Horizontal transfer of plasmids encoding antimicrobial resistance and virulence determinants has been instrumental in Staphylococcus aureus evolution, including the emergence of community-associated methicillin-resistant S. aureus (CA-MRSA). In the early 1990s, the first CA-MRSA strain isolated in Western Australia (WA), WA-5, encoded cadmium, tetracycline, and penicillin resistance genes on plasmid pWBG753 (∼30 kb).
KEYWORDS: CA-MRSA, IS257, MITEs, plasmid, Staphylococcus aureus, Tn552, trimethoprim, conjugation, mupirocin, resolvase
ABSTRACT
Horizontal transfer of plasmids encoding antimicrobial resistance and virulence determinants has been instrumental in Staphylococcus aureus evolution, including the emergence of community-associated methicillin-resistant S. aureus (CA-MRSA). In the early 1990s, the first CA-MRSA strain isolated in Western Australia (WA), WA-5, encoded cadmium, tetracycline, and penicillin resistance genes on plasmid pWBG753 (∼30 kb). WA-5 and pWBG753 appeared only briefly in WA; however, fusidic acid resistance plasmids related to pWBG753 were also present in the first European CA-MRSA isolates at the time. Here, we characterize a 72-kb conjugative plasmid, pWBG731, present in multiresistant WA-5-like clones from the same period. pWBG731 was a cointegrant formed from pWBG753 and a pWBG749 family conjugative plasmid. pWBG731 carried mupirocin, trimethoprim, cadmium, and penicillin resistance genes. The stepwise evolution of pWBG731 likely occurred through the combined actions of IS257, IS257-dependent miniature inverted-repeat transposable elements (MITEs), and the BinL resolution system of the β-lactamase transposon Tn552. An evolutionarily intermediate ∼42-kb nonconjugative plasmid, pWBG715, possessed the same resistance genes as pWBG731 but retained an integrated copy of the small tetracycline resistance plasmid pT181. IS257 likely facilitated the replacement of pT181 with conjugation genes on pWBG731, thus enabling autonomous transfer. Like conjugative plasmid pWBG749, pWBG731 also mobilized nonconjugative plasmids carrying oriT mimics. It seems likely that pWBG731 represents the product of multiple recombination events between the WA-5 pWBG753 plasmid and other mobile genetic elements present in indigenous community-associated methicillin-sensitive S. aureus (CA-MSSA) isolates. The molecular evolution of pWBG731 saliently illustrates how diverse mobile genetic elements can together facilitate rapid accrual and horizontal dissemination of multiresistance in S. aureus CA-MRSA.
INTRODUCTION
Staphylococcus aureus is an opportunistic human and animal pathogen that causes nosocomial and community-associated infections, ranging from noninvasive soft tissue abscesses to life-threatening sepsis, pneumonia, bacteremia, and endocarditis (1–4). The increasing emergence of antimicrobial resistance in both hospital and community settings is a threat to global health. While methicillin-resistant S. aureus (MRSA) isolates were once limited to hospital-associated infections (HA-MRSA), in the last 3 decades, distinct community-associated methicillin-resistant S. aureus (CA-MRSA) strains have proliferated and overtaken HA-MRSA as a dominant source of infection. Methicillin resistance and often other resistance loci are carried by the chromosomally integrated staphylococcal cassette chromosome mec (SCCmec) element, although additional antimicrobial resistance genes in CA-MRSA are generally harbored on circular double-stranded DNA plasmids. In S. aureus, it is estimated that more than 90% of isolates possess at least one plasmid, and of these, ∼79% carry a plasmid of >20 kb, often encoding multiple resistance and virulence determinants (5). Horizontal transfer of these larger plasmids, and associated multiresistance, may occur via conjugation, conjugative mobilization, and/or transduction. This rapid and ongoing resistance evolution reduces available treatment options for S. aureus infections.
The first MRSA isolate, known as “classic MRSA,” appeared in hospitals in the 1960s. By the mid-1970s, large global outbreaks of multiresistant HA-MRSA occurred in hospitals, where these and other HA-MRSA lineages largely remain endemic (6). In contrast, in Western Australia (WA), hospitals were largely unscathed by HA-MRSA, aside from a single outbreak in 1982 by HA-MRSA related to sequence type 239 (ST239), which was also present in eastern parts of Australia during the same period (7). In 1990 to 1992, the first cases of MRSA found outside hospitals in Australia were documented for patients originating in the far north of WA (8). These earliest CA-MRSA isolates, referred to as WA-MRSA (8) and later as WA-5 (9), were distinct from previously characterized HA-MRSA isolates, and the vast majority of WA-5 isolates carried resistance determinants for cadmium, penicillin, and tetracycline on an ∼30-kb plasmid.
