The global spread of multidrug-resistant enterobacteria warrants new strategies to combat these pathogens. One possible approach is the reconsideration of “old” antimicrobials, which remain effective after decades of use. Synthetic 5-nitrofurans such as furazolidone, nitrofurantoin, and nitrofurazone are such a class of antimicrobial drugs.
KEYWORDS: AhpF, Escherichia coli, drug resistance mechanism, furazolidone, genome analysis, nitrofurans, nitrofurantoin, novel nitroreductase
ABSTRACT
The global spread of multidrug-resistant enterobacteria warrants new strategies to combat these pathogens. One possible approach is the reconsideration of “old” antimicrobials, which remain effective after decades of use. Synthetic 5-nitrofurans such as furazolidone, nitrofurantoin, and nitrofurazone are such a class of antimicrobial drugs. Recent epidemiological data showed a very low prevalence of resistance to this antimicrobial class among clinical Escherichia coli isolates in various parts of the world, forecasting the increasing importance of its uses to battle antibiotic-resistant enterobacteria. However, although they have had a long history of clinical use, a detailed understanding of the 5-nitrofurans’ mechanisms of action remains limited. Nitrofurans are known as prodrugs that are activated in E. coli by reduction catalyzed by two redundant nitroreductases, NfsA and NfsB. Furazolidone, nevertheless, retains relatively significant antibacterial activity in the nitroreductase-deficient ΔnfsA ΔnfsB E. coli strain, indicating the presence of additional activating enzymes and/or antibacterial activity of the unreduced form. Using genome sequencing, genetic, biochemical, and bioinformatic approaches, we discovered a novel 5-nitrofuran-activating enzyme, AhpF, in E. coli. The discovery of a new nitrofuran-reducing enzyme opens new avenues for overcoming 5-nitrofuran resistance, such as designing nitrofuran analogues with higher affinity for AhpF or screening for adjuvants that enhance AhpF expression.
INTRODUCTION
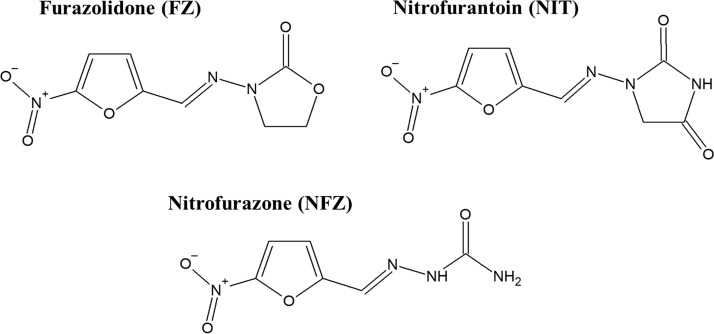
The widespread global emergence of multidrug-resistant enterobacteria warrants novel strategies to combat these pathogens (1, 2). One of the promising approaches is the reconsideration of “old” antimicrobials, which remain effective after decades of use. Synthetic 5-nitrofurans are such a class of antimicrobial drugs (Fig. 1). Typical examples of this group are furazolidone (FZ) used for treating bacterial diarrhea and giardiasis (3) and as a component in combinatorial therapy for Helicobacter pylori infections (4), nitrofurantoin (NIT) for urinary tract infections, and nitrofurazone (NFZ) for skin infections (3). Recent epidemiological data illustrated that the prevalence of resistance to 5-nitrofurans among clinical Escherichia coli isolates remains very low in various parts of the world, including Peru (5, 6), Mexico (7), the United Kingdom (8), Denmark (9), Germany (10), France (11), Iran (12), and China (13) to name a few, heralding the increasing importance of 5-nitrofurans to combat antibiotic-resistant enterobacteria.
FIG 1.
Molecular structures of the 5-nitrofuran antimicrobial agents.
Though the 5-nitrofuran agents have had a long history of clinical use since their introduction in the 1940s and 1950s (14), the knowledge about their mode of action is far from complete. Taking E. coli as a model organism, 5-nitrofurans are prodrugs that require reductive activation mediated by two type I oxygen-insensitive nitroreductases, NfsA and NfsB, in a redundant manner to exert their antibacterial effects (15–18). The minor nitroreductase NfsB is a 24-kDa flavoprotein which catalyzes the reduction of 5-nitrofuran prodrugs into nitroso and hydroxylamino-substituted products using both NADH and NADPH as reducing equivalents (19, 20). The transfer of electrons from NAD(P)H to 5-nitrofuran catalyzed by NfsB occurs via a ping-pong bi-bi mechanism, where the electron donor (NADPH or NADH) reduces the flavin mononucleotide (FMN) cofactor of the NfsB enzyme (ping), which in turn reduces the 5-nitrofuran substrate (pong). Overall, two reactants, NAD(P)H and 5-nitrofuran, give rise to two products, NAD(P)+ and the nitroso derivative (two-two or bi-bi). The flavoprotein NfsA (27 kDa) is NADPH dependent and has a dominant role in activating 5-nitrofuran drugs in E. coli, sharing the same ping-pong bi-bi mechanism of reduction with NfsB (21). The final product of NfsA-catalyzed reduction, however, remains uncharacterized. It is still uncertain what the reactive intermediates of 5-nitrofuran activation by NfsA or NfsB responsible for antibacterial effects are and what their cellular targets are. Diverse effects have been reported, including the triggering of DNA lesions, induction of oxidative stress, and inhibition of the biosynthesis of RNAs and proteins (22–26). However, it is unknown whether these macromolecules (DNA, RNA, and proteins) are directly modified by the reactive intermediates derived from 5-nitrofurans or whether the cellular machineries that carry out replication, transcription, and translation are the primary targets.
Oxygen-sensitive nitroreductase activity (or type II) was also detected in E. coli extracts by using biochemical assays in 1979 (27). This activity involves the one-electron reduction of the 5-nitrofuran drug intro a nitro anion free radical, which is oxidized back into the initial prodrug by oxygen, with generation of superoxide as a result. However, the gene(s) responsible for this activity and the extent of the contribution by this pathway to 5-nitrofuran activation have not been identified.
FZ retains relatively significant antibacterial activity in the nitroreductase-deficient ΔnfsA ΔnfsB E. coli strain, indicating the presence of additional activating enzymes and/or antibacterial activity of the unreduced FZ. Using genome sequencing, genetic, biochemical, and bioinformatic approaches, we identified a new enzyme, AhpF, that plays a role in activating FZ and two closely related drugs, NIT and NFZ, in E. coli.
RESULTS
Mutations in ahpF associated with enhanced FZ resistance.
Guided by the observation that FZ retained relatively significant antibacterial activity in the ΔnfsA ΔnfsB nitroreductase-deficient E. coli K-12 strain, we hypothesized that there are alternative activation enzyme(s) present in E. coli and/or that the unreduced form of FZ has E. coli-inhibitory properties. To examine these hypotheses, 15 independent E. coli spontaneous mutants were selected from the ΔnfsA ΔnfsB nitroreductase-deficient strain at the FZ concentration (40 μg/ml) that kills the parental strain. It is worth noting that among three FZ concentrations we used for selecting resistant mutants (40, 48, and 56 μg/ml), no colonies were observed on the plates containing 48 or 56 μg/ml, even after 48 h of incubation. The mutation rate in the ΔnfsA ΔnfsB cultures to form colonies on FZ-selective agar plates (40 μg/ml) was calculated to be 8.93 × 10−9 per cell per generation (95% confidence interval, 6.51 × 10−9 to 1.17 × 10−8).
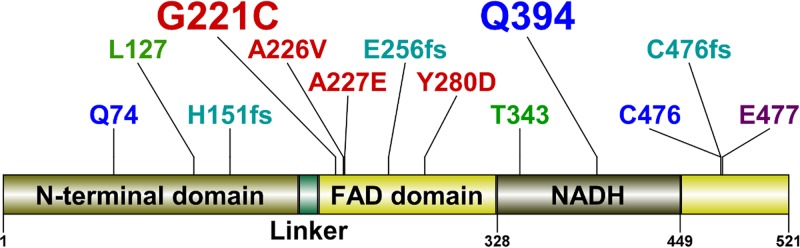
All the E. coli mutants had the same MICFZ of 20 μg/ml, which was higher than that of the parental strain (16 μg/ml) in an agar dilution assay. Genomic DNA of the FZ-resistant mutants was extracted and sequenced using the Illumina MiSeq platform as described in Materials and Methods. Comparative genome sequence analyses identified changes in a single gene, ahpF, in all the mutants. The changes in ahpF included 13 different mutations (Table 1, Fig. 2). The ahpF mutations in these FZr mutants were further confirmed by analyzing the size and sequence of ahpF-specific PCR products (see Fig. S1 in the supplemental material).
TABLE 1.
List of ahpF variants found in the E. coli mutants having increased FZ resistance
| Mutant | Location of the mutation | Predicted mutational change |
|---|---|---|
| FZ08a | 1426_1432delTGCGAAA | Frameshift downstream of Cys476 |
| FZ10a | C1180T | Stop gained, Gln394→stop codon |
| FZ11a | IS1 insertion after the 1,029th nucleotide | Loss of function |
| FZ12a | C680A | Missense, Ala227Glu |
| FZ13a | 1430_1447delAAACCAACGTGAAAGGCG | In-frame deletion, Glu477_Gly482del |
| FZ14a | 451delC | Frameshift downstream of His151 |
| FZ15a | 766delG | Frameshift downstream of Glu256 |
| FZ16a | G661T | Missense, Gly221Cys |
| FZ17a | T838G | Missense, Tyr280Asp |
| FZ18a | C1428A | Stop gained, Cys476→stop codon |
| FZ19b | IS1 insertion after the 380th nucleotide | Loss of function |
| FZ20b | C220T | Stop gained, Gln74→stop codon |
| FZ21b | C677T | Missense, Ala226Val |
| FZ22b | C1180T | Stop gained, Gln394→stop codon |
| FZ23b | G661T | Missense, Gly221Cys |
Mutations were determined using the whole-genome sequencing and ahpF-specific Sanger sequencing.
Mutations were determined using Sanger sequencing of ahpF gene only.
FIG 2.
Mutations in the AhpF protein in FZ-resistant mutants. The AhpF protein consists of four regions: The N-terminal domain (1 to 196), a linker (197 to 209), the FAD domain (210 to 327 and 450 to 521), and the NADH domain (328 to 449). The types of mutations are described by colors: red, missense mutation; blue, nonsense mutation; green, IS1 insertion; aqua, frameshift; purple, in-frame deletion of six codons. The frequency of mutations is described by the size of the annotations: small, 1; large, 2.
