterPure Yeast DNA purification kit. Special multiplex primers were used to amplify mutant-specific molecular barcodes and to attach unique experiment-specific barcodes to each compound-treated pool (19). Illumina MiSeq was used to sequence the barcodes. The barcode counts for each yeast deletion mutant in the presence of G14 were compared to those under DMSO control conditions to determine sensitivity or resistance of individual strains in the pool (the chemical-genetic interaction score) (15). A Bonferroni-corrected hypergeometric distribution test was used to search for significant enrichment of GO terms among the top 10 sensitive and resistant deletion mutants (39). Correlation of the chemical-genetic interaction profile of G14 with the yeast genetic interaction network was performed by a bioinformatics program (36, 38).
UPR activation assay.
S. cerevisiae yeast strain YMJ003 (ura3Δ::UPRE-GFP-TEF2pr-RFP-MET-URA3 his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 LYS+ can1Δ::STE2pr-spHIS5 lyp1Δ::STE3pr-LEU2 CYH2) was used. This strain has a green fluorescent protein (GFP) tagged to inositol-requiring enzyme 1 (IRE1), activating transcription factor 6 (ATF6), and protein kinase RNA-like endoplasmic reticulum kinase (PERK) genes and was used for UPR analysis (40). A colony was cultivated in 2 ml of synthetic defined medium lacking Trp (SD-Trp) for 18 h at 30°C and 150 rpm. The suspension was diluted 1:10 in SD tryptophan drop-out (SD DO-Trp) medium and incubated for 3 h (log phase) at 30°C and 150 rpm. One milliliter of suspension was diluted in 1 ml of SD DO-Trp medium with 7 μg/ml G14 or in DMSO for 3 and 24 h at 30°C with shaking (150 rpm). At each time point the suspension was centrifuged at 10,000 rpm for 1 min, and the yeast cells were observed by fluorescence microscopy; the mean fluorescence intensity (MFI) was calculated using ImageJ software (NIH).
P. lutzii conditions.
The maintenance and the inoculum of P. lutzii strain 01 (ATCC MYA-826) were performed according to Scorzoni et al. (41).
Evaluation of N-glycosylation in P. lutzii.
A suspension of 106 P. lutzii cells/ml in brain heart infusion (BHI) broth was treated with a final concentration of 0.5 μg/ml G14 for 3 days at 37°C and 150 rpm. DMSO in the same proportion was used as a control. The fungal suspension was centrifuged at 4,000 rpm at 4°C for 10 min, and the obtained pellet was used in the following experiments.
(i) Enzyme activity measurement of N-glycosylated enzymes.
The pellet was washed twice with phosphate-buffered saline (PBS) containing 1 mM phenylmethylsulfonyl fluoride. Cells were disrupted by sonication, followed by centrifugation (10,000 rpm) for 15 min at 4°C for the crude protein extract obtained from the supernatant. Protein concentration was quantified by bicinchoninic acid assay (Pierce Chemical Co.), with bovine serum albumin as the standard (42). Activity of α-(1,4)-amylase (α-amylase) was assayed by monitoring starch hydrolysis (43). One unit of α-(1,4)-amylase activity was described as the amount of enzyme required to hydrolyze 0.1 mg of starch/min. Activity of α-(1,3)-glucanase was determined using α-glucan from Aspergillus niger as the substrate (26). One unit of α-(1,3)-glucanase activity was defined as the amount of enzyme required to produce 1 μM reducing sugars/min. N-Acetyl-β-d-glucosaminidase (NAGase) activity was verified by monitoring the rate of formation of ρ-nitrophenol from ρ-nitrophenol-β-N-acetylglucosamine (ρNPGlcNAc; Sigma) (25, 44). One unit of N-acetyl-β-d-glucosaminidase activity was defined as the amount of protein required to produce 1 μM ρ-nitrophenol/min at 37°C. The enzyme activity values correspond to the mean values of at least three replicates.
(ii) Evaluation of glycosylated components of fungal cell wall.