Sequence analysis of a representative WA-5 strain, WBG7583, revealed it to be of ST8, with an SCCmec element of type IVa (2B) (SCCmec IVa [2B]) (9). The 30,047-bp plasmid pWBG753 (5) carries a Tn552-like β-lactamase region and an integrated copy of the tetracycline resistance plasmid pT181, flanked by directly repeated copies of the insertion sequence IS257. Interestingly, after the outbreak of WA-5 CA-MRSA, this particular strain-and-plasmid combination was not documented again. A follow-up study investigating CA-MRSA and community-associated methicillin-sensitive S. aureus (CA-MSSA) carriage in the same regions of WA in 1995 to 1996 (9) did not identify any additional ST8 CA-MRSA isolates. Instead, a variety of CA-MRSA isolates of ST1, ST5, ST45, ST73, and ST78 were isolated, along with the likely CA-MSSA progenitors of the now-dominant Australian CA-MRSA ST93-SCCmec IVa (2B) lineage (10). The majority of CA-MRSA isolates in this study (9), like WA-5, carried SCCmec type IVa (2B) (apart from two ST45 isolates carrying SCCmec type V [5C2]) and resistance determinants for penicillin and cadmium, but plasmids in these strains were diverse and did not resemble pWBG753 or carry tetracycline resistance determinants (5). Furthermore, interrogation of a 2003–2004 collection of 4,099 CA-MRSA isolates from WA revealed that only 0.66% (31) were WA-5, and only 2 of these exhibited tetracycline resistance. This again suggests that the original WA-5 CA-MRSA isolates appeared only briefly before being supplanted by SCCmec IVa (2B) CA-MRSA isolates of diverse lineages with a spectrum of STs similar to those of the indigenous CA-MSSA populations (11).
Among the diverse CA-MRSA isolates discovered in the 1995–1996 follow-up study of WA CA-MRSA were strains harboring a new family of conjugative plasmids, named the pWBG749 family. These included pWBG749 and pWBG745 from ST5 CA-MRSA (SCCmec IVa [2B]). The pWBG749 family of conjugative plasmids is genetically distinct from the well-characterized pSK41/pGO1 family, members of which were first isolated in North America in the mid-1970s (12, 13). pWBG749 conjugatively mobilizes a range of large, nonconjugative, multiresistance plasmids also present in CA-MRSA/CA-MSSA strains isolated in WA in 1990 to 1995 (14). Unlike the classical model of conjugative mobilization, where plasmids encode their own relaxase gene and a distinct oriT, pWBG749 mobilizes plasmids carrying a clone, or “mimic,” of the pWBG749 oriT sequence.
When WA-5 was first isolated, additional ST8 CA-MRSA and CA-MSSA isolates with high-level resistance to mupirocin were isolated (15–17). Again, these strains originated in the northern part of WA, where mupirocin was extensively used in the treatment of staphylococcal skin infections. The majority of isolates harbored an ∼42-kb nonconjugative plasmid named pWBG715, which encoded resistance to mupirocin, penicillin, trimethoprim, cadmium, and tetracycline. One characterized isolate, WBG8101, carried transferrable mupirocin and trimethoprim resistance on what appeared to be an ∼75-kb conjugative plasmid (15–17). To gain further insight into resistance plasmid evolution in some of these earliest CA-MRSA isolates, we present the complete sequence of the conjugative multiresistance plasmid pWBG731. pWBG731 is a cointegrant of two plasmids, the WA-5 pWBG753 plasmid and a conjugative plasmid resembling pWBG745. Copies of IS257 mediated the cointegration of the two plasmids and likely played a role in the acquisition of mupirocin and trimethoprim resistance determinants and the loss of a cointegrated tetracycline resistance plasmid, pT181. We also demonstrate that pWBG731 has an oriT specificity distinct from that of pWBG749 and recognizes the same spectrum of oriT sequences as the recently characterized conjugative plasmid pC02 (18).
RESULTS
Sequencing of pWBG731.
To isolate pWBG731 in a strain devoid of phage, other plasmids, and integrative and conjugative elements, pWBG731 was conjugated from its clinical host WBG8101 into the streptomycin- and novobiocin-resistant RN4220 derivative WBG4515 (19), producing strain WBG10514. Conjugative transfer of pWBG731 from WBG10514 to the rifampin- and fusidic acid-resistant RN4220 derivative WBG541 (19) occurred with a mean frequency of 6.7 × 10−5 exconjugants per donor (averaged from six replicate experiments, with a standard deviation of 5.8 × 10−5 exconjugants per donor). To obtain a complete sequence of pWBG731, paired-end short-read sequencing was initially carried out using plasmid DNA extracted from WBG10514. However, the pWBG731 sequence could not be unambiguously assembled into fewer than five contigs due to the presence of several near-identical copies of the 790-bp insertion sequence IS257. Attempts to close the pWBG731 sequence using PCR primers specific for sequences flanking the IS257 insertions were unsuccessful; unexpectedly, all PCRs produced ∼800-bp products regardless of the primer combination, and sequencing of each product revealed near-identical IS257 sequences flanked by sequences from each contig. This obvious artifact was likely produced through chimeric PCR product formation during amplification and hybridization of distinct but near-identical IS257 sequences (20). To resolve the structure of pWBG731, long-read PacBio SMRT cell sequencing was carried out. SMRT cell subreads (mean length of 2.3 kb) were assembled using the long-read assembler Canu, producing an initial 80-kb circular contig. Following the removal of overlapping contig ends and polishing with paired-end short-read sequences, a final circular 72,158-bp contig was produced, with final mapped-read mean coverage depths of 1,329-fold (σ = 211) for long-read and 1,220-fold (σ = 521) for short-read sequences.