Given that all the mutants had the same MICFZ and some carried major interruptions to the coding sequence (an IS1 insertion in mutants FZ11 and FZ19, nonsense mutation in FZ10 and FZ22, and frameshift mutations in FZ14 and FZ15), all the ahpF mutations reported in this study were expected to result in a dysfunctional AhpF protein. This inference is also supported by the fact that the mutated residues (Gly221, Ala226, Ala227, and Tyr280) are highly conserved among the AhpF homologues according to analyses of evolutionary conservation using two software packages, Consurf (28) and SIFT (29) (see Fig. S2). Notably, the SIFT software takes into consideration the physical properties of amino acid residues in homologous sequence analyses to predict the impact of an amino acid substitution on the protein function, either tolerated or deleterious (30). In this case, all four mutations (Gly221Cys, Ala226Val, Ala227Glu, and Tyr280Asp) were predicted by SIFT to be deleterious to AhpF function with high confidence.
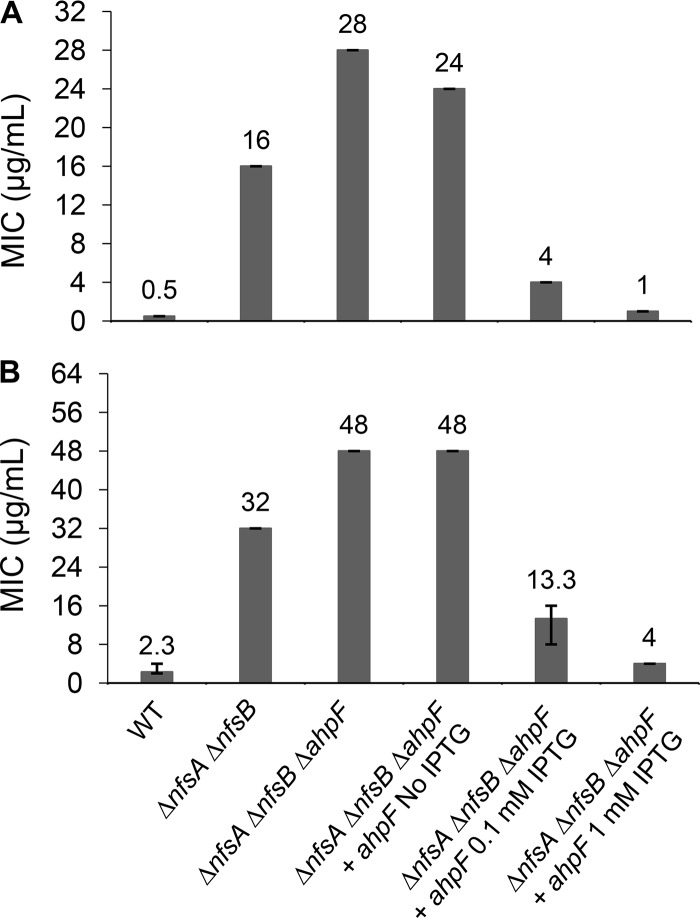
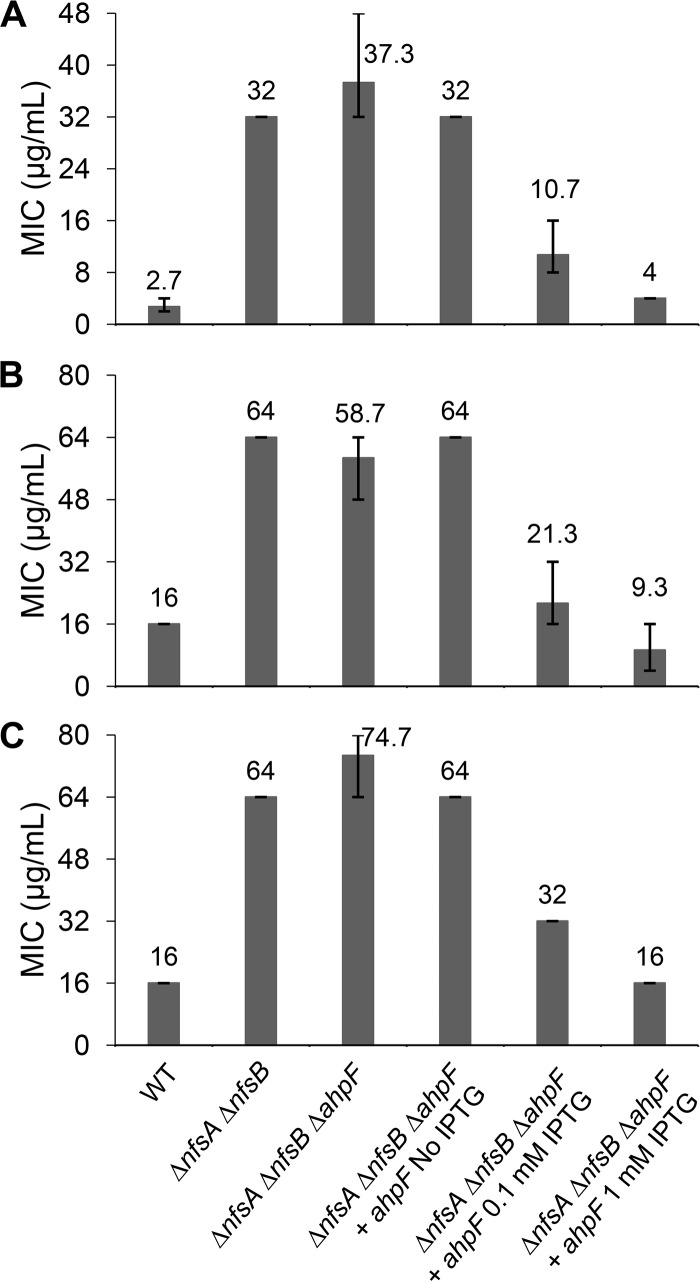
To confirm that a loss of AhpF function was the cause of increased resistance to FZ, a ΔahpF mutation was introduced into the parental ΔnfsA ΔnfsB strain by P1 transduction, using the Keio strain JW0599 as a donor, followed by the removal of the Km cassette as described in Materials and Methods to obtain a triple ΔnfsA ΔnfsB ΔahpF mutant. The FZ sensitivity was examined using the broth microdilution and agar dilution assays. In agreement with the findings from the genomic analyses of the spontaneous FZ-resistant mutants, deletion of ahpF in the ΔnfsA ΔnfsB background led to an increase in the FZ MICs in both assays, from 16 μg/ml for the parental strain to 28 μg/ml for the ΔnfsA ΔnfsB ΔahpF strain in the agar plate assay and from 32 μg/ml to 48 μg/ml in the liquid assay (Fig. 3). Complementation of the ΔahpF mutation by expression of ahpF from a high-copy-number plasmid, pCA24N::ahpF, induced by IPTG (isopropyl-β-d-thiogalactopyranoside) in the ΔnfsA ΔnfsB ΔahpF strain not only restored FZ sensitivity but also increased it dramatically beyond the level of the ΔnfsA ΔnfsB strain and close to the level of the FZ sensitive wild-type strain (nfsA+ nfsB+) (Fig. 3). Taken together, these findings show that AhpF plays a role in FZ activation, in which FZ sensitivity is positively correlated with the amount of ahpF in the cell.
FIG 3.
Confirmation of the AhpF role in FZ activation using knockout mutants and complementation. The FZ susceptibility of the ΔahpF mutant in the nfsAB null background and AhpF-overexpressing strain via an agar dilution assay (A) and broth microdilution assay (B). The E. coli K-12 strain BW25113 was used as the wild-type strain. Expression of the ahpF gene was induced from a chimeric T5-lac promoter of high-copy-number plasmid pCA24N::ahpF by 0.1 mM or 1 mM IPTG. MIC was defined as the minimal FZ concentration that inhibited the visible colony formation in an agar dilution assay (A) or that caused 90% growth inhibition in a broth microdilution assay (B). The MIC values and error bars represent the means and ranges from at least three independent experiments.
Effect of ahpF on susceptibility of E. coli to NIT and NFZ.
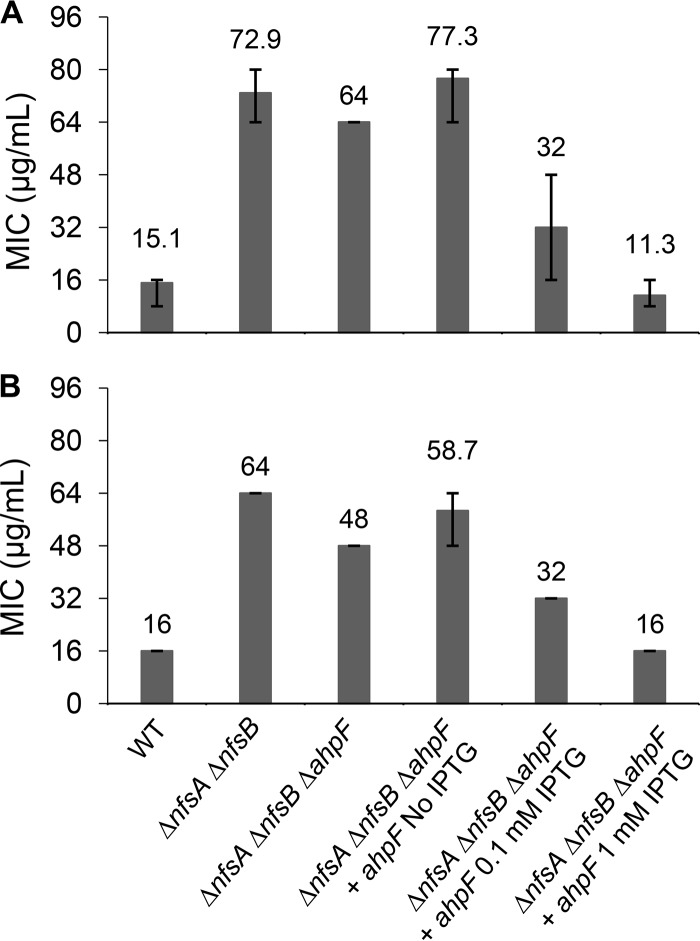
To examine the cross-resistance of ahpF deletion in the nitroreductase-deficient strain (ΔnfsA ΔnfsB) to NIT and NFZ, a broth microdilution assay was performed for these two 5-nitrofuran antibacterials. It is interesting to note that deletion of ahpF conferred a modest increase in sensitivity to NIT and NFZ (Fig. 4). That effect was reverted when the AhpF deficiency in the ΔnfsA ΔnfsB ΔahpF triple mutant was complemented by a low level of AhpF expression from the pCA24N::ahpF plasmid (in the absence of IPTG). Nonetheless, AhpF overexpression upon IPTG induction (0.1 mM or 1 mM) in the ΔnfsA ΔnfsB ΔahpF triple mutant lowered the MICNIT and MICNFZ of the complemented strain to the level of the wild-type (nfsA+ nfsB+ ahpF+) strain, where all nitrofuran-activating nitroreductases are present (Fig. 4). Taken together, all these observations strongly suggest that AhpF catalyzes activation of not only FZ but also NIT and NFZ.