Fungal suspensions with and without G14 treatment were incubated separately with 100 μg/ml of concanavalin A (ConA), conjugated to Alexa Fluor 488 (Invitrogen), and 100 μg/ml of calcofluor white (CFW; Sigma) at 37°C for 30 min as described by de Curcio et al. (28). The fungal cells were washed, suspended in PBS, and submitted to analysis by a BD FACSCanto I flow cytometer. The mean fluorescence intensity (MFI) of 10,000 cells in the fluorescein isothiocyanate (FITC) channel was calculated. ConA-labeled fungal cells were positive for glycosylated proteins. The CFW-labeled cells were analyzed under a fluorescence microscope. A total of 300 cells (triplicate) were analyzed using ImageJ software (NIH) measuring the MFI of CFW to determine the chitin amount, which is N-glycosylated-enzyme dependent.
(iii) Differential gene expression analysis.
RNA from P. lutzii with and without G14 treatment was extracted using the TRIzol method (Invitrogen). cDNA synthesis was performed using RevertAid H Minus Reverse Transcriptase (Fermentas) according to the manufacturer’s instructions using 1 μg of total RNA. The relative expression levels of the α-(1,3)-glucanase (agn), N-acetyl-β-d-glucosaminidase (nag1), α-(1,4)-amylase (amy), inositol requiring enzyme-1 (ire1), and transcription factor (hac1) genes were analyzed (Table 2). α-Tubulin (tub) was used as the housekeeping gene. The reactions were carried out using 1 μl of cDNA, 10 μl of Maximum SYBR green/ROX qPCR Master Mix (2×) (Qiagen), and 0.5 μM each primer. The PCR conditions were as follows: an initial temperature of 95°C for 1 min, followed by 40 cycles of 5 s at 95°C and 60°C for 32 s. The signal emission corresponding to a single product was confirmed by melt curve analysis. Reactions were performed in triplicate with an Applied Biosystems 7500 cycler (Applied Biosystems). Data were analyzed using the 2-ΔΔCT method (where CT is threshold cycle) (45).
Influence of G14 on P. lutzii.
A total of 106 fungal cells/ml in BHI broth were treated with 0.5 μg/ml G14 in a 96-well plate at 37°C in 5% CO2 for 24 and 48 h. The fungal cell was analyzed by MTT assay, bud formation, trypan blue assay, and CFU enumeration. After the treatment, trypan blue reagent (Sigma) was added to the cells, and the percentage of nonblue cells was measured in a total of 100 cells. Also 100 cells were used to analyze bud formation. For this, cells were separated in two groups: (i) cells with only one or without any buds and (ii) cells with more than one bud. In the CFU enumeration assay, an aliquot of 100 μl was diluted (1:100) in PBS and plated on solid BHI medium supplemented with 4% fetal calf serum, 5% P. brasiliensis (strain 192)-spent culture medium, 1.5% glucose, and gentamicin (40 mg/ml) and incubated at 37°C for 15 days, and colonies were counted (46). For the MTT assay, the 96-well plate was centrifuged for 2 min at 5,000 rpm. The supernatant was carefully removed, and 10 μl of MTT solution (5 mg/ml in PBS) plus 40 μl of PBS was added and incubated at 37°C in 5% CO2 for 4 h. The formazan precipitate was solubilized with 100 μl/well of pure DMSO. The supernatant was transferred to other plate and read by spectrophotometer at 570 nm (690-nm reference).
Influence of G14 on P. lutzii-A549 cell interaction.
The human lung epithelial cells (A549) were obtained from Banco de Células do Rio de Janeiro (BCRJ). All the cultivation conditions were performed according to BCRJ instructions. A total of 106 cells/ml were plated in a 96-well plate in monolayer formation at 37°C in 5% CO2. Cells were challenged with 106 P. lutzii cells/ml labeled with 0.5 mg/ml of fluorescein isothiocyanate (FITC; Sigma) for 6 h at 37° C in 5% CO2. The supernatant was removed, and the infection was washed with PBS and treated with 0.5 μg/ml G14 in Dulbecco’s modified Eagle’s medium (DMEM) for 24 h at 37°C in 5% CO2. After the incubation, the interaction was analyzed by flow cytometry and CFU enumeration as described below.
(i) Flow cytometry analysis.