Structure and evolution of pWBG731.
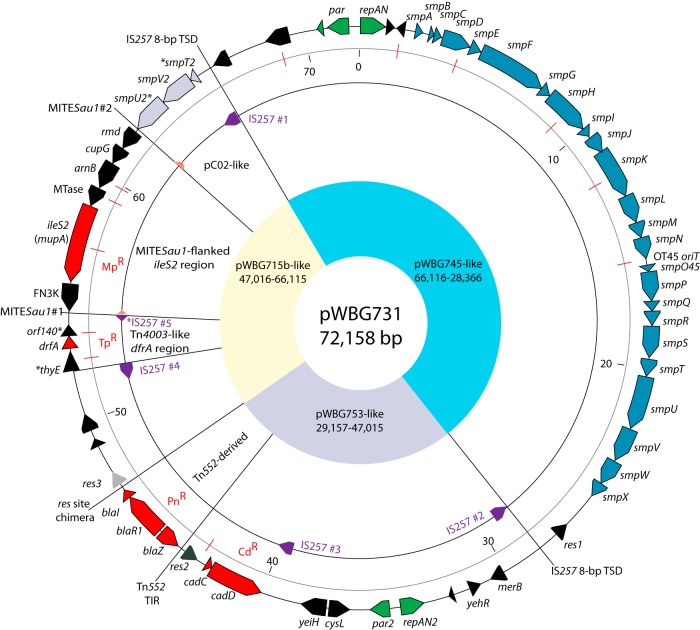
Analysis of the pWBG731 sequence revealed it to be a mosaic comprising parts of at least two staphylococcal plasmids, several additional DNA segments, and five copies of IS257 (IS431) (21) (Fig. 1). A 17,865-bp region was derived from a plasmid closely related to pWBG753, which, as mentioned above, carries genes for resistance to cadmium and a remnant of a Tn552-like β-lactamase transposon (5, 8). A 34,401-bp region was almost identical to the pWBG749 family conjugative plasmids pWBG745 and pBRZ01 (22) and encompassed a contiguous conjugation gene cluster, smpA-smpX (5, 9, 22, 23) (Fig. 1). The pWBG745-like plasmid was flanked by identical, directly repeated copies of IS257 (IS257#1 and IS257#2) (Fig. 1) with adjacent 8-bp target-site duplications (TSDs), 5′-TAATCAAA-3′, of the pWBG745-like sequence, indicating that cointegration likely resulted from the transposition of an IS257 copy residing in the pWBG753-like plasmid into the pWBG745-like plasmid. Consistent with this notion, pWBG753 contains an identical copy of IS257 in a location corresponding to that of IS257#1 in pWBG731. However, in pWBG753, this element instead borders a cointegrated copy of a small tetracycline resistance plasmid related to pT181. Another identical IS257 copy is present at the other end of pT181, along with a flanking 8-bp TSD of pT181 sequence, 5′-AAACAAAA-3′. The observed arrangements of these IS257 copies and TSDs on pWBG753 and pWBG731 are consistent with nonresolved replicative transposition events mediated by IS257#1/IS257#2 present on progenitors of pWBG753 and/or pWBG731. Transposition and cointegration of the pWBG745-like plasmid occurred within the 3′ end of a putative gene of unknown function, whereas transposition into pT181 was within the repC replication initiation gene.
FIG 1.
The 72-kb conjugative multiresistance plasmid pWBG731. Map of the pWBG731 sequence highlighting sequence features. The outer ring shows positions of predicted genes and key cis-acting sequences likely involved in its evolutionary construction. Asterisks before or after a gene name indicate a 5′ or 3′ gene truncation, respectively. The gray ring shows the sequence ruler (in kilobase pairs), and positions of ClaI sites (corresponding to ClaI-digested DNA in Fig. 4B) are shown as red lines. The next inner ring highlights positions of IS257 and MITESau1 elements. All IS257 copies except IS257#5 carry an intact transposase gene. Yellow, blue, and gray sectors indicate regions nearly identical to those present on plasmids pWBG745, pWBG753, and pWBG715. TIR, terminal inverted repeat; MTase, predicted class I SAM-dependent methyltransferase domain-encoding gene.
The pWBG745- and pWBG753-like regions each carried genes encoding RepA_N-type theta-replication initiator proteins (24). Either or both could facilitate the replication of pWBG731 since they both possess all the sequence features important for replication initiation of this replicon type in staphylococci, including Rep box repeats within the rep coding sequence for initiator binding and candidate promoters for structured antisense RNA and rep leader mRNA, which mediate copy number control (25). Moreover, both replication regions are identical to those of the autonomously replicating plasmids pWBG753 and pWBG745 (5, 8, 9). Interestingly, the pC02 conjugative plasmid, another large cointegrant plasmid formed from a pWBG749 family conjugative plasmid and one or more multiresistance plasmids (18), also carries genes coding for two functional RepA_N-type replication initiators and, additionally, two functional pWBG749 family oriT sites. Remarkably, following conjugative transfer of pC02, relaxase-mediated recombination occasionally splits pC02 at the oriT sites to produce two smaller autonomously replicating plasmids in recipient cells (18). A similar phenomenon is observed for pWBG749-mediated mobilization of the multiple-oriT-carrying plasmid pWBG762 (23). These observations suggest that carriage of multiple genes coding for RepA_N replication initiators by cointegrant plasmids is tolerated in S. aureus.