FIG 4.
Confirmation of the AhpF role in nitrofurantoin and nitrofurazone activation using knockout mutants and complementation. Susceptibility to nitrofurantoin (A) and nitrofurazone (B) of the ΔahpF mutant in the nfsAB null background and AhpF-overexpressing strain via a broth microdilution assay. Expression of the ahpF gene was induced from a chimeric T5-lac promoter of high-copy-number plasmid pCA24N::ahpF by 0.1 mM or 1 mM IPTG. The E. coli K-12 strain BW25113 was used as the wild-type strain. MIC was defined as the minimal drug concentration that caused 90% growth inhibition. The MIC values and error bars represent the means and ranges from at least three independent experiments.
We also determined the MICs for the ahpF deletion and complemented strains under anaerobic conditions and found similar patterns in MIC changes for all three 5-nitrofuran drugs (Fig. 5) as those under aerobic conditions. The only exception was NFZ, for which the ΔnfsA ΔnfsB ΔahpF triple mutant had a mildly increased MICNFZ in comparison to that of the ΔnfsA ΔnfsB double mutant parent; the opposite of that was observed under aerobic conditions (Fig. 5C versus 4B).
FIG 5.
Susceptibility to 5-nitrofurans using a broth microdilution assay under anaerobic conditions. (A) Furazolidone; (B) nitrofurantoin; (C) nitrofurazone. Expression of the ahpF gene was induced from a chimeric T5-lac promoter of high-copy-number plasmid pCA24N::ahpF by 0.1 mM or 1 mM IPTG. The E. coli K-12 strain BW25113 was used as the wild-type strain. MIC was defined as the minimal drug concentration that caused 90% growth inhibition. The MIC values and error bars represent the means and ranges from at least three independent experiments.
In vitro activity of AhpF.
To verify the ability of AhpF protein to catalyze the reduction of 5-nitrofurans (FZ, NIT, and NFZ), His-tagged AhpF protein was produced, purified, and assayed in a reaction using 5-nitrofuran and NADH as reactants. The protein was expressed in the ΔahpC E. coli strain from a high-copy-number ASKA plasmid, pCA24N::ahpF (31). The rationale for ahpC deletion in the expression host was to prevent the copurification of AhpC along with AhpF, since these two proteins have been shown to form the multimeric complex AhpC10AhpF2 (32). The presence of AhpC in the protein extract might sequester the AhpF protein, precluding its hypothesized nitrofuran reductase activity.
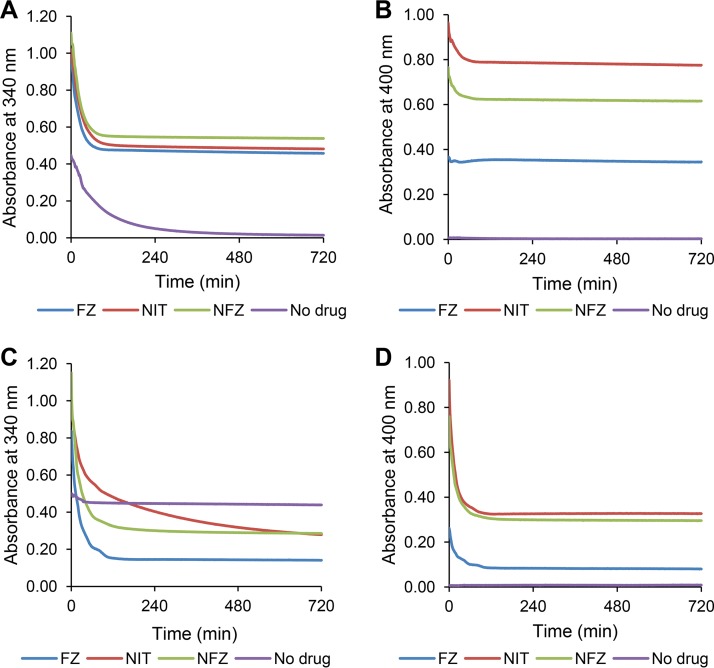
The nitroreductase assay (Fig. 6) was performed in the presence of 5 μg/ml of nickel-nitrilotriacetic acid (Ni-NTA) affinity-purified AhpF protein, NADH, and one of the three 5-nitrofurans (FZ, NIT, and NFZ) at equal amounts (0.1 mM). The absorbance at 400 nm was used to solely monitor the decrease in the concentration of 5-nitrofurans, whereas the absorbance at 340 nm (maximum for NADH) was used to monitor the decrease in the concentration of NADH and 5-nitrofurans simultaneously, given that these two substrates have overlapping absorbances at this wavelength and there was no suitable wavelength where only NADH could be detected. It should be noted that oxidation of NADH in the reactions without any 5-nitrofuran drugs was used as a reference to indicate electron transfer from NADH to the three redox centers of AhpF (the flavin adenine dinucleotide [FAD] cofactor and two disulfide bridge active centers) (33) and ultimately oxygen, due to the oxidase activity of this enzyme (34). In the presence of FZ, NIT, or NFZ, the initial reaction velocity monitored via the decrease in the absorbance at 340 nm (0.01533, 0.01314, or 0.01526 absorbance units [AU]/min, respectively) was significantly higher than that in the sample without 5-nitrofurans (0.00422 AU/min; P < 0.001) (Fig. 6A). Continuous monitoring of the reaction over 12 h showed that the absorbance at 340 nm (measure of NADH oxidation) in nitrofuran-containing samples stopped decreasing after 1.5 h, whereas the absorbance in nitrofuran-free samples continued to decrease throughout the time period of the assay (Fig. 6A). Spectral analysis at the endpoint of the experiment (12 h) showed that the residual absorbance at 340 nm in all samples was exclusively from the 5-nitrofurans (see Fig. S3A, C, E). This indicates that NADH was used up more rapidly in the presence of 5-nitrofurans. By subtracting the contribution of 5-nitrofurans from the absorbance at 340 nm, we calculated that the initial rates of NADH oxidation were 3.64, 2.74, and 2.43 μM/min for the reaction mixtures containing FZ, NIT, and NFZ, respectively, compared with that of the no-drug reference control that was 1.08 μM/min.
FIG 6.
In vitro AhpF versus NfsB nitroreductase assay under aerobic conditions. Purified His-tagged AhpF 5 μg/ml (A and B) or NfsB 1 μg/ml (C and D) was combined with furazolidone (FZ), nitrofurantoin (NIT), or nitrofurazone (NFZ) in the presence of NADH as the reducing cofactor. A reaction without 5-nitrofurans was included as a reference (denoted as a no-drug control) to monitor change of the absorbance at 340 nm due to oxidation of the cofactor NADH by the oxidase activity of AhpF. Each data point represents the mean value from the triplicate measurements.
Notably, the absorbance at 400 nm (monitoring 5-nitrofurans) was unchanged for FZ and decreased modestly for NIT and NFZ (∼16% and 15%, respectively), even after 12 h (Fig. 6B and Fig. S3A, C, and E). This is in contrast with the 5-nitrofuran reduction reaction catalyzed by the well-established nitroreductase NfsB in which, under the same conditions and substrate stoichiometry (1:1), more than half of the initial 5-nitrofuran was reduced (Fig. 6D and Fig. S3B, D, F).
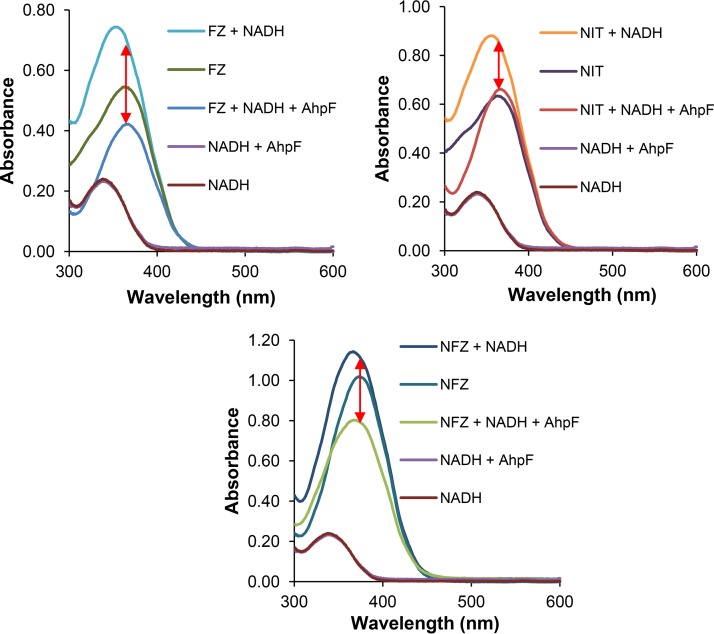
To eliminate the oxygen that appears to serve as an electron sink in the AhpF-catalyzed reaction, we repeated the nitroreductase assay under strict anaerobic conditions. The absorbance spectra from 300 to 600 nm of the reaction mixtures and controls were examined at the end of the assay, after 21 h of incubation at 25°C. Under these conditions, the absorbances between control reactions that contained NADH/AhpF and those containing NADH (in the absence of 5-nitrofurans) over the analyzed spectrum range were identical, showing that no NADH had been oxidized by the AhpF enzyme in the absence of oxygen (Fig. 7) and thus validating the anaerobic condition of the assay. There was an overall decrease in the intensities of the absorbance from 300 to 600 nm between the no-enzyme control (nitrofuran plus NADH) and the reaction sample (nitrofuran plus NADH plus AhpF) for all three drugs, (Fig. 7, red arrows). Given that no molecular oxygen was involved, this difference in the absorbance spectra can be completely attributed to the reaction between the 5-nitrofuran drug and NADH. In conclusion, this analysis shows that the AhpF indeed catalyzes the reduction of 5-nitrofurans.
