This study was performed to evaluate the clinical impacts of putative risk factors in patients with Staphylococcus aureus bloodstream infections (BSIs) through a prospective, multicenter, observational study. All 567 patients with S. aureus BSIs that occurred during a 1-year period in six general hospitals were included in this study. Host- and pathogen-related variables were investigated to determine risk factors for the early mortality of patients with S. aureus BSIs.
KEYWORDS: Staphylococcus aureus, bloodstream infection, clonal complex 5, toxic shock syndrome toxin
ABSTRACT
This study was performed to evaluate the clinical impacts of putative risk factors in patients with Staphylococcus aureus bloodstream infections (BSIs) through a prospective, multicenter, observational study. All 567 patients with S. aureus BSIs that occurred during a 1-year period in six general hospitals were included in this study. Host- and pathogen-related variables were investigated to determine risk factors for the early mortality of patients with S. aureus BSIs. The all-cause mortality rate was 15.0% (85/567) during the 4-week follow-up period from the initial blood culture, and 76.5% (65/85) of the mortality cases occurred within the first 2 weeks. One-quarter (26.8%, 152/567) of the S. aureus blood isolates carried the tst-1 gene, and most (86.2%, 131/152) of them were identified to be clonal complex 5 agr type 2 methicillin-resistant S. aureus (MRSA) strains harboring staphylococcal cassette chromosome mec type II, belonging to the New York/Japan epidemic clone. A multivariable logistic regression showed that the tst-1 positivity of the causative S. aureus isolates was associated with an increased 2-week mortality rate both in patients with S. aureus BSIs (adjusted odds ratio [aOR], 1.62; 95% confidence interval [CI], 0.90 to 2.88) and in patients with MRSA BSIs (aOR, 2.61; 95% CI, 1.19 to 6.03). Two host-related factors, an increased Pitt bacteremia score and advanced age, as well as a pathogen-related factor, carriage of tst-1 by causative MRSA isolates, were risk factors for 2-week mortality in patients with BSIs. Careful management of patients with BSIs caused by the New York/Japan epidemic clone is needed to improve clinical outcomes.
INTRODUCTION
Staphylococcus aureus is the most common bacterial pathogen among Gram-positive cocci, causing both community-acquired and nosocomial bloodstream infections (BSIs). S. aureus BSIs result in high morbidity and mortality rates (1–3), and the underlying comorbidities of patients and initial disease severities are associated with the clinical prognoses of patients with S. aureus BSIs (4, 5). In addition, bacterial factors, including antimicrobial resistance and virulence factors, have also been suggested to be possible risk factors affecting the clinical prognoses of patients.
Methicillin-resistant S. aureus (MRSA) is clinically resistant to all beta-lactam antimicrobials, except for anti-MRSA cephalosporins, due to the production of penicillin-binding protein 2a, which has a low affinity for beta-lactams. The administration of inadequate antimicrobials for MRSA has been regarded as a risk factor for the early mortality of patients with BSIs, though this is debatable (6). Vancomycin MIC creep, i.e., decreased susceptibility to vancomycin within the susceptible range of BSI-causing MRSA strains, has also been proposed to be a risk factor for early mortality in patients (7, 8).
Exotoxins of S. aureus, including staphylococcal enterotoxins (SEs) and toxic shock syndrome toxin 1 (TSST-1), or cytotoxic exotoxins, including Panton-Valentine leukocidin (PVL), are categorized as superantigens (9). A murine model study demonstrated that TSST-1 triggers a T cell-dependent shock syndrome resulting in high lethality by stimulating the release of cytokines, including interleukin-6 and tumor necrosis factors (10, 11); however, the clinical impacts of TSST-1 production by bacterial pathogens in patients with S. aureus BSIs have not been proven to date due to the lack of clinical evidence.
We conducted a comprehensive analysis that included host-, pathogen-, and antimicrobial treatment-related variables to reveal the clinical impacts of putative risk factors on early mortality in patients with S. aureus BSIs through a prospective, multicenter, observational study focusing on the bacterial factors of S. aureus isolates.
RESULTS
Patient characteristics.
Among the 569 patients with laboratory-confirmed S. aureus BSIs, 2 patients were excluded from the study due to discharge before diagnosis. Finally, a total of 567 patients were included in this study (Fig. 1). The median age of the patients was 68.0 years (interquartile range [IQR], 55.0 to 78.0 years), and 58.0% (329/567) were male (Table 1). Diabetes mellitus (21.9%, n = 124) was the most common underlying comorbidity, followed by end-stage renal diseases (21.3%, n = 121), cardiovascular diseases (18.3%, n = 104), and malignancies (15.3%, n = 87). The median value of the Charlson comorbidity index was 2 (IQR, 1 to 3). One-half (52.2%, n = 296) of the cases were hospital-originated (HO) infections, and 27.0% (n = 153) of the patients were hospitalized in intensive care units (ICUs). Critical illness on the day of the initial blood culture was observed in 23.6% (n = 134) of the patients.
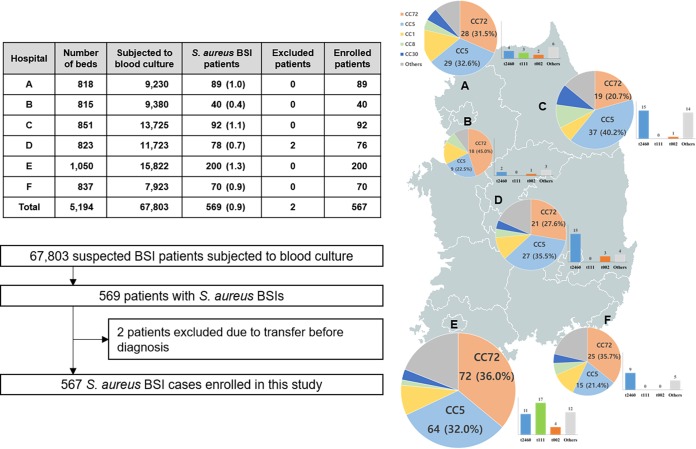
FIG 1.
Geographical distribution of S. aureus BSI cases included in this study. The number of beds, patients subjected to blood culture, and S. aureus BSI cases are indicated in the table. The pie charts indicate the proportions of the major sequence types of the S. aureus blood isolates, and the relative size of the pie chart indicates the number of S. aureus BSI episodes. The bar charts indicate the numbers of CC5 tst-1-positive MRSA isolates grouped according to the spa types in each hospital.
TABLE 1.