Predicted partitioning (par) systems of distinct types are divergently coded from each repAN gene on pWBG731; such systems improve the efficiency of plasmid inheritance during cell division (26). The pWBG753-related par2 gene was similar to the unusual pSK1 par gene (5). Interestingly, the type Ib par system on the pWBG745-like region is more closely related to that found on pWBG749 (97% nucleotide identity) than to that found on pWBG745 (90% nucleotide identity). A repAN-par gene pairing similar to that for pWBG731 was identified on the nonconjugative plasmid pWBG762 (96% nucleotide identity over both the repAN and par genes), illustrating possible recombination and exchange between repAN-par gene combinations on these plasmids.
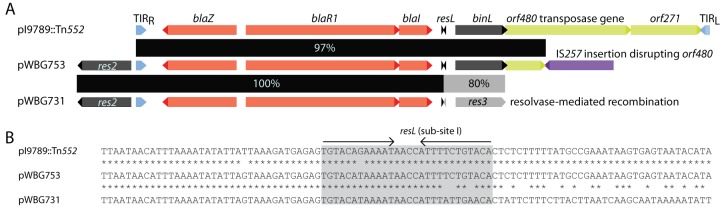
The β-lactamase transposon Tn552 and sequences derived from it are almost ubiquitous in modern S. aureus isolates. In addition to genes for β-lactamase production and its regulation (blaZ-blaR1-blaI), Tn552 carries the binL serine resolvase gene and its target DNA site resL (27–29). BinL-mediated recombination between resL sites on two copies of Tn552 resolves cointegrate intermediates produced during replicative transposition (27). S. aureus plasmids themselves often carry homologous resolution systems (5), which similarly resolve plasmid dimers formed during plasmid replication or via homologous recombination between plasmid copies. Tn552 belongs to a group of transposons that target resolution sites, so it is often found upstream of res genes in staphylococcal plasmids and integrating conjugative elements (ICEs) (26), and DNA inversions and deletions can occur between Tn552 resL sites and plasmid resolution sites (29). The Tn552-like element in pWBG731 is located upstream of the res2 gene, next to subsite II of that gene’s resolution site at a position equivalent to those of previously characterized Tn552 insertion sites (30) and identical to that in pWBG753. While the Tn552-like region on pWBG753 shares ∼97% identity with Tn552 (GenBank accession number X52734), it is incomplete due to truncation by an IS257 insertion in the transposase gene orf480 (Fig. 2). Alignment of the pWBG731 Tn552-like region revealed further truncation, probably attributable to the action of resolvases. The Tn552 regions on pWBG753 and pWBG731 are identical up until the resL resolution site but diverge beyond the expected resolvase cleavage site within subsite I, such that a chimeric res site is present upstream of a unique resolvase gene, res3, on pWBG731. Thus, during the evolution of pWBG731 from a pWBG753-like progenitor, a resolvase-mediated recombination event between the binL and res3 resolution sites appears to have resulted in the deletion of sequences to the right of pWBG753 subsite I, including binL, an orf480 remnant, and a truncating IS257 element.
FIG 2.
Tn552-derived regions on pWBG753 and pWBG731. (A) Gene maps of the Tn552 and Tn552-like sequences on pI9789, pWBG753, and pWBG731, showing the positions of the right and left terminal inverted repeats (TIRR and TIRL, respectively), resL sites, and other genes or insertion sequences. (B) Sequence alignment of the resL sites on the same three plasmids. The region of the resL site is the main recombination site of the resL region “subsite I.” Accessory subsites II and III are identifiable upstream of the binL and res3 coding sequences.
Downstream of the chimeric res3 res site on pWBG731 is a genetically complex 18,304-bp segment extending through to IS257#1. The region between IS257#4 and IS257#5 shares 99% identity to the IS257-flanked composite transposon-like structure Tn4003 containing the dfrA trimethoprim resistance gene but lacks the third IS257 copy and an intervening segment usually found in Tn4003-like elements (31, 32). Additionally, a 192-bp deletion adjacent to IS257#4 has removed the 5′ end of the thyE gene upstream of dfrA. Similar IS257-flanked deletions have been described previously and can moderate the level of trimethoprim resistance conferred, since they alter the hybrid promoter that drives dfrA transcription (33). Tn4003 was first described in the multiresistance plasmid pSK1, which was also prevalent in Australian S. aureus isolates in the 1980s. Such structures were subsequently recognized as cointegrated remnants of a pSK639-like trimethoprim resistance plasmid (34).