FIG 7.
AhpF nitroreductase assay under oxygen-free conditions. The reaction mixture contained purified His-tagged AhpF (5 μg/ml), 5-nitrofuran (0.1 mM; furazolidone [FZ], nitrofurantoin [NIT], or nitrofurazone [NFZ]), and NADH (0.1 mM). The absorbance spectrum was measured after 21 h of incubation at 25°C. Each data point represents the mean value from the triplicate measurements. The red arrows indicate the change in the absorbance caused by AhpF-catalyzed reaction between 5-nitrofuran drugs and NADH.
In silico docking of FZ to the active site of AhpF protein.
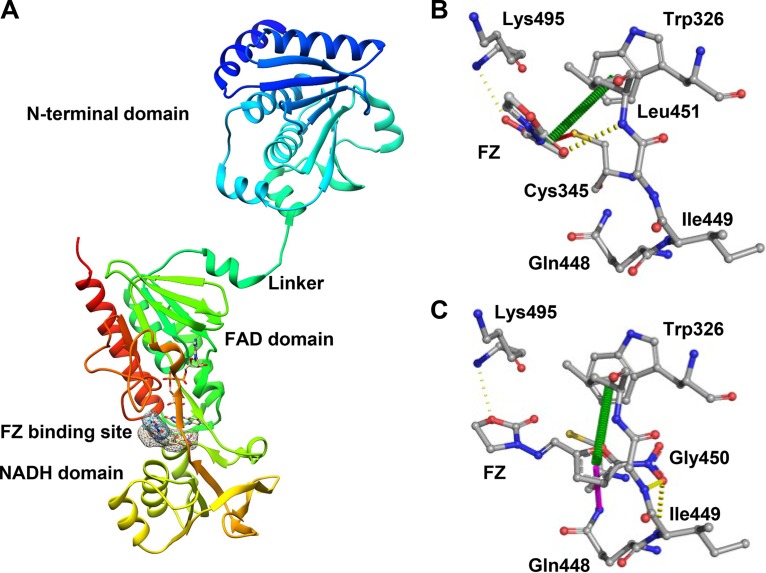
To gain a better understanding of the nitrofuran-AhpF interaction, we performed in silico docking between FZ and the AhpF enzyme using two different tools, SwissDock (35) and AutoDock Vina (36). According to the SwissDock modeling, the most favorable binding pose of the drug was in the cleft between the FAD and NADH domains, which had the lowest full fitness (−2,728.95 kcal/mol) and binding energy (ΔG = −7.13 kcal/mol) (Fig. 8A). Modeling with the AutoDock Vina tool predicted the same binding site with the free energy as low as −6.9 kcal/mol. The predicted orientations of FZ relative to the protein, however, differed by the two modeling approaches (Fig. 8B and C).
FIG 8.
Docking simulation for AhpF and FZ. (A) The cartoon three-dimensional (3D) structure of AhpF protein with 3 domains (N-terminal domain, FAD domain, and NADH domain) and the simulated FZ binding site. The surface of FZ in the binding site predicted by SwissDock and AutoDock Vina is highlighted. Interaction between FZ and AhpF residues in the binding site. Interactions were calculated and annotated using the Arpeggio server. (B) The binding pose predicted by SwissDock; (C) the binding pose predicted by AutoDock Vina. The interatomic interactions are presented as dashed lines; yellow, hydrogen bonding; red, polar contact; green, π-π interaction; purple, amide-π interaction.
Further calculating the interatomic interactions between FZ and the AhpF residues using the Arpeggio server (37) predicted that FZ interacts with protein residues more strongly in the AutoDock Vina model than in the SwissDock model (Fig. 8B and C). In the AutoDock Vina model, the furan ring of FZ was predicted to interact with the indole group of Trp326 via a π-π interaction and with the amide group of Gln448 via an amide-π interaction. The binding was further stabilized by three hydrogen bonds between FZ and three protein residues Ile449, Gly450, and Lys495 (Fig. 8C). In the SwissDock model, the indole ring of Trp326 was also predicted to play an important role in the interaction with the furan ring of FZ (Fig. 8B). Of note is the polar interaction between the oxygen of the nitro group of the drug and the thiol group of Cys345. In the AhpF protein, the Cys345XXCys348 motif establishes the redox active center, participating in the transfer of electrons from the cofactor FAD to the redox center in the N-terminal domain (38). Such an interaction predicted in the SwissDock model allows us to propose a molecular basis for the reaction: NADH donates electrons to FAD which then reduces the disulfide bridge of Cys345 and Cys348; after that, the thiol group of Cys345 contacts the nitro moiety of FZ in a manner that permits the transfer of an electron(s). We speculate that the FZ binding predicted by AutoDock Vina represents a stable binding mode, while that predicted by SwissDock represents a reactive binding mode once the thiol group of Cys345 is available in its reduced state.
DISCUSSION
Characterization of antibiotic-resistant isolates is an important tool to identify the targets and mechanisms of resistance. All mutants with increased MICs for nitrofurans published prior to our work were isolated from wild-type laboratory K-12 or clinical E. coli strains and most frequently reported to possess mutations in genes encoding the prodrug-activating enzymes NfsA and NfsB (16–18). Recently, Vervoort and coworkers (39) incorporated whole-genome sequencing to analyze nitrofurantoin-resistant E. coli mutants isolated from the wild-type parent, identifying, in addition to nfsA and nfsB mutations, a 12-nucleotide deletion in the ribE gene, encoding lumazine synthase. This is an essential enzyme in the biosynthesis of flavin mononucleotide, which in turn is the cofactor for NfsA and NfsB. Long-term laboratory evolution experiments of E. coli laboratory K-12 strains under the selective pressure of nitrofurantoin also reported various mutations in nfsA and/or nfsB in all nitrofurantoin-selected evolved cultures (40, 41). Notably, in these experiments, other mutations were also detected, such as those in genes mprA, ahpF, and porin-encoding or expression-regulatory genes (ompC, ompR, and envZ), although the underlying mechanisms and the degrees to which these mutations individually contribute to nitrofurantoin resistance have not been studied further. Overwhelmingly, the most frequently isolated 5-nitrofuran resistance-causing mutations in the E. coli wild-type strains have therefore been those that disrupt NfsA and NfsB activity. To eliminate these from our genetic screen, we started from the ΔnfsA ΔnfsB E. coli parental strain and selected 15 independent mutants of increased MICFZ. We further employed whole-genome sequencing to pinpoint the mutations. Using this strategy, we discovered the involvement of a novel enzyme, AhpF, in the activation of FZ. Overexpression of this enzyme decreased the MICs for all three tested nitrofurans under aerobic and anaerobic conditions (Fig. 3, 4, and 5). Similar MICs were obtained under both conditions, suggesting that inside the E. coli cell, 5-nitrofuran activation by AhpF is not affected by general aerobic conditions, possibly due to depletion of oxygen during culture growth. In contrast, in an in vitro enzymatic assay combining only the substrates and AhpF, reduction of 5-nitrofurans was oxygen sensitive, resulting in 5-nitrofuran reduction only in the absence of oxygen (Fig. 6 and 7; see also Fig. S3 in the supplemental material).
AhpF is a peroxiredoxin reductase, which together with the peroxiredoxin AhpC, forms the bacterial antioxidant alkyl hydroperoxide reductase AhpCF. The molecular structure of this system and its catalytic mechanism were solved recently. Briefly, AhpF transfers electrons from the electron donor NADH via the redox-active sites of its C-terminal domain and then via the disulfide redox-active center within the N-terminal domain to the oxidized AhpC, which subsequently reduces hydrogen peroxide, organic hydroperoxide, and peroxynitrite (38, 42, 43). Given its dedicated antioxidant function, it is expected that disruption of ahpF would cause an increase in antibiotic susceptibility and its overexpression would confer enhanced antibiotic resistance in general by ameliorating oxidative stress that is generally associated with the effects of a number of antibiotics. For example, it has been shown that ahpF overexpression protects E. coli from aminoglycoside-mediated protein aggregation (44) or from killing by bactericidal antibiotics such as ampicillin, gentamicin, and norfloxacin (45). In contrast to these antibiotics, deletion of the ahpF gene protects E. coli from FZ, while AhpF overexpression majorly increases susceptibility to all three tested 5-nitrofurans, in agreement with a dominant 5-nitrofuran activation role over its protective role under oxidative stress.
Notably, all FZ-resistant mutants isolated from a ΔnfsA ΔnfsB E. coli strain had an MIC of 20 μg/ml, while the complete deletion of ahpF in the ΔnfsA ΔnfsB background resulted in an MIC as high as 28 μg/ml in an agar dilution assay (Fig. 3A). The difference between disruption/point mutants and complete deletion of ahpF could be rationalized based on the published findings that ahpC mRNA is stabilized by RNase III-mediated cleavage in the intergenic region of the bi-cistronic ahpC-ahpF mRNA (46). Deletion of the ahpF open reading frame (ORF) sequence is expected to change the RNase III target and interfere in the processing, resulting in a decrease in the stability of the ahpC transcript and thus in its abundance. Given the antioxidant role of AhpC, lowering its amount in the cell upon the complete deletion of the ahpF sequence may confer increased resistance to FZ in addition to the level caused by absence of the nitroreductase AhpF.
Deletion of ahpF in the ΔnfsA ΔnfsB background caused decreased resistance to NFZ and NIT under aerobic conditions (Fig. 4). This phenomenon can be rationalized as follows: AhpF has opposing dual functions, acting alone to activate nitrofurans, and acting in complex with AhpC to counteract the oxidative stress imposed by 5-nitrofurans (26, 38). Integration of these two roles dictates the net effect of ahpF deletion on nitrofuran susceptibility. Depending on the degree of oxidative stress induced by 5-nitrofurans, the rate of AhpF-mediated 5-nitrofuran reduction and the toxicity triggered by the reduction product, different effects on resistance to the three drugs (FZ, NIT, and NFZ) were observed upon ahpF deletion.