Host-, treatment-, and pathogen-related factors of patients with S. aureus bloodstream infectionsa
| Variable | Value for: |
P value | ||
|---|---|---|---|---|
| All patients (n = 567, 100%) | Patients infected with MRSA (n = 303, 53.4%) | Patients infected with MSSA (n = 264, 46.6%) | ||
| Patients | ||||
| Median (IQR) age (yr) | 68.0 (55.0–78.0) | 70.0 (57.0–78.0) | 67.0 (52.5–77.0) | 0.041 |
| No. (%) male patients | 329 (58.0) | 174 (57.4) | 155 (58.7) | 0.823 |
| No. (%) of patients with the following comorbidity: | ||||
| Malignancy | 87 (15.3) | 43 (14.2) | 44 (16.7) | 0.717 |
| Diabetes mellitus | 124 (21.9) | 66 (21.8) | 58 (22.0) | 0.999 |
| Cardiovascular disease | 104 (18.3) | 58 (19.1) | 46 (17.4) | 0.676 |
| Cerebrovascular disease | 80 (14.1) | 57 (18.8) | 23 (8.7) | 0.001 |
| Liver cirrhosis | 28 (4.9) | 15 (5.0) | 13 (4.9) | 0.999 |
| End-stage renal disease | 121 (21.3) | 72 (23.8) | 49 (18.6) | 0.160 |
| Median (IQR) Charlson comorbidity index | 2 (1–3) | 2 (1–3) | 2 (1–3) | 0.353 |
| No. (%) of patients with: | ||||
| HO infection | 296 (52.2) | 203 (67.0) | 93 (35.2) | <0.001 |
| ICU admission | 153 (27.0) | 99 (32.7) | 54 (20.5) | 0.001 |
| Polymicrobial infection | 34 (6.0) | 18 (5.9) | 16 (6.1) | 0.999 |
| No. (%) of patients with the following source of bloodstream infection: | 0.599 | |||
| Primary | 166 (29.3) | 87 (28.7) | 79 (30.0) | |
| Infective endocarditis | 13 (2.3) | 7 (2.3) | 6 (2.3) | |
| Non-catheter-related endovascular | 4 (0.7) | 2 (0.7) | 2 (0.8) | |
| Unknown | 149 (26.3) | 78 (25.7) | 71 (26.9) | |
| Secondary | 360 (63.5) | 191 (63.0) | 169 (64.0) | |
| Skin and soft tissue | 111 (19.6) | 59 (19.5) | 52 (19.7) | |
| Pulmonary | 91 (16.0) | 50 (16.5) | 41 (15.5) | |
| Bone and joint | 71 (12.5) | 30 (9.9) | 41 (15.5) | |
| Urinary tract | 56 (9.9) | 33 (10.9) | 23 (8.7) | |
| Others | 31 (5.5) | 19 (6.3) | 12 (4.5) | |
| Catheter related | 41 (7.2) | 25 (8.3) | 16 (6.1) | |
| Laboratory findings | ||||
| Median (IQR) white blood cell count (103 cells/μl) | 11.7 (8.2–16.1) | 11.5 (8.4–15.6) | 11.7 (7.7–17.0) | 0.754 |
| Median (IQR) platelet count (103 cells/μl) | 186 (112.5–272) | 200 (108–289) | 173.5 (115–267) | 0.177 |
| Median (IQR) C-reactive protein concn (μg/ml) | 10.8 (3.7–18.6) | 10.6 (3.7–17.9) | 11.0 (3.7–19.4) | 0.595 |
| No. (%) of patients with: | ||||
| Acute kidney injury | 56 (9.9) | 28 (9.2) | 18 (6.8) | 0.368 |
| Critical illness (Pitt score ≥ 4) | 134 (23.6) | 74 (24.4) | 60 (22.7) | 0.708 |
| Infectious disease consultation | 318 (56.1) | 190 (62.7) | 128 (48.5) | 0.001 |
| 14-day mortality | 65 (11.5) | 35 (11.6) | 30 (11.4) | 0.999 |
| No. (%) of patients with appropriate antimicrobial treatment | ||||
| Empirical | 295 (52.0) | 98 (32.3) | 197 (74.6) | <0.001 |
| Revised definitive | 434 (76.5) | 200 (66.0) | 234 (88.6) | <0.001 |
| No. (%) of patients infected with pathogens with the following characteristics: | ||||
| Strain type | <0.001 | |||
| CC72 | 183 (32.3) | 113 (37.3) | 70 (26.5) | |
| CC5 | 181 (31.9) | 156 (51.5) | 25 (9.5) | |
| CC1 | 58 (10.2) | 8 (2.6) | 50 (18.9) | |
| CC8 | 26 (4.6) | 14 (4.6) | 12 (4.5) | |
| CC30 | 25 (4.4) | 2 (0.7) | 23 (8.7) | |
| Others | 94 (16.6) | 10 (3.3) | 84 (31.8) | |
| Virulence factors | ||||
| sea | 57 (10.1) | 5 (1.7) | 52 (19.7) | <0.001 |
| seb | 1 (0.2) | 0 (0) | 1 (0.4) | 0.945 |
| sec | 141 (24.9) | 134 (44.2) | 7 (2.7) | <0.001 |
| sed | 6 (1.1) | 5 (1.7) | 1 (0.4) | 0.287 |
| see | 0 (0) | 0 (0) | 0 (0) | 0.101 |
| seg | 394 (69.5) | 270 (89.1) | 124 (47.0) | <0.001 |
| seh | 23 (4.1) | 5 (1.7) | 18 (6.8) | 0.004 |
| sei | 88 (15.5) | 63 (20.8) | 25 (9.5) | <0.001 |
| sej | 5 (0.9) | 4 (1.3) | 1 (0.4) | 0.456 |
| pvl | 13 (2.3) | 11 (3.6) | 2 (0.8) | 0.046 |
| tst-1 | 152 (26.8) | 133 (43.9) | 19 (7.2) | <0.001 |
| agr type | <0.001 | |||
| Type 1 | 321 (56.6) | 135 (44.6) | 186 (70.5) | |
| Type 2 | 183 (32.3) | 154 (50.8) | 29 (11.0) | |
| Type 3 | 57 (10.1) | 14 (4.6) | 43 (16.3) | |
| Type 4 | 6 (1.1) | 0 (0) | 6 (2.3) | |
| SCCmec | ||||
| Type II | 166 (29.3) | 166 (54.8) | ||
| Type IV | 132 (23.3) | 132 (43.6) | ||
| Other types | 5 (0.9) | 5 (1.6) | ||
Boldface indicates statistically significant characteristics.
The 567 S. aureus BSI cases were categorized into three groups by the source of BSI: primary (29.3%, n = 166), secondary (63.5%, n = 360), and catheter-related (7.2%, n = 41) BSI groups. The primary BSI group included 13 infective endocarditis and 4 non-catheter-related endovascular infection cases. The most common primary site of infection causing secondary BSIs was skin and soft tissue (19.6%, n = 111), followed by the respiratory tract (16.0%, n = 91), bone and joint (12.5%, n = 71), and the urinary tract (9.9%, n = 56). More than half (56.1%, n = 318) of the patients with S. aureus BSIs were sent for a consultation with infectious disease specialists. An appropriate antimicrobial regimen was administered to 52.0% (n = 295) of the patients as an empirical treatment and to 76.5% (n = 434) as a definitive treatment.
Mortality dynamics of patients with S. aureus BSIs.
The all-cause mortality rate of the patients with S. aureus BSIs for 4 weeks from the initial blood culture was 15.0% (85/567). The mortality dynamics of the patients with S. aureus BSIs showed a power-law tendency curve of cumulative survival over the 4-week follow-up period. The rate of death/number of subjects at risk was 8.0% (45/561) in the first week, and the rate decreased to 3.9% (20/511) in the second week, to 1.4% (7/491) in the third week, and to 2.6% (13/484) in the fourth week. More than three-quarters (76.5%, 65/85) of the deaths during the 4-week follow-up period occurred within the first 2 weeks; therefore, the 2-week mortality was determined to be a primary endpoint for early mortality to assess putative risk factors affecting BSI-related clinical outcomes in patients with S. aureus BSIs.