IS257#5 was itself truncated by a copy of the 148-bp insertion sequence-like element ISLE39. ISLE39 and the related element ISLE49 contain the same terminal-inverted-repeat sequences as IS257 but lack a transposase gene (Fig. 3). They have been identified on staphylococcal plasmids flanking the quaternary ammonium compound resistance genes qacB and smr (qacC) (35, 36), and similar elements in the pathogenicity island SaPIbov2 flank the biofilm-associated protein gene (bap) (37). These sequences represent “miniature inverted-repeat transposable elements” (MITEs), and the copies on pWBG731 (which are identical to ISLE39) have now been designated MITESau1 in the ISfinder database (https://www-is.biotoul.fr) to clarify the nature of these elements. MITEs have been largely overlooked because of their small size, but their abundance and significance in both prokaryotic and eukaryotic genomes are increasingly being recognized (38–40). Consistent with this, NCBI BLASTN searches revealed 70 staphylococcal entries containing one or more complete copies of MITESau1-like elements in the nonredundant database and 1,293 entries in the refseq_genomes database (NCBI searches conducted on 28 July 2019).
FIG 3.
Alignment of MITESau1-like elements. Shown is an alignment (M-Coffee [60]) of distinct MITESau1-like sequences from pWBG731, pSW49 (GenBank accession number AM040730.1), pWBG759 (GenBank accession number GQ900401.1), SaPIbov2 (GenBank accession number AY220730.1), and pAvX (GenBank accession number MH785253.1). Nonidentical nucleotides are shown in the alignment with MITESau1. The terminal inverted repeats of the elements are indicated by horizontal arrows, and the ends of pWBG731 IS257#1 are presented at the top for comparison, with nonmatching nucleotides shown in lowercase type. MITESau1 of pWBG731 is identical to ISLE39 (GenBank accession number AF535086.1).
MITESau1#1 (see above) and a directly repeated and identical copy, MITESau1#2, flank the 7.8-kb mupirocin resistance region containing the ileS2 (mupA) gene in pWBG731 (Fig. 1). ileS2 encodes an alternate isoleucine-tRNA ligase and is present on numerous S. aureus plasmids, usually flanked by copies of IS257. Plasmid-borne ileS2 regions on diverse plasmids appear to have originated from a common ancestral sequence. Flanking regions often contain genes likely reflecting the context of ileS2 prior to its capture and repurposing as a staphylococcal mupirocin resistance locus. Some plasmids, such as pGO400 and pUSA03, carry only the ileS2 sequence, suggesting that over time, nonessential DNA surrounding ileS2 has been effectively pruned away during evolution (41). Notably, BLASTN searches of the region from MITESau1#2 to ileS2 on pWBG731 revealed it to be the largest region detected so far, containing three additional genes, suggesting that it may represent one of the earliest “unpruned” evolutionary configurations. Intriguingly, the segment downstream of ileS2 in the S. aureus plasmid pV030-8 (GenBank accession number EU366902) extends further than on pWBG731 but lacks any MITESau1 sequences. Thus, the staphylococcal ileS2 region appears to have had a complex evolutionary history involving both IS257 and MITESau1.
Sandwiched between IS257#1 and MITESau1#2 is a region encoding a second set of homologues of the conjugation cluster genes smpT, smpV, and smpU. The smpT2 gene is 5′ truncated by IS257#1, and smpU2 is 3′ truncated by MITESau1#2, leaving only smpV2 intact. The smp cluster gene order for these same genes on pWBG749 and pWBG745 and the main smp cluster on pWBG731 is smpT-smpU-smpV. The distinct smpT-smpV-smpU gene order present in this second copy on pWBG731 is also present on pC02 and putative pWBG749 family conjugative plasmids in S. aureus MO408 (GenBank accession number AIWO01000029) and Staphylococcus epidermidis VCU120 (GenBank accession number NZ_AHLC01000011) (23). The closest DNA match of this second smpT-smpV-smpU cluster, at 70 to 80% nucleotide identity, was to the pC02 conjugation gene cluster (42). SmpU shares sequence similarity with DNA topoisomerase III, a protein commonly encoded by diverse conjugative plasmids (43). While SmpT and SmpV genes are conserved on pWBG749 family plasmids, they share no primary sequence similarity with characterized proteins, so it is unclear if the capture of this region on pWBG731 holds any significance for conjugation or if it is merely a remnant of past recombination events. Nevertheless, the presence of a pC02-like region on pWBG731 suggests that more distantly related members of pWBG749 family conjugative plasmids, or at least parts of them, occasionally cross paths and recombine.
pWBG715 represents an evolutionary intermediate between pWBG753 and pWBG731.