The in vitro nitroreductase assay of purified AhpF provided a hint of the mechanism of 5-nitrofuran reduction by this enzyme. The monitoring of two substrates, 5-nitrofuran and NADH, showed that NADH became oxidized, while the 5-nitrofuran concentration stayed largely unchanged during the time course of the enzymatic experiment when the reaction was carried out under aerobic conditions, i.e., in the presence of oxygen (Fig. 6 and S3). This is a characteristic of the futile reduction cycle catalyzed by the type II oxygen-sensitive nitroreductases as reported by Peterson et al. (27). These type II oxygen-sensitive nitroreductases catalyze the one-electron reduction of the nitroaromatic prodrugs to result in a nitro anion free radical, which in the presence of oxygen, is subsequently oxidized back to the nitro group while reducing oxygen to generate superoxide. Our AhpF enzymatic assays under aerobic conditions showed that NADH oxidation occurred in the absence of 5-nitrofurans, due to the direct reduction of oxygen, but its rate increased in the presence of 5-nitrofurans. The concentration of 5-nitrofurans, on the other hand, did not change, in agreement with the futile redox cycle of 5-nitrofurans. In contrast, in our oxygen-free assays, NADH was oxidized only in the presence of 5-nitrofurans, which in turn remained reduced in the absence of oxygen. This again is in agreement with the observed properties of the type II oxygen-sensitive nitroreductases. Detailed kinetic studies of the AhpF enzyme and determination of the reduced product under aerobic and anaerobic conditions are warranted to shed more light on AhpF-catalyzed 5-nitrofuran reduction mechanism.
In addition to the enzymatic assay, we described in silico simulation of the FZ-AhpF interaction by using two software packages, AutoDock Vina and SwissDock. Both of these predicted the binding of FZ in the cleft near the C-terminal disulfide bridge center between the FAD and NADH binding sites (Fig. 8). However, the predicted FZ orientation within the binding site differs substantially between the two models obtained using AutoDock Vina and SwissDock. Further experimental approaches, such as solving the cocrystal structure of the AhpF enzyme with the drug and/or enzyme structure-function analysis of engineered mutant enzymes, are required to improve the interaction model. This is important, since a reliable enzyme/drug interaction model may facilitate the rational design of 5-nitrofuran analogues which are activated by AhpF with a greater efficiency than the existing commercial 5-nitrofurans. These analogues could potentially be employed effectively against 5-nitrofuran-resistant pathogenic E. coli clinical isolates that all have been identified to date to be nfsA and/or nfsB loss-of-function mutants.
The selection of mutants with increased FZ resistance in the absence of NfsA and NfsB did not result in any mutations of putative 5-nitrofuran targets, reflecting the high probability that once activated, the reactive intermediate(s) of 5-nitrofurans attacks multiple cellular components promiscuously rather than having specific targets. Alternatively, the drug targets are essential in a manner that no resistance-causing mutations are allowed. It is also noteworthy that 5-nitrofurans still possess some antibacterial effect in the ΔnfsA ΔnfsB ΔahpF E. coli triple mutant. It is therefore conceivable that E. coli possesses other activation enzymes and/or that the nonreduced forms of 5-nitrofurans have antibacterial properties.
Our discovery of a new 5-nitrofuran-activating enzyme in E. coli, AhpF, provides opportunities for the development of novel strategies for 5-nitrofuran-based antibacterial therapies. Screening for small molecules to upregulate the expression/availability of AhpF and designing 5-nitrofuran analogues with high affinity for AhpF are promising approaches to discover novel antibacterial candidates to counteract pathogenic E. coli isolates that are resistant to current commercial 5-nitrofurans (FZ, NIT, and NFZ) due to the nfsA and nfsB mutation.
MATERIALS AND METHODS
E. coli strains, growth condition, and antibiotics.
All E. coli strains and plasmids used in this study are listed in Table 2. Mutations of the Keio single-gene deletion E. coli collection containing the FLP recombination target (FRT)-flanked Kanr marker (47) were introduced into E. coli recipient strains by P1 transduction according to the standard procedure (48). The FRT-flanked Kanr cassette was then removed using the FLP-mediated recombination as previously described (49). The plasmid pCA24N::ahpF was purified from the E. coli strain JW0599 of the ASKA collection (31) using the ChargeSwitch-Pro Plasmid Miniprep kit (Thermo Fisher Scientific) and then chemically transformed (50) into the ΔnfsA ΔnfsB ΔahpF strain for the complementation assay or ΔahpC strain for AhpF production.
TABLE 2.
E. coli strains and plasmids used in this study
| Strain or plasmid | Genotype or description | Source |
|---|---|---|
| Strains | ||
| BW25113 | rrnB3 ΔlacZ4787 hsdR514 Δ(araBAD)567 Δ(rhaBAD)568 rph-1 | 47 |
| K2479 | BW25113 ΔnfsA ΔnfsB | This study |
| K2506 | BW25113 ΔnfsA ΔnfsB ΔahpF | This study |
| K2511 | BW25113 ΔnfsA ΔnfsB ΔahpF pCA24N::ahpF | This study |
| K2526 | BL21 pCA24N::nfsB | This study |
| K2528 | BW25113 ΔahpC pCA24N::ahpF | This study |
| Plasmids | ||
| pCP20 | Apr, Cmr, FLP+, λ cI857+, λ pR Repts; For removal of kan markersby FLP-mediated site-specific recombination | 77 |
| pCA24N::ahpF | Cmr; lacIq, pCA24N PT5-lac::ahpF | 31 |
| pCA24N::nfsB | Cmr; lacIq, pCA24N PT5-lac::nfsB | 31 |
E. coli was grown in 2× yeast extract-tryptone (YT) medium (BD Difco) at 37°C with shaking at 200 rpm. For the preparation of exponentially growing cells, an overnight culture was diluted 100-fold and incubated until it reached an optical density at 600 nm (OD600) of approximately 0.1 to 0.4. This cell suspension was then diluted to a desirable density depending on the specific purpose. Antibacterials used in this study were purchased from Goldbio, apart from nitrofurazone, which was purchased from Sigma.
Isolating FZ-resistant mutants.
FZ-resistant mutants were isolated from spontaneous mutations in E. coli overnight populations. The ΔnfsA ΔnfsB strain (K2479) was used as the parental strain for selection. Twenty independent overnight cultures were prepared. Each cell culture (100 μl) was mixed with 2.5 ml of molten soft agar (2× YT, 0.5% agarose) and then poured onto 2× YT plates containing 40 μg/ml, 48 μg/ml, or 56 μg/ml of FZ. The agar was allowed to solidify. The plates were examined after 24 h and 48 h of incubation at 37°C. Colonies formed on these plates were subcultured onto 2× YT agar and incubated overnight at 37°C. Only one colony was collected from each culture to minimize the chance of repeatedly isolating the same mutation. Putative resistant mutants were clonally purified and examined for increased MICFZ by the agar dilution assay, giving rise to 15 true FZ-resistant mutants.
Fluctuation assay.
Twenty-seven parallel cultures, each 100 μl, were prepared on a 96-well plate (polystyrene; Jet Biofil) at the starting inoculum of 105 CFU/ml. The cultures were incubated at 37°C for 24 h with vigorous shaking. Three cultures were used to determine the bacterial concentration (and thus the number of plated cells per culture) by plating 10-fold serial dilutions on nonselective agar plates. Each of the remaining 24 cultures (100 μl) was mixed with 2.5 ml of molten soft agar (2× YT, 0.5% agarose) and poured onto selective agar plates containing FZ (40 μg/ml). After 48 h of incubation at 37°C, the colonies formed on the selective agar were counted. The most probable number of mutations per culture (m) and its 95% confidence interval were calculated using the newton.LD() and confint.LD() functions, respectively, of the rSalvador package v1.7 (51) in the R environment (v.3.4.4) (52). The mutation rate (μ) was estimated as m/Nt, in which Nt is the number of plated cells per culture (4.93 × 108).
Genomic comparative analysis.
The genomic DNA of FZ-resistant mutants and the parental strain (ΔnfsA ΔnfsB E. coli strain) were purified using the UltraClean Microbial DNA Isolation kit according to the manufacturer’s instructions (Qiagen). The DNA samples were then submitted to the Massey Genome Service (New Zealand Genomics Ltd., Massey University, Palmerston North, New Zealand) for whole-genome sequencing using Illumina TruSeq Nano DNA library preparation and 2 × 250-base paired-end v2 sequencing chemistry on the Illumina MiSeq sequencing platform. The raw reads were trimmed to a quality cutoff value of Q30 (equivalent to error probability P = 0.001) and any short-length reads (<25 base by default) were removed using SolexaQA++ v3.1.7.1 (53). The DNA sequence data generated resulted in a theoretical genome coverage that was at least 40× based on the E. coli strain BW25113 genome size. The trimmed reads were aligned with the reference E. coli strain BW25113 genome (ASM75055v1 from Ensembl [54]) with Bowtie2 v2.3.2 using the very-sensitive mode (55). The resulting sam files were then converted to bam files using SAMtools v1.5 (56), and variant calling was carried out with freebayes v1.0.2 (57) using the default parameters, except ploidy was set to 1 (-p 1). The variants were functionally annotated using SnpEff v4.3p (58). The ahpF mutations were mapped to the corresponding protein domains using the visualization software DOG v2.0.1 (59).
To identify structural variations in the genomes of FZ-resistant mutants, the unmapped reads were extracted using SAMtools v1.5 (56) and then assembled to generate contigs using SPAdes v3.9.0 with the “careful” option (60). The resulting contigs were compared with the E. coli reference genome BW25113 (accession no. CP009273.1) (61), using the website platform NCBI Nucleotide BLAST 2.7.0+ (62), to determine the boundaries where the structure variations occurred.
ahpF sequence analysis.
Genomic DNA of FZ-resistant isolates was extracted by water boiling as previously described (63). The PCRs were performed in 50-μl mixtures containing 10 μl of 5× TaKaRa PrimeSTAR PCR buffer, 0.2 mM each deoxynucleoside triphosphate (dNTP), 0.2 μM ahpF forward primer (5′-AGGTGAAGCAACTCTGGCTC-3′), 0.2 μM ahpF reverse primer (5′-GCAACCCATCGATTTCGACC-3′), 0.5 μl of PrimeSTAR HS DNA polymerase (TaKaRa Bio USA), and 5 μl of the DNA extract. The PCR conditions included an initial denaturation at 94°C for 30 s, followed by 30 cycles of denaturation at 98°C for 10 s, annealing at 55°C for 5 s, and extension at 72°C for 2 min. The PCR products were analyzed by agarose gel electrophoresis as previously described (64).