The supernatant was carefully removed, and 0.065 μM Alexa Fluor 647-phalloidin (ThermoFisher Scientific) solution prepared in cold 4% paraformaldehyde (Sigma) with 0.5% Triton X-100 (Amersham Biosciences) was added to the cells for 20 min. The cells were recovered in PBS and submitted to analysis by BD FACSCanto I flow cytometer. A total of 10,000 events were acquired from Alexa Fluor 647-positive events (allophycocyanin [APC] filter for the host cell population). FITC-positive events (FITC filter for the fungal cell population) were analyzed from the APC-positive population. This relation reveals the percentage of fungal cells interacting with host cells, termed the interaction rate. To the remaining suspension, 200 μg/ml trypan blue (Sigma) was added and incubated for 10 min at room temperature to quench FITC fluorescence of externally adhered fungal cells. Trypan blue reagent is able to decrease the fluorescence intensity of FITC, and it is not capable of reaching the intracellular host compartment, thus differentiating intracellular and adhered fungal cells (47). A total of 10,000 events were acquired from APC-positive events gate, obtaining a rate of fungal cells (FITC positive) internalized in the host cells. For the estimation of adhered cells, the internalized fungal cell rate was subtracted from interaction rate.
(ii) CFU enumeration.
The supernatant was discarded, and the nonadherent fungal cells were removed with PBS washes. Subsequently, 15 μg/ml of ketoconazole (Sigma) in DMEM was added for 1 h at 37°C to kill only the adhered fungi, followed by PBS washes to remove them. The A549 cells were lysed with 100 μl of sterile MilliQ water (32), and the suspension was plated on solid BHI medium supplemented as described previously and incubated at 37°C for 15 days for colony counts (46). The CFU count obtained indicates fungal cells that were internalized.
Anti-P. lutzii activity of macrophage due to G14 modulation.
RAW 264.7 macrophages were obtained from BCRJ, and all the cultivation conditions were performed according to BCRJ instructions. The macrophage suspension was adjusted to 106 cells/ml, and 100 μl was plated in a 96-well plate in monolayer formation at 37°C in 5% CO2. After macrophage monolayer formation, the supernatant was carefully removed, and cells were infected with 100 μl at 106 P. lutzii cells/ml in DMEM or treated with 0.2 mg/ml lipopolysaccharide (LPS) in DMEM (positive control) or only DMEM (negative control). The plates were incubated at 37°C in 5% CO2 for 6 h. After this the supernatant was removed, and the monolayer was carefully washed with PBS and treated with 0.5 μg/ml G14 for 24 h at 37°C in 5% CO2. Nitrite production was evaluated using a Griess assay (12, 48). For this, the supernatants were transferred to another plate and mixed 1:1 with 100 μl/well fresh solution of N-(1-naphthyl) ethylenediamine dihydrochloride (NEED; 0.1%) and sulfanilamide (1%, vol/vol) The reaction was read by spectrophotometer at 540 nm. The viability of P. lutzii cells internalized by macrophages was determined by CFU analysis. This assay was conducted as described above.
Statistical analyses.
Graphs and statistical analyzes were performed with GraphPad Prism, version 5 (La Jolla, CA, USA), using a t test.
ACKNOWLEDGMENTS
This work was supported by Rede Nacional de Métodos Alternativos, Conselho Nacional de Pesquisa e Desenvolvimento (RENAMA-CNPq) (403586/2012-7), Fundação de Amparo à Pesquisa do Estado de São Paulo (FAPESP) (2016/17048-4 to C.M.M., 2015/03700-9 to M.J.S.M.-G., 2015/14023-8 to H.C.D.O., and 2013/10917-9 to L.S.), Coordenação de Aperfeiçoamento de Pessoal de Nível Superior (CAPES), Programa de Doutorado Sanduíche no Exterior (PDSE), Programa de Apoio ao Desenvolvimento Científico da Faculdade de Ciências Farmacêuticas da UNESP (PADC/FCF), and Pró-Reitora de Pesquisa da UNESP (PROPe/UNESP).