Bacterial characteristics.
Among the 567 S. aureus blood isolates, the most common strain type was clonal complex 72 (CC72) (32.3%, n = 183), followed by CC5 (31.9%, n = 181), CC1 (10.2%, n = 58), CC8 (4.6%, n = 26), and CC30 (4.4%, n = 25). By agr typing, agr type 1 was the dominant genotype, being found for 56.6% (n = 321) of the S. aureus isolates, followed by agr type 2 (32.3%, n = 183). More than half (53.4%, 303/567) of the S. aureus isolates exhibited resistance to cefoxitin; i.e., they had the MRSA phenotype. Nonsusceptibility to erythromycin (65.0% versus 20.8%, P < 0.001) and clindamycin (55.4% versus 4.6%, P < 0.001) was more frequently identified in MRSA isolates than in methicillin-susceptible S. aureus (MSSA) isolates (see Table S1 in the supplemental material). All S. aureus isolates exhibited susceptibility to both glycopeptides and daptomycin. All of the MRSA isolates harbored the mecA gene, and no MRSA isolate harbored the mecC gene. Most of the MRSA isolates carried staphylococcal cassette chromosome mec (SCCmec) type II (54.8%, 166/303) or SCCmec type IV (43.6%, n = 132).
The most frequently identified gene among the nine genes for SEs was seg (69.5%, 394/567), followed by sec (24.9%, n = 141) and sei (15.5%, n = 88), while see was not identified in any S. aureus blood isolates. The pvl gene was identified in only 2.3% (n = 13) of the S. aureus isolates. Notably, 26.8% (n = 152) of S. aureus isolates harbored the tst-1 gene. Quantitative real-time PCR (RT-PCR) experiments demonstrated that the relative expression levels of tst-1 in the S. aureus isolates ranged from 0.17 to 7.08 compared with the level of expression in reference strain ATCC 700699.
Clonal traits of MRSA blood isolates.
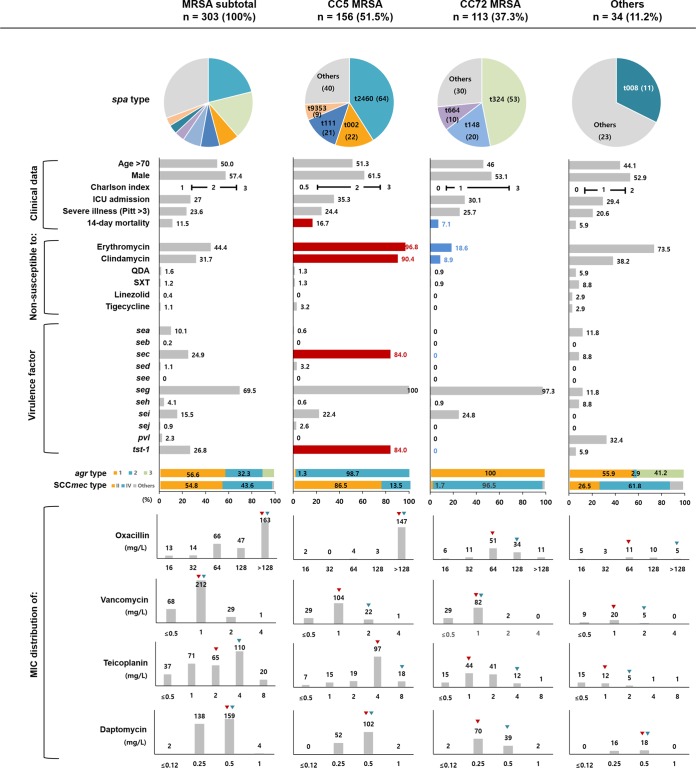
Multilocus sequence typing (MLST) revealed that two major clones, CC5 (51.5%, 156/303) and CC72 (37.3%, n = 113), were found for the majority of the MRSA blood isolates (Fig. 2). The most common spa types of CC5 MRSA isolates were t2460 (41.0%, 64/156), followed by t002 (14.1%, n = 22), t111 (13.5%, n = 21), and t9353 (5.8%, n = 9), while the CC72 MRSA isolates were mostly identified to be t324 (46.9%, 53/113), t148 (17.7%, n = 20), and t664 (8.9%, n = 10). The CC5 MRSA blood isolates mostly belonged to agr type 2 (98.7%, 154/156), and all CC72 MRSA blood isolates belonged to agr type 1. Most (86.5%, 135/156) of the CC5 MRSA blood isolates harbored SCCmec type II, while all but four (96.5%, 109/113) of the CC72 MRSA blood isolates harbored SCCmec type IV. Furthermore, CC5 and CC72 exhibited distinct clonal features in terms of antimicrobial resistance and virulence profiles. The CC5 MRSA blood isolates (MIC50, >128 μg/ml; MIC90, >128 μg/ml) exhibited higher oxacillin MICs than the CC72 isolates (MIC50, 64 μg/ml; MIC90, 128 μg/ml). Nonsusceptibility to erythromycin (96.8% versus 18.6%) and clindamycin (90.4% versus 8.9%) was more frequently observed in the CC5 MRSA isolates than in the CC72 MRSA isolates. The CC5 MRSA isolates frequently carried the virulence genes sec (84.0%, 131/156) and tst-1 (84.0%, 131/156), while none of the CC72 MRSA isolates carried them. The SmaI macrorestriction banding patterns of all the tst-1-positive CC5 MRSA isolates found by pulsed-field gel electrophoresis (PFGE) clustered the isolates as a clade with the New York/Japan epidemic clone ATCC 700699 with similarity coefficients of >85% (Fig. S1).
FIG 2.
Distribution of host- and pathogen-related factors in patients with MRSA bloodstream infections stratified according to strain type. The pie charts indicate the proportions of major spa types. Horizontal bar charts indicate the proportion of isolates for each variable, and colored bar charts and numbers indicate significant differences in proportions between CC5 and CC72 MRSA isolates (red, more prevalent; blue, less prevalent). Vertical bar charts indicate the MIC distributions of oxacillin, vancomycin, teicoplanin, and daptomycin, and red and green triangles represent the MIC50 and MIC90, respectively.
Risk factors for early mortality in patients with S. aureus BSIs.