The ahpF amplicons were cleaned up using ChargeSwitch-Pro PCR Clean-Up kit (Invitrogen) according to the manufacturer’s instructions and submitted to the Massey Genome Service (Massey University, Palmerston North, New Zealand) for DNA Sanger sequencing using BigDye Terminator v3.1. The primers used for sequencing included ahpF forward primer, ahpF reverse primer, and ahpF internal forward primer (5′-GTTCACCTCGCTGGTACTGG-3′). The low-quality bases of raw sequences were trimmed until the average quality of 20 bases was over 30 using Chromas v2.6.4 (Technelysium Pty Ltd.). The trimmed sequence of the ahpF amplicon was then aligned with the E. coli reference genome BW25113 (accession no. CP009273.1) using the website platform NCBI Nucleotide BLAST 2.7.0+ (62) to determine the mutations in the gene ahpF.
In silico analysis of missense mutations in ahpF.
The effect of missense mutations in the gene ahpF in FZ-resistant E. coli mutants was predicted by the SIFT web server (29) using the UniRef 90 database with default parameters and the ConSurf web server with default parameters (28).
Antimicrobial susceptibility assays.
The antimicrobial susceptibility of E. coli K-12 strains to 5-nitrofurans was examined using the agar dilution and broth microdilution assays as previously described (65, 66). The range of drug concentrations tested included 32, 28, 24, 20, 16, 12, 8, 4, 2, 1, 0.5, 0.25, 0.125, and 0 μg/ml in the agar dilution assay and 80, 64, 48, 32, 16, 8, 4, 2, 1, 0.5, and 0.25 μg/ml in the broth microdilution assay. In ahpF-complemented strains, expression of ahpF was under the control of a chimeric PT5-lac promoter of a high-copy-number plasmid, pCA24N::ahpF, and induced by 0.1 mM or 1 mM IPTG. For the broth microdilution assay under anaerobic conditions, the plates were incubated in a BBL GasPak 150 anaerobic jar (Becton, Dickinson) containing three EZ GasPak sachets with an oxygen indicator.
Production and purification of His-tagged AhpF and NfsB proteins.
The His-tagged AhpF and NfsB proteins were expressed from the high-copy-number plasmids pCA24N::ahpF and pCA24N::nfsB (31) in the E. coli strains K2528 and K2526, respectively. The E. coli culture was grown to reach an OD600 of approximately 0.6 and then induced with 1 mM IPTG at 37°C for 4 h. The cells were harvested by centrifugation at 4,000 × g for 15 min at 4°C. The pellet was stored at −20°C until being used for cell lysis and protein purification.
The affinity purification of His-tagged AhpF from cell lysate was performed using a Ni-NTA Spin kit according to the manufacturer’s instructions, with some modifications (Qiagen). First, the cell pellet harvested from 50 ml of the cell culture was suspended in 3,000 μl lysis buffer (NPI20; 50 mM NaH2PO4, 300 mM NaCl, 20 mM imidazole, pH 8.0) containing 1 mg/ml lysozyme (Boehringer Mannheim) and frozen at −80°C. The suspension was then thawed at room temperature, 2 μl of Benzonase endonuclease at 10 units/μl (Sigma) was added, and the mixture was incubated at 4°C on a tube roller. The lysis mixture was sonicated two times, each session lasted for 2 min, including alternate 1-s on/off pulses at the power 2 using the microtip of a Virsonic 600 ultrasonic cell disruptor. The cell lysate was then centrifuged at 12,000 × g for 30 min at 4°C.
The Ni-NTA spin column was equilibrated with 600 μl NPI20 buffer and centrifuged at 890 × g for 2 min. Next, 600 μl of cell lysate was loaded into the Ni-NTA spin column and centrifuged at 270 × g for 5 min. Following that, the Ni-NTA spin column was washed four times with 600 μl of NPI50 (50 mM NaH2PO4, 300 mM NaCl, 50 mM imidazole, pH 8.0) and centrifuged at 890 × g for 2 min. A His-tagged protein (AhpF or NfsB) was eluted by loading 200 μl of NPI500 buffer (50 mM NaH2PO4, 300 mM NaCl, 500 mM imidazole, pH 8.0) into the Ni-NTA spin column and centrifuging at 890 × g for 2 min. The elution step was performed four times and the eluates were pooled. The exchange to Tris buffer (pH 7.4, 50 mM) and further removal of small unwanted proteins were performed using a Vivaspin ultrafiltration device 2 (GE Healthcare) with the cutoff size of 100 kDa for AhpF eluates and 10 kDa for NfsB eluates, according to the manufacturer’s instructions. Purity of the protein extract was analyzed using SDS-PAGE, followed by Coomassie blue staining (67, 68), and densitometric analyses were performed with ImageJ software v1.51k (69) (see Fig. S4 in the supplemental material).
Protein quantification assay.
The quantity of the protein was determined using a Coomassie (Bradford) protein assay kit according to the manufacturer’s instructions (Thermo Scientific).
Nitrofuran reductase assay.
The enzymatic assay for His-tagged AhpF or NfsB protein extract was performed on a 96-well plate (polystyrene; Jet Biofil) with the total volume of 200 μl containing 0.1 mM 5-nitrofuran (FZ, NIT, or NFZ) and 0.1 mM NADH in 50 mM Tris-HCl buffer (pH 7.4). The activity was determined in the presence of 5 μg/ml of the AhpF enzyme or 1 μg/ml of the NfsB enzyme. The wells without 5-nitrofurans were used as references to monitor the oxidation of NADH by the oxidase activity of AhpF. The wells containing no protein extract were used as negative controls. Each reaction was performed in triplicates. The reaction was initiated by adding the enzyme. The progress of the reaction was monitored by measuring absorbances at 340 nm and 400 nm every 1 min for 12 h at 25°C using a Multiskan GO microplate spectrophotometer (Thermo Scientific). The absorbance spectrum from 300 to 600 nm was recorded at the end of the experiment.
For the nitroreductase assay under anaerobic conditions, the same protocol was applied with some changes. The Tris buffer (7.4) and water were gassed with oxygen-free carbon dioxide and placed in an anaerobic chamber (Coy Laboratory) overnight before the experiment to remove dissolved oxygen (O2). All the pipetting steps were performed in an anaerobic chamber. The assay plate was then placed in an anaerobic jar and incubated at 25°C for 21 h. The absorbance spectrum from 300 to 600 nm was recorded at the end of the experiment.
Linear regression and comparison of the initial reaction rate were performed using the emmeans package v.1.3.3 (70) in the R statistical environment (v.3.5.3) (52).
In silico docking of furazolidone to the AhpF protein.
For the SwissDock server (35, 71), a blind docking simulation was implemented using ready-to-dock FZ ligand data file from ZINC database (accession number ZINC113418) (72) and the AhpF structural data file from Protein Data Bank (PDB identifier [ID] 4O5Q) (38). The process was performed using the ACCURATE mode with the flexibility set to 3 Å. The generated docking poses between AhpF and FZ were visualized using UCSF Chimera v1.13 (73). Briefly, the binding poses which had steric clash between FZ and the cofactor FAD were purged. The hydrogen bonds were then annotated between the ligand and the protein residues; the binding poses with less than 1 hydrogen bond were removed. The remaining binding poses were then ranked according to the full fitness score and the number of hydrogens between FZ and AhpF residues.
For the AutoDock Vina, the PDBQT files for AhpF (from pdb file 4O5Q) and FZ (from mol2 file ZINC113418) were generated using AutoDock Tools (ADT) as previously described (74). First, a blind docking protocol was performed for AhpF and FZ using QuickVina-W (75) with an exhaustiveness of 24 and the number of modes at 50 (spacing, 1 Å; x = 68, y = 66, z = 124; grid box center, −38.786, −27.238, 28.694). This step was performed three times. The resulting poses were used to determine the most likely binding region of the AhpF protein. After that, the docking of FZ into AhpF was executed in a smaller grid box (spacing, 1 Å; x = 40, y = 40, z = 40; grid box center, −36.936, −33.981, 49.29) with an exhaustiveness of 24 and the number of modes at 50 using AutoDock Vina (36). This local docking was performed 10 times until the most favorable binding poses converged.
The most favorable binding poses obtained from the SwissDock and AutoDock Vina simulations were analyzed using the Arpeggio web server, which calculates and visualizes the interaction between the ligand and protein residues (37). The interactions were visualized using PyMOL v1.8.4.0 (76).
Supplementary Material
ACKNOWLEDGMENTS
We thank the Genetics Strains Research Center, National Institute of Genetics, Japan, for providing the ASKA collection and the Massey Genome Service for the excellent genome sequencing service. We also thank Catrina Olivera for providing the E. coli strain K2526 and Prya Soni for technical assistance with anaerobic chamber experiments. The Keio Collection was purchased from Dharmacon via Thermo Fisher (Australia).
This work was funded by a Callaghan Innovation PhD scholarship, Massey University, New Zealand Pharmaceuticals Ltd., and a Carmine Family donation.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/AAC.00868-19.