REFERENCES
- 1.Shikanai-Yasuda MA, Mendes RP, Colombo AL, Queiroz-Telles F, Kono ASG, Paniago AMM, Nathan A, Valle A, Bagagli E, Benard G, Ferreira MS, Teixeira MM, Silva-Vergara ML, Pereira RM, Cavalcante RS, Hahn R, Durlacher RR, Khoury Z, Camargo ZP, Moretti ML, Martinez R. 2017. Brazilian guidelines for the clinical management of paracoccidioidomycosis. Rev Soc Bras Med Trop 50:715–740. doi: 10.1590/0037-8682-0230-2017. [DOI] [PubMed] [Google Scholar]
- 2.Martinez R. 2015. Epidemiology of paracoccidioidomycosis. Rev Inst Med Trop S Paulo 57(Suppl 19):11–20. doi: 10.1590/S0036-46652015000700004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.San-Blas G, Urbina JA, Marchan E, Contreras LM, Sorais F, San-Blas F. 1997. Inhibition of Paracoccidioides brasiliensis by ajoene is associated with blockade of phosphatidylcholine biosynthesis. Microbiology 143:1583–1586. doi: 10.1099/00221287-143-5-1583. [DOI] [PubMed] [Google Scholar]
- 4.Thomaz L, Apitz-Castro R, Marques AF, Travassos LR, Taborda CP. 2008. Experimental paracoccidioidomycosis: alternative therapy with ajoene, compound from Allium sativum, associated with sulfamethoxazole/trimethoprim. Med Mycol 46:113–118. doi: 10.1080/13693780701651681. [DOI] [PubMed] [Google Scholar]
- 5.Martins CVB, da Silva DL, Neres ATM, Magalhães TFF, Watanabe GA, Modolo LV, Sabino AA, de Fátima A, de Resende MA. 2009. Curcumin as a promising antifungal of clinical interest. J Antimicrob Chemother 63:337–339. doi: 10.1093/jac/dkn488. [DOI] [PubMed] [Google Scholar]
- 6.de Sá NP, Cisalpino PS, Tavares LC, Espíndola L, Pizzolatti MG, Santos PC, de Paula TP, Rosa CA, de Souza DG, Santos DA, Johann S. 2015. Antifungal activity of 6-quinolinyl N-oxide chalcones against Paracoccidioides. J Antimicrob Chemother 70:841–845. doi: 10.1093/jac/dku427. [DOI] [PubMed] [Google Scholar]
- 7.Johann S, Cisalpino PS, Watanabe GA, Cota BB, de Siqueira EP, Pizzolatti MG, Zani CL, de Resende MA. 2010. Antifungal activity of extracts of some plants used in Brazilian traditional medicine against the pathogenic fungus Paracoccidioides brasiliensis. Pharm Biol 48:388–396. doi: 10.3109/13880200903150385. [DOI] [PubMed] [Google Scholar]
- 8.de Paula e Silva ACA, Costa-Orlandi CB, Gullo FP, Sangalli-Leite F, de Oliveira HC, Silva J. d F d, Scorzoni L, Pitangui N. d S, Rossi SA, Benaducci T, Wolf VG, Regasini LO, Petrônio MS, Silva DHS, Bolzani VS, Fusco-Almeida AM, Mendes-Giannini MJS. 2014. Antifungal activity of decyl gallate against several species of pathogenic fungi. Evid Based Complement Alternat Med 2014:506273. doi: 10.1155/2014/506273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gullo FP, Sardi JC, Santos VA, Sangalli-Leite F, Pitangui NS, Rossi SA, de Paula E Silva AC, Soares LA, Silva JF, Oliveira HC, Furlan M, Silva DH, Bolzani VS, Mendes-Giannini MJ, Fusco-Almeida AM. 2012. Antifungal activity of maytenin and pristimerin. Evid Based Complement Alternat Med 2012:340787. doi: 10.1155/2012/340787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Calvo TR, Lima ZP, Silva JS, Ballesteros KV, Pellizzon CH, Hiruma-Lima CA, Tamashiro J, Brito AR, Takahira RK, Vilegas W. 2007. Constituents and antiulcer effect of Alchornea glandulosa: activation of cell proliferation in gastric mucosa during the healing process. Biol Pharm Bull 30:451–459. doi: 10.1248/bpb.30.451. [DOI] [PubMed] [Google Scholar]
- 11.Treviño-Cueto B, Luis M, Contreras-Esquivel JC, Rodríguez R, Aguilera A, Aguilar CN. 2007. Gallic acid and tannase accumulation during fungal solid state culture of a tannin-rich desert plant (Larrea tridentata Cov.). Bioresour Technol 98:721–724. doi: 10.1016/j.biortech.2006.02.015. [DOI] [PubMed] [Google Scholar]