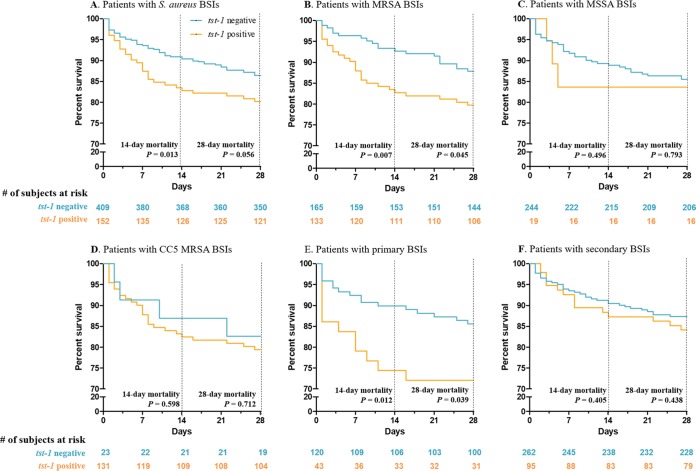
The results of univariable logistic regression determined that five host-related variables, including age (per year; odds ratio [OR], 1.04; 95% confidence interval [CI], 1.02 to 1.06), underlying cardiovascular diseases (OR, 2.03; 95% CI, 1.11 to 3.58), Charlson comorbidity index (per point; OR, 1.17; 95% CI, 1.02 to 1.34), ICU admission (OR, 2.84; 95% CI, 1.67 to 4.82), and critical illness on the day of the initial blood culture (OR, 3.53; 95% CI, 2.07 to 6.02), were significantly associated with an increased 2-week mortality rate in patients with S. aureus BSIs (Table 2). Two bacterial factors, tst-1 positivity (OR, 1.99; 95% CI, 1.15 to 3.38) and causative pathogens of agr type 2 (OR, 3.16; 95% CI, 1.48 to 7.37), were also risk factors for 2-week mortality. The factor agr type 2 was omitted from the multivariable analyses due to a high multicollinearity with tst-1 positivity. A multivariable analysis for the patients with S. aureus BSIs revealed that advanced age (per year; adjusted OR [aOR], 1.05; 95% CI, 1.02 to 1.07), Charlson comorbidity index (per score; aOR, 1.24; 95% CI, 1.06 to 1.44), ICU admission (aOR, 2.38; 95% CI, 1.32 to 4.29), and critical illness (aOR, 3.82; 95% CI, 2.12 to 6.93) were independent risk factors for 2-week mortality with statistical significance, and the tst-1 positivity (aOR, 1.62; 95% CI, 0.90 to 2.88) of the causative pathogens was also associated with an increased 2-week mortality rate with borderline significance. Similar findings were observed in patients with MRSA BSIs. A multivariable analysis in patients with MRSA BSIs showed that tst-1 positivity (aOR, 2.61; 95% CI, 1.19 to 6.03) was an independent risk factor when analyzed with three host-related variables exhibiting P values of <0.05 in the univariable analyses. Kaplan-Meier curves stratified according to tst-1 positivity showed significant differences in 2-week mortality as a primary endpoint both in patients with S. aureus BSIs (log-rank test, P = 0.013; Fig. 3A) and in patients with MRSA BSIs (log-rank test, P = 0.002; Fig. 3B) but not in patients with MSSA BSIs (log-rank test, P = 0.496; Fig. 3C). In the survival analyses for the CC5 MRSA BSI patients stratified according to tst-1 positivity, the 2-week mortality of the patients with CC5 tst-1-positive MRSA BSIs (17.6%, 23/131) was higher than that of the patients with CC5 tst-1-negative MRSA BSIs (12.0%, 3/25); however, statistical significance was not observed due to the low number of CC5 tst-1-negative MRSA cases (Fig. 3D). By the source of BSI, the difference in the 2-week mortalities stratified according to tst-1 positivity was more prominent in patients with primary S. aureus BSIs (log-rank test, P = 0.012; Fig. 3E) than in those with secondary S. aureus BSIs (log-rank test, P = 0.405; Fig. 3F).
TABLE 2.
Host-, treatment-, and pathogen-related factors affecting 2-week mortality of patients with S. aureus bloodstream infectionsc
| Variablea | Patients with S. aureus BSIs |
Patients with MRSA BSIs |
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Nonsurvivors (n = 65, 11.5%) | Survivors (n = 502, 88.5%) | Univariable analysis |
Multivariable analysis |
Nonsurvivors (n = 35) | Survivors (n = 268) | Univariable analysis |
Multivariable analysis |
|||||
| OR (95% CI) | P value | OR (95% CI) | P value | OR (95% CI) | P value | OR (95% CI) | P value | |||||
| Patients | ||||||||||||
| Median (IQR) age (yr) | 75.0 (67.0–81.0) | 68.0 (54.0–77.0) | 1.04 (1.02–1.06) | <0.001 | 1.05 (1.02–1.07) | <0.001 | 78.0 (74.5–81.5) | 68.0 (56.0–77.0) | 1.07 (1.03–1.11) | <0.001 | 1.08 (1.04–1.13) | 0.001 |
| No. (%) of male patients | 41 (63.1) | 288 (57.4) | 1.27 (0.75–2.19) | 0.381 | 22 (62.9) | 152 (56.7) | 1.29 (0.63–2.74) | 0.490 | ||||
| No. (%) of patients with the following comorbidity: | ||||||||||||
| Malignancy | 14 (21.5) | 73 (14.5) | 1.52 (0.79–2.76) | 0.183 | 7 (20.0) | 36 (13.4) | 1.61 (0.61–3.79) | 0.299 | ||||
| Diabetes mellitus | 12 (18.5) | 112 (22.3) | 0.79 (0.39–1.48) | 0.481 | 6 (17.1) | 60 (22.4) | 0.72 (0.26–1.70) | 0.481 | ||||
| Cardiovascular disease | 19 (29.2) | 85 (16.9) | 2.03 (1.11–3.58) | 0.018 | 1.05 (0.52–2.00) | 0.928 | 7 (20.0) | 51 (19.0) | 1.06 (0.41–2.45) | 0.891 | ||
| Cerebrovascular disease | 9 (13.8) | 71 (14.1) | 0.98 (0.43–1.97) | 0.948 | 6 (17.1) | 51 (19.0) | 0.88 (0.32–2.10) | 0.788 | ||||
| Liver cirrhosis | 1 (1.5) | 6 (1.2) | 1.29 (0.07–7.73) | 0.814 | 1 (2.9) | 4 (1.5) | 1.94 (0.10–13.60) | 0.558 | ||||
| End-stage renal disease | 15 (23.1) | 106 (21.1) | 1.12 (0.59–2.03) | 0.717 | 12 (34.3) | 60 (22.4) | 1.81 (0.83–3.79) | 0.124 | ||||
| Median (IQR) Charlson per point | 2 (0–4) | 2 (0–3) | 1.17 (1.02–1.34) | 0.018 | 1.24 (1.06–1.44) | 0.008 | 2 (1–3) | 1 (0–3) | 1.12 (0.92–1.35) | 0.221 | ||
| No. (%) of patients with ICU admission | 31 (47.7) | 122 (24.3) | 2.84 (1.67–4.82) | <0.001 | 2.38 (1.32–4.29) | 0.004 | 18 (51.4) | 81 (30.2) | 2.44 (1.20–5.02) | 0.014 | 2.14 (0.97–4.73) | 0.060 |
| No. (%) of patients with the following primary site of BSI | ||||||||||||
| Primary, endovascular | 23 (35.4) | 143 (28.5) | 1 | 13 (37.1) | 74 (27.6) | 1 | ||||||
| Secondary | 37 (56.9) | 323 (64.3) | 0.71 (0.41–1.26) | 0.232 | 18 (51.4) | 173 (64.6) | 0.59 (0.28–1.29) | 0.179 | ||||