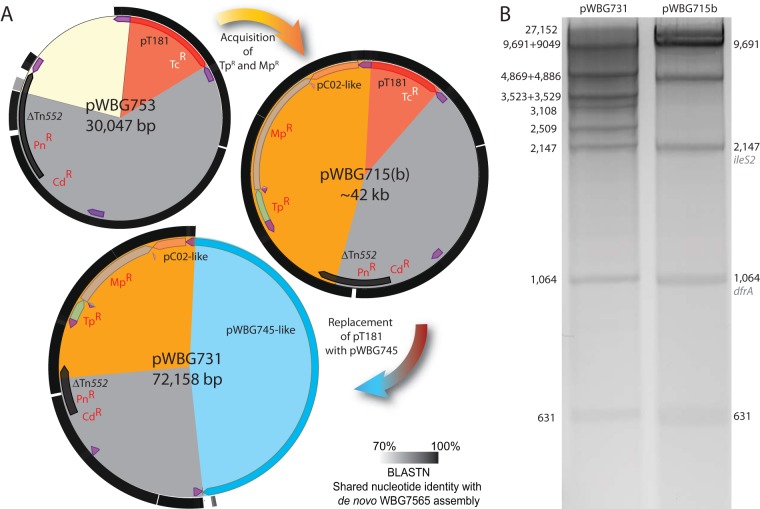
At the time of pWBG731 isolation in the 1990s, numerous patients carried CA-MRSA strains containing an ∼41.4-kb nonconjugative mupirocin resistance plasmid (17), typified by pWBG715. pWBG715 conferred the same resistances as pWBG731 but additionally mediated tetracycline resistance. The similarities between resistance profiles of pWBG731 and pWBG715 and the sequence similarity between pWBG731 and pWBG753 led us to suspect that pWBG715 might represent an evolutionary intermediate between pWBG753 and pWBG731 that had retained an integrated copy of pT181. Indeed, the predicted size of a pWBG731-like plasmid with the pWBG745-related region swapped for pT181 was 42,989 bp. Unfortunately, pWBG715 was lost from WBG7569 during storage, but a related isolate, WBG7565, was available that carried a plasmid with an identical restriction profile, named pWBG715b here (17). WBG7565 was sequenced using Illumina short-read sequencing. De novo assembly and in silico multilocus sequence typing (MLST) profiling confirmed that WBG7565, like WA-5, was of ST8 (44, 45). Mapping of the assembled reads of pWBG715b revealed contigs matching with ∼99% nucleotide identity to the entire pWBG731 plasmid except for the conjugation cluster region, consistent with the hypothesis that pWBG715b represented an intermediate plasmid lacking the cointegrated pWBG745-like plasmid (Fig. 4A). The WBG7565 contigs also matched across the entire pWBG753 plasmid except for the region downstream of the Tn552-like resL site. Importantly this confirmed that WBG7565 likely carried an IS257-flanked copy of pT181 identical to that on pWBG753. We next constructed a mock reference sequence for pWBG715, where we replaced the pWBG745-related region between IS257#1 and IS257#2 with the corresponding pT181-like region of pWBG753. The WBG7565 contigs mapped almost entirely over this sequence (Fig. 3A). Finally, comparison of electrophoresed ClaI-digested pWBG731 and pWBG715b supported the in silico-predicted fragment sizes for both pWBG731 and the mock pWBG715b sequence (Fig. 4B). In summary, it seems likely that pWBG731 evolved in a stepwise fashion from pWBG753 and pWBG715. A pWBG753-like plasmid likely first acquired genes for resistance to mupirocin and trimethoprim from one or more sources in events mediated by the IS257 transposase and a Tn552-like resolution system, to produce a pWBG715-like plasmid. Following this, the pT181-like region of pWBG715 was then replaced with a pWBG745-like conjugation gene cluster, producing pWBG731.
FIG 4.
Stepwise evolution of pWBG753, pWBG715, and pWBG731. (A) Plasmid maps of sequenced plasmids pWBG753 and pWBG731 and map for the predicted sequence of pWBG715 (not to scale relative to each other). The outermost rings on each plasmid map indicate percent nucleotide identity to contigs from a BLASTN query of the de novo sequence assembly of WBG7565, which carries pWBG715b, against each plasmid sequence. The gray sectors indicate the pWBG753 backbone conserved on all three plasmids, the yellow region on pWBG753 represents the region replaced with the orange region on pWBG715, and the red sector indicates the pT181 region on pWBG715 that was replaced by the blue pWBG745-like conjugative plasmid on pWBG731. (B) Agarose electrophoresis of ClaI-digested pWBG731 (left) and pWBG715b (right) DNA. Fragment sizes for pWBG731 are indicated on the left, and recognizable corresponding fragments in the pWBG715 digest are indicated on the right.
pWBG731 mobilizes plasmids carrying OT45 and OTUNa oriT sequences.
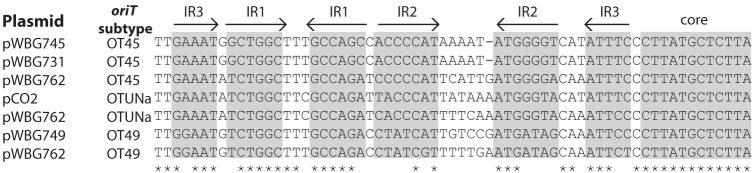
There are several lineages of pWBG749 families, and their oriT sequences have diverged into at least five subtypes (23). Many nonconjugative plasmids carry multiple oriT sequences of different subtypes, suggesting that their acquisition might enable mobilization by members of different pWBG749 family lineages. Nonconjugative plasmid pWBG762, for instance, carries oriT sequences of subtypes OT49, OT45, and OTUNa (14). pWBG749 carries the OT49 oriT and mobilizes only recombinant plasmids carrying an OT49 oriT mimic (23). Since pWBG731 carried the OT45 oriT (Fig. 5), we predicted that pWBG731 would mobilize only plasmids carrying the OT45 oriT mimic but not plasmids carrying OT49 or OTUNa oriT mimics (23). Individual pLI50 plasmids carrying each of the three pWBG762-derived oriT sequences were introduced into RN4220 by electroporation, and pWBG731 was subsequently introduced into each strain by conjugation. The resulting exconjugants were used as donors in conjugation experiments to detect the mobilization of each pLI50 plasmid to WBG4515. Additionally, donor strains carrying the same pLI50 clones, but with pWBG749e (erythromycin resistance-marked pWBG749 [14]), were used as donors for comparison. As expected, pWBG731 was not able to mobilize the pLI50 plasmid carrying the pWBG762 OT49 oriT mimic. However, pWBG731 mobilized pLI50 carrying the OT45 oriT sequence and, interestingly, also mobilized pLI50 carrying the OTUNa oriT at a similar rate (Table 1). While this result was somewhat unexpected, a similar result was recently documented for pC02, which harbors an OTUNa oriT and mobilizes plasmids carrying both OTUNa and OT45 oriT mimics (18).