- 12.Lopes FC, Calvo TR, Vilegas W, Carlos IZ. 2005. Inhibition of hydrogen peroxide, nitric oxide and TNF-alpha production in peritoneal macrophages by ethyl acetate fraction from Alchornea glandulosa. Biol Pharm Bull 28:1726–1730. doi: 10.1248/bpb.28.1726. [DOI] [PubMed] [Google Scholar]
- 13.Lopes FC, Rocha A, Pirraco A, Regasini LO, Siqueira JR, Silva DH, Bolzani VS, Carlos IZ, Soares R. 2011. Alchornea glandulosa ethyl acetate fraction exhibits antiangiogenic activity: preliminary findings from in vitro assays using human umbilical vein endothelial cells. J Med Food 14:1244–1253. doi: 10.1089/jmf.2010.0204. [DOI] [PubMed] [Google Scholar]
- 14.Leal PC, Mascarello A, Derita M, Zuljan F, Nunes RJ, Zacchino S, Yunes RA. 2009. Relation between lipophilicity of alkyl gallates and antifungal activity against yeasts and filamentous fungi. Bioorg Med Chem Lett 19:1793–1796. doi: 10.1016/j.bmcl.2009.01.061. [DOI] [PubMed] [Google Scholar]
- 15.Parsons AB, Lopez A, Givoni IE, Williams DE, Gray CA, Porter J, Chua G, Sopko R, Brost RL, Ho CH, Wang J, Ketela T, Brenner C, Brill JA, Fernandez GE, Lorenz TC, Payne GS, Ishihara S, Ohya Y, Andrews B, Hughes TR, Frey BJ, Graham TR, Andersen RJ, Boone C. 2006. Exploring the mode-of-action of bioactive compounds by chemical-genetic profiling in yeast. Cell 126:611–625. doi: 10.1016/j.cell.2006.06.040. [DOI] [PubMed] [Google Scholar]
- 16.Ho CH, Piotrowski J, Dixon SJ, Baryshnikova A, Costanzo M, Boone C. 2011. Combining functional genomics and chemical biology to identify targets of bioactive compounds. Curr Opin Chem Biol 15:66–78. doi: 10.1016/j.cbpa.2010.10.023. [DOI] [PubMed] [Google Scholar]
- 17.Fung SY, Sofiyev V, Schneiderman J, Hirschfeld AF, Victor RE, Woods K, Piotrowski JS, Deshpande R, Li SC, de Voogd NJ, Myers CL, Boone C, Andersen RJ, Turvey SE. 2014. Unbiased screening of marine sponge extracts for anti-inflammatory agents combined with chemical genomics identifies girolline as an inhibitor of protein synthesis. ACS Chem Biol 9:247–257. doi: 10.1021/cb400740c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Williams DE, Dalisay DS, Patrick BO, Matainaho T, Andrusiak K, Deshpande R, Myers CL, Piotrowski JS, Boone C, Yoshida M, Andersen RJ. 2011. Padanamides A and B, highly modified linear tetrapeptides produced in culture by a Streptomyces sp. isolated from a marine sediment. Org Lett 13:3936–3939. doi: 10.1021/ol2014494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Smith AM, Heisler LE, Mellor J, Kaper F, Thompson MJ, Chee M, Roth FP, Giaever G, Nislow C. 2009. Quantitative phenotyping via deep barcode sequencing. Genome Res 19:1836–1842. doi: 10.1101/gr.093955.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Krishnan K, Askew DS. 2014. Endoplasmic reticulum stress and fungal pathogenesis. Fungal Biol Rev 28:29–35. doi: 10.1016/j.fbr.2014.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Elbein AD. 1987. Inhibitors of the biosynthesis and processing of N-linked oligosaccharide chains. Annu Rev Biochem 56:497–534. doi: 10.1146/annurev.bi.56.070187.002433. [DOI] [PubMed] [Google Scholar]
- 22.Jakob CA, Burda P, Roth J, Aebi M. 1998. Degradation of misfolded endoplasmic reticulum glycoproteins in Saccharomyces cerevisiae is determined by a specific oligosaccharide structure. J Cell Biol 142:1223–1233. doi: 10.1083/jcb.142.5.1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lodish H, Berk A, Zipursky SL. 2000. Protein glycosylation in the ER and Golgi complex, section 17.7 In Lodish H, Berk A, Zipursky SL, Matsudaira P, Baltimore D, Darnell J (ed), Molecular cell biology, 4th ed W. H. Freeman, New York, NY. [Google Scholar]
- 24.Free SJ. 2013. Fungal cell wall organization and biosynthesis. Adv Genet 81:33–82. doi: 10.1016/B978-0-12-407677-8.00002-6. [DOI] [PubMed] [Google Scholar]