| Catheter associated | 5 (7.7) | 36 (7.2) | 0.86 (9.28–2.27) | 0.781 | 4 (11.4) | 21 (7.8) | 1.08 (0.28–3.44) | 0.897 | ||||
| No. (%) of patients with critical illness (Pitt score ≥ 4) | 31 (47.7) | 103 (20.5) | 3.53 (2.07–6.02) | <0.001 | 3.82 (2.12–6.93) | <0.001 | 16 (45.7) | 58 (21.6) | 3.05 (1.46–6.31) | 0.003 | 3.32 (1.46–7.60) | 0.040 |
| No. (%) of patients with ID consultation | 31 (47.7) | 287 (57.2) | 0.68 (0.41–1.15) | 0.149 | 21 (60.0) | 169 (63.1) | 0.88 (0.43–1.84) | 0.725 | ||||
| No. (%) of patients with the following treatment: | ||||||||||||
| Empirical | 40 (61.5) | 255 (50.8) | 1.55 (0.92–2.66) | 0.105 | 13 (37.1) | 85 (31.7) | 1.27 (0.60–2.62) | 0.519 | ||||
| Revised definitive | 48 (73.8) | 386 (76.9) | 0.85 (0.48–1.57) | 0.586 | 21 (60.0) | 179 (66.8) | 0.75 (0.36–1.56) | 0.426 | ||||
| No. (%) of patients infected with pathogens with the following characteristics: | ||||||||||||
| Nonsusceptibility | ||||||||||||
| Cefoxitin (MRSA) | 35 (53.8) | 268 (53.4) | 1.02 (0.61–1.72) | 0.944 | 35 (100) | 268 (100) | ||||||
| Erythromycin | 31 (47.7) | 221 (44.0) | 1.16 (0.69–1.95) | 0.576 | 26 (74.3) | 171 (63.8) | 1.64 (0.76–3.83) | 0.225 | ||||
| Clindamycin | 25 (38.5) | 155 (30.9) | 1.40 (0.81–2.37) | 0.218 | 24 (68.6) | 144 (53.7) | 1.88 (0.90–4.13) | 0.101 | - | |||
| QDA | 0 (0) | 9 (1.8) | 0 (0) | 5 (1.9) | ||||||||
| SXT | 1 (1.5) | 5 (1.0) | 1.29 (0.07–7.73) | 0.814 | 0 (0) | 6 (2.2) | ||||||
| Linezolid | 0 (0) | 2 (0.4) | 0 (0) | 1 (0.4) | ||||||||
| Tigecycline | 1 (1.5) | 5 (1.0) | 1.55 (0.08–9.83) | 0.690 | 1 (2.9) | 5 (1.9) | 1.55 (0.08–9.97) | 0.694 | ||||
| Virulence factor | ||||||||||||
| sea | 5 (7.7) | 52 (10.4) | 0.72 (0.24–1.72) | 0.503 | 1 (2.9) | 4 (1.5) | 1.94 (0.10–13.60) | 0.558 | ||||
| seb | 0 (0) | 1 (0.2) | 0 (0) | 0 (0) | ||||||||
| sec | 22 (33.8) | 119 (23.8) | 1.64 (0.93–2.83) | 0.079 | 20 (57.1) | 114 (42.5) | 1.80 (0.89–3.73) | 0.105 | ||||
| sed | 1 (1.5) | 5 (1.0) | 1.55 (0.08–9.83) | 0.690 | 1 (2.9) | 4 (1.5) | 1.94 (0.10–13.60) | 0.558 | ||||
| seg | 48 (73.8) | 346 (68.9) | 1.27 (0.72–2.34) | 0.418 | 0 (0) | 0 (0) | 4.61 (0.94–83.33) | 0.139 | ||||
| seh | 3 (4.6) | 20 (4.0) | 1.17 (0.27–3.53) | 0.808 | 0 (0) | 5 (1.9) | ||||||
| sei | 10 (15.4) | 78 (15.5) | 0.99 (0.46–1.94) | 0.974 | 8 (22.9) | 55 (20.5) | 1.15 (0.47–2.56) | 0.749 | ||||
| sej | 1 (1.5) | 4 (0.8) | 1.95 (0.10–13.40) | 0.555 | 1 (2.9) | 3 (1.1) | 2.60 (0.13–20.94) | 0.414 | ||||
| PVL | 0 (0) | 13 (2.6) | 0 (0) | 11 (4.1) | ||||||||
| tst-1 | 28 (43.1) | 134 (26.7) | 1.99 (1.15–3.38) | 0.012 | 1.62 (0.90–2.88) | 0.104 | 25 (71.4) | 115 (42.9) | 2.75 (1.34–5.94) | 0.007 | 2.61 (1.19–6.03) | 0.019 |
| agr type 2b | 31 (47.7) | 152 (30.3) | 2.10 (1.24–3.54) | 0.005 | 26 (74.3) | 128 (47.8) | 3.16 (1.48–7.37) | 0.005 | ||||
Variables exhibiting statistical significance for 2-week mortality either in patients with S. aureus BSIs or in patients with MRSA BSIs are presented.
agr type 2 was omitted from multivariable regressions, because it showed multicollinearity with tst-1 positivity.
Abbreviations: BSI, bloodstream infection; ID, infectious disease; MRSA, methicillin-resistant S. aureus; QDA, quinupristin-dalfopristin; SXT, trimethoprim-sulfamethoxazole; PVL, Panton-Valentine leukocidin.
FIG 3.
Survival analyses of patients with S. aureus bloodstream infections stratified according to tst-1 positivity.
More than half (51.3%, 78/152) of the S. aureus isolates harboring tst-1 exhibited higher expression levels of TSST-1 than reference strain ATCC 700699, and the 2-week mortality rate of patients infected by the high-level producers (19.2%, 15/78) was higher than that of patients infected by the low-level producers (14.9%, 11/74), although the difference in 2-week mortality was not statistically significant (Fig. S2).
DISCUSSION
Through a prospective, multicenter, observational study including all patients with S. aureus BSIs that occurred in six general hospitals over a 1-year period, the clinical impacts of the putative risk factors on early mortality were evaluated. A total of 567 patients with S. aureus BSIs were included in this study, and 15.0% (85/567) of them died within the 4-week follow-up period. Most (76.5%, 65/85) of the deaths occurred within the first 2 weeks of the follow-up period, which is similar to the mortality dynamics of patients with Klebsiella pneumoniae BSIs (82.8%, 77/93) or with Escherichia coli BSIs (78.6%, 110/140) in our previous clinical studies based on the same cohort (12, 13). Therefore, the 2-week mortality could be applied as a primary endpoint to estimate the number of S. aureus BSI-related deaths.
A multivariable analysis showed that the four host-related variables, including advanced age, increased Charlson comorbidity index, ICU admission, and critical illness on the day of the initial blood culture, were independent risk factors for the 2-week mortality of patients with S. aureus BSIs. These observations were understandable because these four patient-related variables are associated with the disease severity of BSIs directly and/or indirectly and they were common risk factors for early mortality in patients with S. aureus BSIs in many previous studies (2, 4, 14, 15).