REFERENCES
- 1.Iredell J, Brown J, Tagg K. 2016. Antibiotic resistance in Enterobacteriaceae: mechanisms and clinical implications. BMJ 352:h6420. doi: 10.1136/bmj.h6420. [DOI] [PubMed] [Google Scholar]
- 2.Theuretzbacher U. 2017. Global antimicrobial resistance in Gram-negative pathogens and clinical need. Curr Opin Microbiol 39:106–112. doi: 10.1016/j.mib.2017.10.028. [DOI] [PubMed] [Google Scholar]
- 3.Vass M, Hruska K, Franek M. 2008. Nitrofuran antibiotics: a review on the application, prohibition and residual analysis. Vet Med (Praha) 53:469–500. doi: 10.17221/1979-VETMED. [DOI] [Google Scholar]
- 4.Zhuge L, Wang Y, Wu S, Zhao RL, Li Z, Xie Y. 2018. Furazolidone treatment for Helicobacter Pylori infection: a systematic review and meta-analysis. Helicobacter 23:e12468. doi: 10.1111/hel.12468. [DOI] [PubMed] [Google Scholar]
- 5.Medina AM, Rivera FP, Pons MJ, Riveros M, Gomes C, Bernal M, Meza R, Maves RC, Huicho L, Chea-Woo E, Lanata CF, Gil AI, Ochoa TJ, Ruiz J. 2015. Comparative analysis of antimicrobial resistance in enterotoxigenic Escherichia coli isolates from two paediatric cohort studies in Lima, Peru. Trans R Soc Trop Med Hyg 109:493–502. doi: 10.1093/trstmh/trv054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Palma N, Pons MJ, Gomes C, Mateu J, Riveros M, Garcia W, Jacobs J, Garcia C, Ochoa TJ, Ruiz J. 2017. Resistance to quinolones, cephalosporins and macrolides in Escherichia coli causing bacteraemia in Peruvian children. J Glob Antimicrob Resist 11:28–33. doi: 10.1016/j.jgar.2017.06.011. [DOI] [PubMed] [Google Scholar]
- 7.Lagunas-Rangel FA. 2018. Antimicrobial susceptibility profiles of bacteria causing urinary tract infections in Mexico: single-centre experience with 10 years of results. J Glob Antimicrob Resist 14:90–94. doi: 10.1016/j.jgar.2018.03.004. [DOI] [PubMed] [Google Scholar]
- 8.Bryce A, Costelloe C, Wootton M, Butler CC, Hay AD. 2018. Comparison of risk factors for, and prevalence of, antibiotic resistance in contaminating and pathogenic urinary Escherichia coli in children in primary care: prospective cohort study. J Antimicrob Chemother 73:1359–1367. doi: 10.1093/jac/dkx525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cordoba G, Holm A, Hansen F, Hammerum AM, Bjerrum L. 2017. Prevalence of antimicrobial resistant Escherichia coli from patients with suspected urinary tract infection in primary care, Denmark. BMC Infect Dis 17:670. doi: 10.1186/s12879-017-2785-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hitzenbichler F, Simon M, Holzmann T, Iberer M, Zimmermann M, Salzberger B, Hanses F. 2018. Antibiotic resistance in E. coli isolates from patients with urinary tract infections presenting to the emergency department. Infection 46:325–331. doi: 10.1007/s15010-018-1117-5. [DOI] [PubMed] [Google Scholar]
- 11.Bouxom H, Fournier D, Bouiller K, Hocquet D, Bertrand X. 2018. Which non-carbapenem antibiotics are active against extended-spectrum beta-lactamase-producing Enterobacteriaceae? Int J Antimicrob Agents 52:100–103. doi: 10.1016/j.ijantimicag.2018.03.014. [DOI] [PubMed] [Google Scholar]
- 12.Shahbazi S, Asadi Karam MR, Habibi M, Talebi A, Bouzari S. 2018. Distribution of extended-spectrum beta-lactam, quinolone and carbapenem resistance genes, and genetic diversity among uropathogenic Escherichia coli isolates in Tehran, Iran. J Glob Antimicrob Resist 14:118–125. doi: 10.1016/j.jgar.2018.03.006. [DOI] [PubMed] [Google Scholar]
- 13.Zhang X, Zhang Y, Wang F, Wang C, Chen L, Liu H, Lu H, Wen H, Zhou T. 2018. Unravelling mechanisms of nitrofurantoin resistance and epidemiological characteristics among Escherichia coli clinical isolates. Int J Antimicrob Agents 52:226–232. doi: 10.1016/j.ijantimicag.2018.04.021. [DOI] [PubMed] [Google Scholar]
- 14.Chamberlain RE. 1976. Chemotherapeutic properties of prominent nitrofurans. J Antimicrob Chemother 2:325–336. doi: 10.1093/jac/2.4.325. [DOI] [PubMed] [Google Scholar]
- 15.McCalla DR, Kaiser C, Green MH. 1978. Genetics of nitrofurazone resistance in Escherichia coli. J Bacteriol 133:10–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Whiteway J, Koziarz P, Veall J, Sandhu N, Kumar P, Hoecher B, Lambert IB. 1998. Oxygen-insensitive nitroreductases: analysis of the roles of nfsA and nfsB in development of resistance to 5-nitrofuran derivatives in Escherichia coli. J Bacteriol 180:5529–5539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sandegren L, Lindqvist A, Kahlmeter G, Andersson DI. 2008. Nitrofurantoin resistance mechanism and fitness cost in Escherichia coli. J Antimicrob Chemother 62:495–503. doi: 10.1093/jac/dkn222. [DOI] [PubMed] [Google Scholar]
- 18.Martínez-Puchol S, Gomes C, Pons MJ, Ruiz-Roldán L, Torrents de la Peña A, Ochoa TJ, Ruiz J. 2015. Development and analysis of furazolidone-resistant Escherichia coli mutants. APMIS 123:676–681. doi: 10.1111/apm.12401. [DOI] [PubMed] [Google Scholar]
- 19.Zenno S, Koike H, Tanokura M, Saigo K. 1996. Gene cloning, purification, and characterization of NfsB, a minor oxygen-insensitive nitroreductase from Escherichia coli, similar in biochemical properties to FRase I, the major flavin reductase in Vibrio fischeri. J Biochem 120:736–744. doi: 10.1093/oxfordjournals.jbchem.a021473. [DOI] [PubMed] [Google Scholar]
- 20.Race PR, Lovering AL, Green RM, Ossor A, White SA, Searle PF, Wrighton CJ, Hyde EI. 2005. Structural and mechanistic studies of Escherichia coli nitroreductase with the antibiotic nitrofurazone. Reversed binding orientations in different redox states of the enzyme. J Biol Chem 280:13256–13264. doi: 10.1074/jbc.M409652200. [DOI] [PubMed] [Google Scholar]
- 21.Zenno S, Koike H, Kumar AN, Jayaraman R, Tanokura M, Saigo K. 1996. Biochemical characterization of NfsA, the Escherichia coli major nitroreductase exhibiting a high amino acid sequence homology to Frp, a Vibrio harveyi flavin oxidoreductase. J Bacteriol 178:4508–4514. doi: 10.1128/jb.178.15.4508-4514.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.McCalla DR. 1979. Nitrofurans, p 176–213. In Hahn FE. (ed), Mechanism of action of antibacterial agents; antibiotics, vol 5/1 Springer, Heidelberg, Germany. [Google Scholar]
- 23.McOsker CC, Fitzpatrick PM. 1994. Nitrofurantoin: mechanism of action and implications for resistance development in common uropathogens. J Antimicrob Chemother 33 Suppl A:23–30. doi: 10.1093/jac/33.suppl_a.23. [DOI] [PubMed] [Google Scholar]
- 24.Bertenyi KK, Lambert IB. 1996. The mutational specificity of furazolidone in the lacI gene of Escherichia coli. Mutat Res 357:199–208. doi: 10.1016/0027-5107(96)00102-9. [DOI] [PubMed] [Google Scholar]
- 25.Ona KR, Courcelle CT, Courcelle J. 2009. Nucleotide excision repair is a predominant mechanism for processing nitrofurazone-induced DNA damage in Escherichia coli. J Bacteriol 191:4959–4965. doi: 10.1128/JB.00495-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mitosch K, Rieckh G, Bollenbach T. 2019. Temporal order and precision of complex stress responses in individual bacteria. Mol Syst Biol 15:e8470. doi: 10.15252/msb.20188470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Peterson FJ, Mason RP, Hovsepian J, Holtzman JL. 1979. Oxygen-sensitive and -insensitive nitroreduction by Escherichia coli and rat hepatic microsomes. J Biol Chem 254:4009–4014. [PubMed] [Google Scholar]
- 28.Ashkenazy H, Abadi S, Martz E, Chay O, Mayrose I, Pupko T, Ben-Tal N. 2016. ConSurf 2016: an improved methodology to estimate and visualize evolutionary conservation in macromolecules. Nucleic Acids Res 44:W344–W350. doi: 10.1093/nar/gkw408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sim NL, Kumar P, Hu J, Henikoff S, Schneider G, Ng PC. 2012. SIFT web server: predicting effects of amino acid substitutions on proteins. Nucleic Acids Res 40:W452–W457. doi: 10.1093/nar/gks539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ng PC, Henikoff S. 2003. SIFT: predicting amino acid changes that affect protein function. Nucleic Acids Res 31:3812–3814. doi: 10.1093/nar/gkg509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kitagawa M, Ara T, Arifuzzaman M, Ioka-Nakamichi T, Inamoto E, Toyonaga H, Mori H. 2005. Complete set of ORF clones of Escherichia coli ASKA library (a complete set of E. coli K-12 ORF archive): unique resources for biological research. DNA Res 12:291–299. doi: 10.1093/dnares/dsi012. [DOI] [PubMed] [Google Scholar]
- 32.Kamariah N, Nartey W, Eisenhaber B, Eisenhaber F, Gruber G. 2016. Low resolution solution structure of an enzymatic active AhpC10:AhpF2 ensemble of the Escherichia coli alkyl hydroperoxide reductase. J Struct Biol 193:13–22. doi: 10.1016/j.jsb.2015.11.004. [DOI] [PubMed] [Google Scholar]
- 33.Dip PV, Kamariah N, Nartey W, Beushausen C, Kostyuchenko VA, Ng TS, Lok SM, Saw WG, Eisenhaber F, Eisenhaber B, Gruber G. 2014. Key roles of the Escherichia coli AhpC C-terminus in assembly and catalysis of alkylhydroperoxide reductase, an enzyme essential for the alleviation of oxidative stress. Biochim Biophys Acta 1837:1932–1943. doi: 10.1016/j.bbabio.2014.08.007. [DOI] [PubMed] [Google Scholar]
- 34.Poole LB, Ellis HR. 1996. Flavin-dependent alkyl hydroperoxide reductase from Salmonella typhimurium. 1. Purification and enzymatic activities of overexpressed AhpF and AhpC proteins. Biochemistry 35:56–64. doi: 10.1021/bi951887s. [DOI] [PubMed] [Google Scholar]