- 25.Dos Reis Almeida FB, Carvalho FC, Mariano VS, Alegre ACP, Silva R. d N, Hanna ES, Roque-Barreira MC. 2011. Influence of N-glycosylation on the morphogenesis and growth of Paracoccidioides brasiliensis and on the biological activities of yeast proteins. PLoS One 6:e29216. doi: 10.1371/journal.pone.0029216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dos Reis Almeida FB, Pigosso LL, de Lima Damasio AR, Monteiro VN, de Almeida Soares CM, Silva RN, Roque-Barreira MC. 2014. α-(1,4)-Amylase, but not α- and β-(1,3)-glucanases, may be responsible for the impaired growth and morphogenesis of Paracoccidioides brasiliensis induced by N-glycosylation inhibition. Yeast 31:1–11. doi: 10.1002/yea.2983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Almeida F, Antonieto AC, Pessoni AM, Monteiro VN, Alegre-Maller AC, Pigosso LL, Pereira M, Soares CM, Roque-Barreira MC. 2016. Influence of N-glycans on expression of cell wall remodeling related genes in Paracoccidioides brasiliensis yeast cells. Curr Genomics 17:112–118. doi: 10.2174/1389202917666151116212705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.de Curcio JS, Silva MG, Silva Bailão MG, Báo SN, Casaletti L, Bailão AM, de Almeida Soares CM. 2017. Identification of membrane proteome of. Future Sci OA 3:FSO232. doi: 10.4155/fsoa-2017-0044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Malhotra JD, Kaufman RJ. 2011. ER stress and its functional link to mitochondria: role in cell survival and death. Cold Spring Harb Perspect Biol 3:a004424. doi: 10.1101/cshperspect.a004424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Carbonell LM. 1967. Cell wall changes during the budding process of Paracoccidioides brasiliensis and Blastomyces dermatitidis. J Bacteriol 94:213–223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Camacho E, Nino-Vega GA. 2017. Paracoccidioides spp.: virulence factors and immune-evasion strategies. Mediators Inflamm 2017:5313691. doi: 10.1155/2017/5313691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mendes-Giannini MJ, Hanna SA, da Silva JL, Andreotti PF, Vincenzi LR, Benard G, Lenzi HL, Soares CP. 2004. Invasion of epithelial mammalian cells by Paracoccidioides brasiliensis leads to cytoskeletal rearrangement and apoptosis of the host cell. Microbes Infect 6:882–891. doi: 10.1016/j.micinf.2004.05.005. [DOI] [PubMed] [Google Scholar]
- 33.Nishikaku AS, Molina RF, Ribeiro LC, Scavone R, Albe BP, Cunha CS, Burger E. 2009. Nitric oxide participation in granulomatous response induced by Paracoccidioides brasiliensis infection in mice. Med Microbiol Immunol 198:123–135. doi: 10.1007/s00430-009-0113-x. [DOI] [PubMed] [Google Scholar]
- 34.Burger E, Nishikaku AS, Gameiro J, Francelin C, Camargo ZP, Verinaud L. 2013. Cytokines expressed in the granulomatous lesions in experimental paracoccidioidomycosis: role in host protective immunity and as fungal virulence factor. J Clin Cell Immunol S1:010. doi: 10.4172/2155-9899.S1-010. [DOI] [Google Scholar]
- 35.Morais MC, Luqman S, Kondratyuk TP, Petronio MS, Regasini LO, Silva DH, Bolzani VS, Soares CP, Pezzuto JM. 2010. Suppression of TNF-α induced NFκB activity by gallic acid and its semi-synthetic esters: possible role in cancer chemoprevention. Nat Prod Res 24:1758–1765. doi: 10.1080/14786410903335232. [DOI] [PubMed] [Google Scholar]