Among the pathogen-related factors, the tst-1 positivity of the bacterial pathogen was associated with an increased 2-week mortality rate in patients with S. aureus BSIs. TSST-1, one of the superantigens secreted by S. aureus isolates, induces massive cytokine production by human T lymphocytes and is associated with staphylococcal toxic shock syndrome (10). The tst-1 gene is located on a mobile pathogenic island, such as SaPI, SaPIn1, SaPIm1, SAPI2, and SaPIbov1 (16, 17), and the expression of TSST-1 is regulated by two factors consisting of the agr and sarA systems (18). In this study, the tst-1 gene was identified in 26.8% (152/567) of all S. aureus blood isolates, and all the tst-1-harboring S. aureus isolates were confirmed to produce TSST-1 by mRNA quantification. The range of tst-1 expression levels of clinical blood isolates relative to that of the reference strain, S. aureus ATCC 700699, was 0.17 to 7.08, exhibiting up to a 41.6-fold difference among the isolates. The patients with TSST-1-producing S. aureus BSIs had accompanying acute kidney injuries more frequently than those with TSST-1-nonproducing S. aureus BSIs (11.8% [18/152] versus 6.7% [28/415], P = 0.073); however, no significant differences in inflammatory markers, including white blood cell counts, platelet counts, and levels of C-reactive protein, were identified between those groups. The 2-week mortality rate of the patients with TSST-1-producing S. aureus BSIs was higher than that of the patients with TSST-1-nonproducing S. aureus BSIs, irrespective of the initial severity of disease in the patients (among the patients with critical illness, 34.9% versus 20.0%; among the patients without critical illness, 10.9% versus 5.8%). This work provides the first clinical evidence that BSIs caused by TSST-1-producing S. aureus strains resulted in a high early mortality in patients.
Notably, most (86.2%, 131/152) of the tst-1-carrying S. aureus isolates were identified to be CC5 MRSA isolates. All CC5 MRSA isolates producing TSST-1 in this study belonged to agr type 2 and harbored SCCmec type II. The PFGE banding patterns of these isolates were similar to the pattern of the New York/Japan epidemic clone (19). Our findings suggest that the tst-1-positive CC5 agr type 2 MRSA strains belonging to the New York/Japan epidemic clone make up a high-risk virulent clone resulting in poor clinical outcomes in patients with BSIs.
There are some limitations to be considered when interpreting the results of our study. First, this study was performed in a single country, South Korea, whose population is relatively homogeneous, and ethnic or racial diversity could not be evaluated. Second, unpredictable compounding factors might exist due to the observational nature of this study. Finally, the possible clinical impacts of the genetic backgrounds of CC5 MRSA isolates, other than TSST-1 production, could not be eliminated due to the low number of cases caused by CC5 tst-1-negative MRSA isolates. Further investigations should be performed.
In conclusion, two host-related factors, advanced patient age and initial critical illness determined by the Pitt bacteremia score, as well as a pathogen-related factor, TSST-1 production, were independent risk factors for the 2-week mortality of the patients with S. aureus BSIs in this study. Furthermore, the New York/Japan epidemic clone, which is endemic in South Korea, is a high-risk clone associated with a poor prognosis of patients with BSIs. Careful management of patients with BSIs caused by the high-risk clone is needed to improve clinical outcomes.
MATERIALS AND METHODS
Study population.
All patients with laboratory-confirmed S. aureus BSIs during a 1-year period from May 2016 to April 2017 in six general hospitals participating in a national antimicrobial resistance surveillance system in South Korea, namely, Kor-GLASS, were enrolled in this study (20). The sentinel hospitals are located in different provinces of the Korean peninsula, and their bed sizes range from 815 to 1,050 (Fig. 1). Among 67,803 patients subjected to blood cultures during the study period, 578 (0.8%) patients were diagnosed with S. aureus BSIs. Putative host-related risk factors, including demographic conditions, underlying comorbidities, the primary site of infection, infectious disease consultation, and antimicrobial treatment regimens, were investigated by reviewing electronic medical records. The Charlson comorbidity index and the Pitt bacteremia score were assessed on the day of the initial blood culture (21, 22). In addition, inflammatory markers of the patients with S. aureus BSIs, including white blood cell counts, platelet counts, and C-reactive protein levels, were investigated on the day of the initial blood culture. The mortality dynamics of patients with S. aureus BSIs were assessed during the 4-week follow-up period, and the clinical outcomes of the patients were evaluated by all-cause mortality rates for the first 2 weeks. Only the first blood isolate from each patient was collected for microbial analyses, and sequential isolates were discarded. Each isolate was inoculated in a cryotube containing 20% (wt/vol) skimmed milk and stored at −80°C in each sentinel hospital, and the cryotubes were transferred to an analysis center for microbiological assessments twice a month. The need for informed consent of the participants for reviewing medical records was waived by the institutional review board of each sentinel hospital due to the purely observational nature of this study.
Definitions.
An HO infection was determined when a patient with a laboratory-confirmed BSI was hospitalized for ≥48 h at the time of initial blood specimen collection for enrichment culture. A polymicrobial BSI was determined when a microorganism other than S. aureus was recovered from the blood culture within 48 h. The sources of BSIs were investigated according to the criteria suggested by the Centers for Disease Control and Prevention (23). The sources of BSIs were categorized into 3 groups: (i) a primary source, including infective endocarditis, noncatheter endovascular, and unknown sources, (ii) a secondary source, including another primary nonendovascular infection focus, and (iii) an endovascular catheter source (24). Critical illness on the day of the initial blood culture was determined as a Pitt bacteremia score of ≥4 (25). Empirical antimicrobial treatment was defined as the administration of an initial antimicrobial regimen in a blind manner without in vitro antimicrobial susceptibility testing results for the BSI-causing microorganism, and definitive antimicrobial treatment was defined as the administration of a revised antimicrobial regimen based on the in vitro antimicrobial susceptibility testing results within 72 h after the initial blood culture. An adequate antimicrobial treatment was defined as the administration of one or more antimicrobials active in vitro against the BSI-causing S. aureus isolate.
Microbiological evaluation.
Bacterial species were identified by the matrix-assisted laser desorption ionization–time-of-flight mass spectrometry technique using a Bruker Biotyper instrument (Bruker Daltonik GmbH, Bremen, Germany), and the identities were confirmed by partial sequence analyses of 16S rRNA. Susceptibility to the antimicrobials cefoxitin, erythromycin, clindamycin, quinupristin-dalfopristin, and trimethoprim-sulfamethoxazole was tested by the disk diffusion method on cation-adjusted Mueller-Hinton agar (Difco Laboratories, Detroit, MI). The MICs of oxacillin, vancomycin, teicoplanin, tigecycline, daptomycin, and linezolid were determined by the broth microdilution method according to Clinical and Laboratory Standards Institute guidelines (26). Calcium-supplemented Mueller-Hinton broth (final concentration of calcium, 50 μg/ml) was used for daptomycin MIC determination. S. aureus ATCC 25923 was used as a quality control strain. The mecA and mecC genes were evaluated by PCR in all isolates exhibiting resistance to cefoxitin, as previously described (27, 28). The SCCmec type was determined by PCR assays (29).