- 35.Grosdidier A, Zoete V, Michielin O. 2011. SwissDock, a protein-small molecule docking web service based on EADock DSS. Nucleic Acids Res 39:W270–W277. doi: 10.1093/nar/gkr366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Trott O, Olson AJ. 2010. AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J Comput Chem 31:455–461. doi: 10.1002/jcc.21334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jubb HC, Higueruelo AP, Ochoa-Montano B, Pitt WR, Ascher DB, Blundell TL. 2017. Arpeggio: a web server for calculating and visualising interatomic interactions in protein structures. J Mol Biol 429:365–371. doi: 10.1016/j.jmb.2016.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Dip PV, Kamariah N, Subramanian Manimekalai MS, Nartey W, Balakrishna AM, Eisenhaber F, Eisenhaber B, Grüber G. 2014. Structure, mechanism and ensemble formation of the alkylhydroperoxide reductase subunits AhpC and AhpF from Escherichia coli. Acta Crystallogr D Biol Crystallogr 70:2848–2862. doi: 10.1107/S1399004714019233. [DOI] [PubMed] [Google Scholar]
- 39.Vervoort J, Xavier BB, Stewardson A, Coenen S, Godycki-Cwirko M, Adriaenssens N, Kowalczyk A, Lammens C, Harbarth S, Goossens H, Malhotra-Kumar S. 2014. An in vitro deletion in ribE encoding lumazine synthase contributes to nitrofurantoin resistance in Escherichia coli. Antimicrob Agents Chemother 58:7225–7233. doi: 10.1128/AAC.03952-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lázár V, Nagy I, Spohn R, Csörgő B, Györkei Á, Nyerges Á, Horváth B, Vörös A, Busa-Fekete R, Hrtyan M, Bogos B, Méhi O, Fekete G, Szappanos B, Kégl B, Papp B, Pál C. 2014. Genome-wide analysis captures the determinants of the antibiotic cross-resistance interaction network. Nat Commun 5:4352. doi: 10.1038/ncomms5352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chevereau G, Dravecka M, Batur T, Guvenek A, Ayhan DH, Toprak E, Bollenbach T. 2015. Quantifying the determinants of evolutionary dynamics leading to drug resistance. PLoS Biol 13:e1002299. doi: 10.1371/journal.pbio.1002299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kamariah N, Manimekalai MS, Nartey W, Eisenhaber F, Eisenhaber B, Gruber G. 2015. Crystallographic and solution studies of NAD+- and NADH-bound alkylhydroperoxide reductase subunit F (AhpF) from Escherichia coli provide insight into sequential enzymatic steps. Biochim Biophys Acta 1847:1139–1152. doi: 10.1016/j.bbabio.2015.06.011. [DOI] [PubMed] [Google Scholar]
- 43.Kamariah N, Eisenhaber B, Eisenhaber F, Gruber G. 2017. Essential role of the flexible linker on the conformational equilibrium of bacterial peroxiredoxin reductase for effective regeneration of peroxiredoxin. J Biol Chem 292:6667–6679. doi: 10.1074/jbc.M117.775858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ling JQ, Cho C, Guo LT, Aerni HR, Rinehart J, Soll D. 2012. Protein aggregation caused by aminoglycoside action is prevented by a hydrogen peroxide scavenger. Mol Cell 48:713–722. doi: 10.1016/j.molcel.2012.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Dwyer DJ, Belenky PA, Yang JH, MacDonald IC, Martell JD, Takahashi N, Chan CTY, Lobritz MA, Braff D, Schwarz EG, Ye JD, Pati M, Vercruysse M, Ralifo PS, Allison KR, Khalil AS, Ting AY, Walker GC, Collins JJ. 2014. Antibiotics induce redox-related physiological alterations as part of their lethality. Proc Natl Acad Sci U S A 111:E2100–E2109. doi: 10.1073/pnas.1401876111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gordon GC, Cameron JC, Pfleger BF. 2017. RNA sequencing identifies new RNase III cleavage sites in Escherichia coli and reveals increased regulation of mRNA. mBio 8:e00128-17. doi: 10.1128/mBio.00128-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Baba T, Ara T, Hasegawa M, Takai Y, Okumura Y, Baba M, Datsenko KA, Tomita M, Wanner BL, Mori H. 2006. Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: the Keio collection. Mol Syst Biol 2:2006.0008. doi: 10.1038/msb4100050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Thomason LC, Costantino N, Court DL. 2007. E. coli genome manipulation by P1 transduction. Curr Protoc Mol Biol Chapter 1:Unit 1 17. [DOI] [PubMed] [Google Scholar]
- 49.Datsenko KA, Wanner BL. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc Natl Acad Sci U S A 97:6640–6645. doi: 10.1073/pnas.120163297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Green R, Rogers EJ. 2013. Transformation of chemically competent E. coli. Methods Enzymol 529:329–336. doi: 10.1016/B978-0-12-418687-3.00028-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zheng Q. 2017. rSalvador: an R package for the fluctuation experiment. G3 (Bethesda) 7:3849–3856. doi: 10.1534/g3.117.300120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.R Core Team. 2018. R: a language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria: https://www.R-project.org/. [Google Scholar]
- 53.Cox MP, Peterson DA, Biggs PJ. 2010. SolexaQA: at-a-glance quality assessment of Illumina second-generation sequencing data. BMC Bioinformatics 11:485. doi: 10.1186/1471-2105-11-485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kersey PJ, Allen JE, Allot A, Barba M, Boddu S, Bolt BJ, Carvalho-Silva D, Christensen M, Davis P, Grabmueller C, Kumar N, Liu Z, Maurel T, Moore B, McDowall MD, Maheswari U, Naamati G, Newman V, Ong CK, Paulini M, Pedro H, Perry E, Russell M, Sparrow H, Tapanari E, Taylor K, Vullo A, Williams G, Zadissia A, Olson A, Stein J, Wei S, Tello-Ruiz M, Ware D, Luciani A, Potter S, Finn RD, Urban M, Hammond-Kosack KE, Bolser DM, De Silva N, Howe KL, Langridge N, Maslen G, Staines DM, Yates A. 2018. Ensembl Genomes 2018: an integrated omics infrastructure for non-vertebrate species. Nucleic Acids Res 46:D802–D808. doi: 10.1093/nar/gkx1011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Langmead B, Salzberg SL. 2012. Fast gapped-read alignment with Bowtie 2. Nat Methods 9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R, 1000 Genome Project Data Processing Subgroup. 2009. The Sequence Alignment/Map format and SAMtools. Bioinformatics 25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Garrison E, Marth G. 2012. Haplotype-based variant detection from short-read sequencing. arXiv 1207.3907 [q-bio.GN] https://arxiv.org/abs/1207.3907.
- 58.Cingolani P, Platts A, Wang Le L, Coon M, Nguyen T, Wang L, Land SJ, Lu X, Ruden DM. 2012. A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff: SNPs in the genome of Drosophila melanogaster strain w1118; iso-2; iso-3. Fly (Austin) 6:80–92. doi: 10.4161/fly.19695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ren J, Wen L, Gao X, Jin C, Xue Y, Yao X. 2009. DOG 1.0: illustrator of protein domain structures. Cell Res 19:271–273. doi: 10.1038/cr.2009.6. [DOI] [PubMed] [Google Scholar]
- 60.Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD, Pyshkin AV, Sirotkin AV, Vyahhi N, Tesler G, Alekseyev MA, Pevzner PA. 2012. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol 19:455–477. doi: 10.1089/cmb.2012.0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Grenier F, Matteau D, Baby V, Rodrigue S. 2014. Complete Genome Sequence of Escherichia coli BW25113. Genome Announc 2:e01038-14. doi: 10.1128/genomeA.01038-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Dashti AA, Jadaon MM, Abdulsamad AM, Dashti HM. 2009. Heat treatment of bacteria: a simple method of DNA extraction for molecular techniques. Kuwait Med J 41:117–122. [Google Scholar]
- 64.Koontz L. 2013. Agarose gel electrophoresis. Methods Enzymol 529:35–45. doi: 10.1016/B978-0-12-418687-3.00004-5. [DOI] [PubMed] [Google Scholar]
- 65.Wiegand I, Hilpert K, Hancock RE. 2008. Agar and broth dilution methods to determine the minimal inhibitory concentration (MIC) of antimicrobial substances. Nat Protoc 3:163–175. doi: 10.1038/nprot.2007.521. [DOI] [PubMed] [Google Scholar]
- 66.Campbell J. 2010. High-throughput assessment of bacterial growth inhibition by optical density measurements. Curr Protoc Chem Biol 2:195–208. doi: 10.1002/9780470559277.ch100115. [DOI] [PubMed] [Google Scholar]
- 67.Brunelle JL, Green R. 2014. Coomassie blue staining. Methods Enzymol 541:161–167. doi: 10.1016/B978-0-12-420119-4.00013-6. [DOI] [PubMed] [Google Scholar]
- 68.Brunelle JL, Green R. 2014. One-dimensional SDS-polyacrylamide gel electrophoresis (1D SDS-PAGE). Methods Enzymol 541:151–159. doi: 10.1016/B978-0-12-420119-4.00012-4. [DOI] [PubMed] [Google Scholar]
- 69.Schneider CA, Rasband WS, Eliceiri KW. 2012. NIH Image to ImageJ: 25 years of image analysis. Nat Methods 9:671–675. doi: 10.1038/nmeth.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Lenth R. 2019. emmeans: estimated marginal means, aka least-squares means, v1.3.3. https://CRAN.R-project.org/package=emmeans.
- 71.Grosdidier A, Zoete V, Michielin O. 2011. Fast docking using the CHARMM force field with EADock DSS. J Comput Chem 32:2149–2159. doi: 10.1002/jcc.21797. [DOI] [PubMed] [Google Scholar]
- 72.Irwin JJ, Sterling T, Mysinger MM, Bolstad ES, Coleman RG. 2012. ZINC: a free tool to discover chemistry for biology. J Chem Inf Model 52:1757–1768. doi: 10.1021/ci3001277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE. 2004. UCSF chimera–a visualization system for exploratory research and analysis. J Comput Chem 25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 74.Forli S, Huey R, Pique ME, Sanner MF, Goodsell DS, Olson AJ. 2016. Computational protein-ligand docking and virtual drug screening with the AutoDock suite. Nat Protoc 11:905–919. doi: 10.1038/nprot.2016.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Hassan NM, Alhossary AA, Mu Y, Kwoh CK. 2017. Protein-ligand blind docking using QuickVina-W with inter-process spatio-temporal integration. Sci Rep 7:15451. doi: 10.1038/s41598-017-15571-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Schrodinger LLC. 2015. The PyMOL molecular graphics system, v1.8. https://github.com/schrodinger/pymol-open-source.
- 77.Cherepanov PP, Wackernagel W. 1995. Gene disruption in Escherichia coli: TcR and KmR cassettes with the option of Flp-catalyzed excision of the antibiotic-resistance determinant. Gene 158:9–14. doi: 10.1016/0378-1119(95)00193-a. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.