- 36.Piotrowski JS, Li SC, Deshpande R, Simpkins SW, Nelson J, Yashiroda Y, Barber JM, Safizadeh H, Wilson E, Okada H, Gebre AA, Kubo K, Torres NP, LeBlanc MA, Andrusiak K, Okamoto R, Yoshimura M, DeRango-Adem E, van Leeuwen J, Shirahige K, Baryshnikova A, Brown GW, Hirano H, Costanzo M, Andrews B, Ohya Y, Osada H, Yoshida M, Myers CL, Boone C. 2017. Functional annotation of chemical libraries across diverse biological processes. Nat Chem Biol 13:982–993. doi: 10.1038/nchembio.2436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Piotrowski JS, Okada H, Lu F, Li SC, Hinchman L, Ranjan A, Smith DL, Higbee AJ, Ulbrich A, Coon JJ, Deshpande R, Bukhman YV, McIlwain S, Ong IM, Myers CL, Boone C, Landick R, Ralph J, Kabbage M, Ohya Y. 2015. Plant-derived antifungal agent poacic acid targets beta-1,3-glucan. Proc Natl Acad Sci U S A 112:E1490–E1497. doi: 10.1073/pnas.1410400112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Piotrowski JS, Simpkins SW, Li SC, Deshpande R, McIlwain SJ, Ong IM, Myers CL, Boone C, Andersen RJ. 2015. Chemical genomic profiling via barcode sequencing to predict compound mode of action. Methods Mol Biol 1263:299–318. doi: 10.1007/978-1-4939-2269-7_23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Boyle EI, Weng S, Gollub J, Jin H, Botstein D, Cherry JM, Sherlock G. 2004. GO::TermFinder–open source software for accessing Gene Ontology information and finding significantly enriched Gene Ontology terms associated with a list of genes. Bioinformatics 20:3710–3715. doi: 10.1093/bioinformatics/bth456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jonikas MC, Collins SR, Denic V, Oh E, Quan EM, Schmid V, Weibezahn J, Schwappach B, Walter P, Weissman JS, Schuldiner M. 2009. Comprehensive characterization of genes required for protein folding in the endoplasmic reticulum. Science 323:1693–1697. doi: 10.1126/science.1167983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Scorzoni L, de Paula e Silva AC, Singulani JL, Leite FS, de Oliveira HC, da Silva RA, Fusco-Almeida AM, Mendes-Giannini MJ. 2015. Comparison of virulence between Paracoccidioides brasiliensis and Paracoccidioides lutzii using Galleria mellonella as a host model. Virulence 6:766–776. doi: 10.1080/21505594.2015.1085277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.dos Reis Almeida FB, de Oliveira LL, Valle de Sousa M, Roque Barreira MC, Hanna ES. 2010. Paracoccin from Paracoccidioides brasiliensis; purification through affinity with chitin and identification of N-acetyl-beta-d-glucosaminidase activity. Yeast 27:67–76. doi: 10.1002/yea.1731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Fuwa H. 1954. A new method for microdetermination of amylase activity by the use of amylose as the substrate. J Biochem 4:583–603. doi: 10.1093/oxfordjournals.jbchem.a126476. [DOI] [Google Scholar]
- 44.Almeida F, Sardinha-Silva A, da Silva TA, Pessoni AM, Pinzan CF, Alegre-Maller ACP, Cecílio NT, Moretti NS, Damásio ARL, Pedersoli WR, Mineo JR, Silva RN, Roque-Barreira MC. 2015. Toxoplasma gondii Chitinase induces macrophage activation. PLoS One 10:e0144507. doi: 10.1371/journal.pone.0144507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Livak KJ, Schmittgen TD. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 46.Singulani JL, Scorzoni L, Lourencetti NMS, Oliveira LR, Conçolaro RS, da Silva PB, Nazaré AC, Polaquini CR, Victorelli FD, Chorilli M, Regasini LO, Fusco Almeida AM, Mendes Giannini M. 2018. Potential of the association of dodecyl gallate with nanostructured lipid system as a treatment for paracoccidioidomycosis: in vitro and in vivo efficacy and toxicity. Int J Pharm 547:630–636. doi: 10.1016/j.ijpharm.2018.06.013. [DOI] [PubMed] [Google Scholar]
- 47.Nogueira SV, Fonseca FL, Rodrigues ML, Mundodi V, Abi-Chacra EA, Winters MS, Alderete JF, de Almeida Soares CM. 2010. Paracoccidioides brasiliensis enolase is a surface protein that binds plasminogen and mediates interaction of yeast forms with host cells. Infect Immun 78:4040–4050. doi: 10.1128/IAI.00221-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Salina AC, Souza TP, Serezani CH, Medeiros AI. 2017. Efferocytosis-induced prostaglandin E2 production impairs alveolar macrophage effector functions during Streptococcus pneumoniae infection. Innate Immun 23:219–227. doi: 10.1177/1753425916684934. [DOI] [PubMed] [Google Scholar]