For virulence assessment, a total of 11 virulence genes for SEs (sea, seb, sec, sed, see, seg, seh, sei, and sej), TSST-1 (tst-1), and PVL (pvl) were assessed by PCR. The expression level of the tst-1 gene of each S. aureus isolate was evaluated by quantitative real-time PCR (RT-PCR) assays. Briefly, S. aureus isolates were cultured in Muller-Hinton broth at 37°C with vigorous agitation until the optical density at 600 nm was 1.5 to 2. Bacterial RNA was extracted using an RNeasy minikit (Qiagen, Hilden, Germany), and cDNA was synthesized using a TOPscript cDNA synthesis kit (Enzynomics, Daejeon, South Korea). Amplification was carried out in a 20-μl final volume containing 10 μl of premix (RbTaq qPCR 2× preMIX; Enzynomics), 1 μl of each primer (for tst-1 or gyrA), 7 μl of distilled water, and 1 μl of cDNA using a CFX96 system (Bio-Rad, Hercules, CA). The reaction conditions were 95°C for 30 s, followed by 40 cycles of 95°C for 10 s, 60°C for 15 s, and 72°C for 15 s. The expression levels of the tst-1 gene were normalized against those of the gyrA gene. The expression level of the tst-1 gene in S. aureus ATCC 700699 was also measured in every batch for comparison. The oligonucleotide sequences of the primers used in this study are summarized in Table S2 in the supplemental material.
For strain typing, MLST experiments were performed by comparing the partial sequences of the seven housekeeping genes, including arc, aroE, glpF, gmk, pta, tpi, and yqiL, to the sequences in the S. aureus MLST database (https://pubmlst.org/saureus/) to determine the allelic types, the sequence types, and the CCs of the S. aureus blood isolates (30). The agr types of the S. aureus isolates were determined by multiplex PCR, as previously described (31). In addition, PCR and sequencing of the staphylococcal protein A gene repeat region were performed to determine the spa types of S. aureus isolates by comparing them to the types on the RidomSpaServer website (http://spa.ridom.de/) as previously described (32). Finally, the SmaI macrorestriction banding patterns of representative CC5 MRSA blood isolates selected by the spa types and virulence profiles were evaluated by PFGE using a CHEF-DRII system (Bio-Rad, Hercules, CA) as previously described (33). S. aureus ATCC 700699 (Mu50, New York/Japan epidemic clone, tst-1 positive), S. aureus ATCC BAA1681 (USA100, tst-1 negative), and S. aureus ATCC BAA1768 (USA800) were used as reference strains for the PFGE experiments.
Statistical analysis.
Statistical analyses were conducted using R software (version 3.4.3; R Development Core Team 2017; http://www.R-project.org/). Differences between groups were determined using the Mann-Whitney U test and Fisher’s exact test for continuous variables and categorical variables, respectively. The odds ratio (OR) of each putative risk factor was calculated by univariable logistic regressions, and adjusted ORs (aORs) were calculated by multivariable logistic regressions with variables exhibiting P values of <0.05 in the univariable analyses. Survival analyses were performed by constructing Kaplan-Meier curves, and differences in mortalities between groups were determined by log-rank tests. All the results of statistical analyses were considered significant when the P values were <0.05.
Supplementary Material
ACKNOWLEDGMENTS
We thank all Kor-GLASS participants for their contribution to the program.
This work was supported by the research program funded by the South Korea Centers for Disease Control and Prevention (grant 2017E4400101).
We have no conflicts of interest to declare.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/AAC.01362-19.
REFERENCES
- 1.Cluff LE, Reynolds RC, Page DL, Breckenridge JL. 1968. Staphylococcal bacteremia and altered host resistance. Ann Intern Med 69:859–873. doi: 10.7326/0003-4819-69-5-859. [DOI] [PubMed] [Google Scholar]
- 2.Julander I. 1985. Unfavourable prognostic factors in Staphylococcus aureus septicemia and endocarditis. Scand J Infect Dis 17:179–187. doi: 10.3109/inf.1985.17.issue-2.09. [DOI] [PubMed] [Google Scholar]
- 3.Wisplinghoff H, Bischoff T, Tallent SM, Seifert H, Wenzel RP, Edmond MB. 2004. Nosocomial bloodstream infections in US hospitals: analysis of 24,179 cases from a prospective nationwide surveillance study. Clin Infect Dis 39:309–317. doi: 10.1086/421946. [DOI] [PubMed] [Google Scholar]
- 4.Lesens O, Methlin C, Hansmann Y, Remy V, Martinot M, Bergin C, Meyer P, Christmann D. 2003. Role of comorbidity in mortality related to Staphylococcus aureus bacteremia: a prospective study using the Charlson weighted index of comorbidity. Infect Control Hosp Epidemiol 24:890–896. doi: 10.1086/502156. [DOI] [PubMed] [Google Scholar]
- 5.Paulsen J, Mehl A, Askim A, Solligård E, Åsvold BO, Damås JK. 2015. Epidemiology and outcome of Staphylococcus aureus bloodstream infection and sepsis in a Norwegian county 1996–2011: an observational study. BMC Infect Dis 15:116. doi: 10.1186/s12879-015-0849-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cosgrove SE, Sakoulas G, Perencevich EN, Schwaber MJ, Karchmer AW, Carmeli Y. 2003. Comparison of mortality associated with methicillin-resistant and methicillin-susceptible Staphylococcus aureus bacteremia: a meta-analysis. Clin Infect Dis 36:53–59. doi: 10.1086/345476. [DOI] [PubMed] [Google Scholar]
- 7.Kalil AC, Van Schooneveld TC, Fey PD, Rupp ME. 2014. Association between vancomycin minimum inhibitory concentration and mortality among patients with Staphylococcus aureus bloodstream infections: a systematic review and meta-analysis. JAMA 312:1552–1564. doi: 10.1001/jama.2014.6364. [DOI] [PubMed] [Google Scholar]
- 8.van Hal SJ, Lodise TP, Paterson DL. 2012. The clinical significance of vancomycin minimum inhibitory concentration in Staphylococcus aureus infections: a systematic review and meta-analysis. Clin Infect Dis 54:755–771. doi: 10.1093/cid/cir935. [DOI] [PubMed] [Google Scholar]
- 9.Dinges MM, Orwin PM, Schlievert PM. 2000. Exotoxins of Staphylococcus aureus. Clin Microbiol Rev 13:16–34. doi: 10.1128/cmr.13.1.16-34.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Miethke T, Duschek K, Wahl C, Heeg K, Wagner H. 1993. Pathogenesis of the toxic shock syndrome: T cell mediated lethal shock caused by the superantigen TSST-1. Eur J Immunol 23:1494–1500. doi: 10.1002/eji.1830230715. [DOI] [PubMed] [Google Scholar]