FIG 5.
Comparison of oriT sequences on pWBG749 family plasmids. Shown is an alignment of the oriT sequences (excluding upstream AR1 to AR3 regions) on conjugative plasmids pWBG745, pWBG731, pC02, and pWBG749, along with oriT mimic sequences of subtypes OT49, OT45, and OTUNa present on pWBG762. Comparison of the IR2 regions reveals the similarity between the IR2 regions of the OT45 and OTUNa oriT sequences in contrast to the IR2 region of OT49 sequences.
TABLE 1.
Mobilization of oriT mimics by pWBG749e and pWBG731
| Construct | Conjugative plasmid | Cloned oriT mimic subtype | Avg transfer frequency (no. of exconjugants/donor) (SD)a
|
|
|---|---|---|---|---|
| Conjugative plasmid | pLI50 clone | |||
| pLI50 | pWBG749e | None | 4.2 × 10−5 (±1.1 × 10−5) | Not detectedb |
| pLI762-49 | pWBG749e | OT49 | 1.6 × 10−5 (±4.3 × 10−6) | 2.1 × 10−6 (±3.7 × 10−7) |
| pLI762-45 | pWBG749e | OT45 | 1.6 × 10−5 (±7.1 × 10−6) | Not detected |
| pLI762-UNa | pWBG749e | OTUNa | 3.3 × 10−5 (±5.5 × 10−6) | Not detected |
| pLI50 | pWBG731 | None | 5.1 × 10−5 (±3.5 × 10−5) | Not detected |
| pLI762-49 | pWBG731 | OT49 | 6.6 × 10−5 (±4.4 × 10−5) | Not detected |
| pLI762-45 | pWBG731 | OT45 | 1.7 × 10−4 (±9.0 × 10−5) | 5.9 × 10−5 (±1.8 × 10−6) |
| pLI762-UNa | pWBG731 | OTUNa | 2.1 × 10−4 (±4.7 × 10−5) | 1.5 × 10−5 (±1.7 × 10−5) |
Per-donor transfer frequencies are the averages from three independent experiments (± standard deviations).
The detection limit was approximately 1 × 10−8 exconjugants per donor.
DISCUSSION
In this work, we described the evolution of one of the largest experimentally confirmed conjugative plasmids in S. aureus. In the last few years, several of the largest documented (>60-kb) conjugative plasmids have been identified in S. aureus, including the 61-kb plasmid pC02 and the 85-kb mecB-carrying plasmid pSAWWU4229_1 (46) (while conjugative transfer for pSAWWU4229_1 has not been demonstrated, it also carries a pWBG749-related conjugation cluster [our unpublished data]). It is possible that these larger conjugative cointegrants may be more prevalent in S. aureus than previously documented and are only now being resolved following improvements in high-throughput long-read sequencing technologies (pC02 and pSAWWU4229_1 were also sequenced using PacBio technology [42, 46]). Given the inherent challenges in assembling larger plasmids carrying repetitive elements from short-read data, it is difficult to speculate if pWBG731-like plasmids are more common in sequence databases than currently appreciated. However, the evolution of pWBG753, pWBG715, and pWBG731 provides remarkable insight into how diverse mobile genetic elements can work together to rapidly repackage multiresistance determinants in a modular fashion and facilitate their subsequent dissemination via conjugation. These three plasmids likely represent only a fraction of the plasmid cointegrants formed, split, and recombined during the evolution of pWBG731.
Around 91% of unique nonconjugative S. aureus plasmids larger than 20 kb carry at least one oriT mimic (47, 48), so the apparent capacity for mobilization via this mechanism is extremely common in S. aureus. The pWBG749 family oriT sequences have, however, diverged, and pWBG749 cannot mobilize plasmids carrying the OT45- or OTUNa-type oriT sequences present on pWBG731 and pC02 plasmids, respectively (Fig. 5) (8). The pWBG731 conjugation gene cluster, which harbors an OT45 oriT, shares only ∼75% nucleotide identity to the conjugation gene cluster of pC02 (18), which harbors an OTUNa oriT. This suggests that the OT45 and OTUNa oriT sequences have evolved within distinct pWBG749 family lineages. Moreover, trees based on the oriT sequences group OT45 and OTUNa oriT sequences into distinct clades (23). Experiments here revealed that pWBG731 mobilizes plasmids carrying either an OT45 or OTUNa oriT but not a plasmid carrying an OT49 oriT, a mobilization profile identical to that of pC02 (18). This suggests that despite the divergence of pC02 and pWBG731, their oriT sequences are functionally equivalent in terms of oriT specificity and mobilization. We previously speculated that the more variable inverted-repeat 2 (IR2) region of each oriT is involved in oriT specificity (23). Reexamination of the OT45 and OTUNa oriT sequences (Fig. 5) highlights that the oriT regions are more similar in their IR2 regions than in IR2 regions on the other oriT subtypes, consistent with this notion. In summary, together with previous mobilization experiments with pWBG749e (23) and pC02 (18), there is now direct experimental evidence for the in trans mobilization of plasmids carrying each of the OT49, OTUNa, and OT45 oriT mimic subtypes. These collectively represent 92% of the pWBG749 family oriT mimics identified on S. aureus plasmids (23, 47).