- 11.Loffler B, Hussain M, Grundmeier M, Brück M, Holzinger D, Varga G, Roth J, Kahl BC, Proctor RA, Peters G. 2010. Staphylococcus aureus Panton-Valentine leukocidin is a very potent cytotoxic factor for human neutrophils. PLoS Pathog 6:e1000715. doi: 10.1371/journal.ppat.1000715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yoon EJ, Choi MH, Park YS, Lee HS, Kim D, Lee H, Shin KS, Shin JH, Uh Y, Kim YA, Shin JH, Jeong SH. 2018. Impact of host-pathogen-treatment tripartite components on early mortality of patients with Escherichia coli bloodstream infection: prospective observational study. EBioMedicine 35:76–86. doi: 10.1016/j.ebiom.2018.08.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kim D, Park BY, Choi MH, Yoon EJ, Lee H, Lee KJ, Park YS, Shin JH, Uh Y, Shin KS, Shin JH, Kim YA, Jeong SH. 2019. Antimicrobial resistance and virulence factors of Klebsiella pneumoniae affecting 30 day mortality in patients with bloodstream infection. J Antimicrob Chemother 74:190–199. doi: 10.1093/jac/dky397. [DOI] [PubMed] [Google Scholar]
- 14.Gasch O, Camoez M, Dominguez MA, Padilla B, Pintado V, Almirante B, Molina J, Lopez-Medrano F, Ruiz E, Martinez JA, Bereciartua E, Rodriguez-Lopez F, Fernandez-Mazarrasa C, Goenaga MA, Benito N, Rodriguez-Baño J, Espejo E, Pujol M, REIPI/GEIH Study Groups. 2013. Predictive factors for mortality in patients with methicillin-resistant Staphylococcus aureus bloodstream infection: impact on outcome of host, microorganism and therapy. Clin Microbiol Infect 19:1049–1057. doi: 10.1111/1469-0691.12108. [DOI] [PubMed] [Google Scholar]
- 15.Minejima E, Delayo V, Lou M, Ny P, Nieberg P, She RC, Wong-Beringer A. 2019. Utility of qSOFA score in identifying patients at risk for poor outcome in Staphylococcus aureus bacteremia. BMC Infect Dis 19:149. doi: 10.1186/s12879-019-3770-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Subedi A, Ubeda C, Adhikari RP, Penadés JR, Novick RP. 2007. Sequence analysis reveals genetic exchanges and intraspecific spread of SaPI2, a pathogenicity island involved in menstrual toxic shock. Microbiology 153:3235–3245. doi: 10.1099/mic.0.2007/006932-0. [DOI] [PubMed] [Google Scholar]
- 17.Novick RP, Christie GE, Penades JR. 2010. The phage-related chromosomal islands of Gram-positive bacteria. Nat Rev Microbiol 8:541–551. doi: 10.1038/nrmicro2393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Andrey DO, Jousselin A, Villanueva M, Renzoni A, Monod A, Barras C, Rodriguez N, Kelley WL. 2015. Impact of the regulators SigB, Rot, SarA and sarS on the toxic shock Tst promoter and TSST-1 expression in Staphylococcus aureus. PLoS One 10:e0135579. doi: 10.1371/journal.pone.0135579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Oliveira DC, Tomasz A, de Lencastre H. 2002. Secrets of success of a human pathogen: molecular evolution of pandemic clones of meticillin-resistant Staphylococcus aureus. Lancet Infect Dis 2:180–189. doi: 10.1016/S1473-3099(02)00227-X. [DOI] [PubMed] [Google Scholar]
- 20.Lee H, Yoon EJ, Kim D, Jeong SH, Shin JH, Shin JH, Shin KS, Kim YA, Uh Y, Park C, Lee KJ. 2018. Establishment of the South Korean national antimicrobial resistance surveillance system, Kor-GLASS, in 2016. Euro Surveill 23(42):pii=1700734 10.2807/1560-7917.ES.2018.23.42.1700734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Charlson ME, Pompei P, Ales KL, MacKenzie CR. 1987. A new method of classifying prognostic comorbidity in longitudinal studies: development and validation. J Chronic Dis 40:373–383. doi: 10.1016/0021-9681(87)90171-8. [DOI] [PubMed] [Google Scholar]
- 22.Hilf M, Yu VL, Sharp J, Zuravleff JJ, Korvick JA, Muder RR. 1989. Antibiotic therapy for Pseudomonas aeruginosa bacteremia: outcome correlations in a prospective study of 200 patients. Am J Med 87:540–546. doi: 10.1016/0002-9343(89)90695-5. [DOI] [PubMed] [Google Scholar]
- 23.National Healthcare Safety Network, Centers for Disease Control and Prevention. 2019. The National Healthcare Safety Network (NHSN): patient safety component manual. National Healthcare Safety Network, Centers for Disease Control and Prevention, Atlanta, GA. [Google Scholar]
- 24.Rose WE, Eickhoff JC, Shukla SK, Pantrangi M, Rooijakkers S, Cosgrove SE, Nizet V, Sakoulas G. 2012. Elevated serum interleukin-10 at time of hospital admission is predictive of mortality in patients with Staphylococcus aureus bacteremia. J Infect Dis 206:1604–1611. doi: 10.1093/infdis/jis552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yu VL, Chiou CC, Feldman C, Ortqvist A, Rello J, Morris AJ, Baddour LM, Luna CM, Snydman DR, Ip M, Ko WC, Chedid MB, Andremont A, Klugman KP, International Pneumococcal Study Group. 2003. An international prospective study of pneumococcal bacteremia: correlation with in vitro resistance, antibiotics administered, and clinical outcome. Clin Infect Dis 37:230–237. doi: 10.1086/377534. [DOI] [PubMed] [Google Scholar]
- 26.Clinical and Laboratory Standards Institute. 2016. M100-S26. Performance standards for antimicrobial susceptibility testing: twenty-sixth informational supplement. Clinical and Laboratory Standards Institute, Wayne, PA. [Google Scholar]
- 27.Milheirico C, Oliveira DC, de Lencastre H. 2007. Update to the multiplex PCR strategy for assignment of mec element types in Staphylococcus aureus. Antimicrob Agents Chemother 51:3374–3377. doi: 10.1128/AAC.00275-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Stegger M, Andersen PS, Kearns A, Pichon B, Holmes MA, Edwards G, Laurent F, Teale C, Skov R, Larsen AR. 2012. Rapid detection, differentiation and typing of methicillin-resistant Staphylococcus aureus harbouring either mecA or the new mecA homologue mecALGA251. Clin Microbiol Infect 18:395–400. doi: 10.1111/j.1469-0691.2011.03715.x. [DOI] [PubMed] [Google Scholar]
- 29.Oliveira DC, de Lencastre H. 2002. Multiplex PCR strategy for rapid identification of structural types and variants of the mec element in methicillin-resistant Staphylococcus aureus. Antimicrob Agents Chemother 46:2155–2161. doi: 10.1128/aac.46.7.2155-2161.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jolley KA, Bray JE, Maiden M. 2018. Open-access bacterial population genomics: BIGSdb software, the PubMLST.org website and their applications. Wellcome Open Res 3:124. doi: 10.12688/wellcomeopenres.14826.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gilot P, Lina G, Cochard T, Poutrel B. 2002. Analysis of the genetic variability of genes encoding the RNA III-activating components Agr and TRAP in a population of Staphylococcus aureus strains isolated from cows with mastitis. J Clin Microbiol 40:4060–4067. doi: 10.1128/JCM.40.11.4060-4067.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Koreen L, Ramaswamy SV, Graviss EA, Naidich S, Musser JM, Kreiswirth BN. 2004. spa typing method for discriminating among Staphylococcus aureus isolates: implications for use of a single marker to detect genetic micro- and macrovariation. J Clin Microbiol 42:792–799. doi: 10.1128/jcm.42.2.792-799.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.McDougal LK, Steward CD, Killgore GE, Chaitram JM, McAllister SK, Tenover FC. 2003. Pulsed-field gel electrophoresis typing of oxacillin-resistant Staphylococcus aureus isolates from the United States: establishing a national database. J Clin Microbiol 41:5113–5120. doi: 10.1128/jcm.41.11.5113-5120.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.