Plasmid cointegrates of the type observed here in pWBG731, pWBG715b, and pWBG753 have been described on numerous occasions, within plasmids and staphylococcal chromosomes (26). Often, these IS257-mediated cointegrations result in the capture of antimicrobial resistance genes, such as tet(K) from pT181, described above. The cointegrative capture of a pWBG745-like plasmid into pWBG731 in this case does not appear to have resulted in the acquisition of any identifiable resistance determinants. Rather, it seems that an existing pWBG753-like multiresistance plasmid has acquired the capacity for conjugative self-transmissibility via this single cointegration event. As such, this represents an interesting counterpoint to the process of IS257-mediated resistance gene accretion evident in pSK41 family conjugative plasmids. In the course of pWBG731 evolution, the cointegrated pT181-like plasmid in pWBG753 has been deleted, presumably as a consequence of homologous recombination between the flanking IS257 copies, either before or after the cointegration of the pWBG745-like plasmid. In addition to the tet(K) resistance gene, this deletion also removed the pre/RSa mobilization system present in pT181. Thus, in this multiresistance plasmid lineage, there has been a swap from the capacity for horizontal transmission by mobilization, dependent on a conjugation system coresident in the same cell, to independent conjugative proficiency. In this regard, it is worth noting that pWBG753 is one of the few S. aureus multiresistance plasmids that lacks any recognizable pWBG749- or pSK41-like oriT mimic (23, 47, 49), which might explain the selective acquisition of mob and/or conjugation genes in this plasmid lineage.
Significant sequence diversity is observed between IS257 copies, which can provide insights into evolutionary pathways. In this regard, it is worth noting that pWBG731 IS257#4 is identical to IS257L of Tn4003, whereas the remnant of IS257#5 is identical to Tn4003 IS257R1 (IS257L and IS257R1 themselves differ by three nucleotide substitutions and a single indel). This is consistent with the similarity between the dfrA trimethoprim resistance region of pWBG731 and Tn4003 on pSK1. More interestingly, pWBG731 IS257#1 and IS257#2 are also identical to Tn4003 IS257R1, possibly suggesting that the replicative transposition of IS257#1, -#2, or -#5 played a role in the incorporation of sequences between IS257#5 and IS257#1, resulting in the capture of the pT181- and pWBG745-like plasmids in this lineage. Unfortunately, any potentially informative TSD has been removed by the truncation of IS257#5 by MITESau1. The copy of IS257 truncating the Tn552 transposase gene in pWBG753 is most similar to IS257#4, differing by only a single nucleotide. Finally, oriented in the opposite orientation to all other IS257 copies on pWBG731, IS257#3 is the most divergent element, differing from IS257#1, IS257#2, and IS257#5 by 15 substitutions and a single indel.
WA-5 marked the beginning of CA-MRSA in WA; however, it was quickly supplanted by locally abundant S. aureus lineages that acquired a near-identical SCCmec IVa (2B). The appearance of the WA-5 plasmids pWBG753, pWBG715, and pWBG731 was similarly brief. pWBG753 shares very little sequence similarity to extant plasmids in sequence databases aside from its Tn552, pT181, and IS257-related sequences. However, it shares 99.8% nucleotide identity over 83% of its length with the fusidic acid resistance plasmid p11819-97 present in ST80 European CA-MRSA. ST80 became the dominant CA-MRSA lineage in Europe and North Africa throughout the 1990s (50, 51). Phylogenetic analyses of European ST80 MRSA isolates dating back to this period suggest that the acquisition of p11819-97 and SCCmec IV occurred immediately prior to their rapid expansion. So while the appearance of WA-5 and pWBG753 was brief in Australia, the related p11819-97 plasmid may have been pivotal in the evolution and dominance of European CA-MRSA during the same period. The cointegrant pWBG731 examined here appears to represent the result of several gene transfer events that occurred between the pWBG753-harboring WA-5 strain and incumbent community-associated S. aureus lineages carrying pWBG749 family plasmids.
MATERIALS AND METHODS
pWBG731 DNA extraction.
A 5-ml tryptic soy broth (TSB) seeder culture (containing 80 μM Cd) was inoculated from a single colony of WBG10514 and grown overnight with shaking at 37°C. From this, 500 μl was used to seed 200 ml of TSB (with Cd), which was grown under the same conditions for 7 h. Cells were harvested by centrifugation, washed in 10 ml Tris-EDTA (TE) buffer, and then pelleted again by centrifugation. The GenElute HP plasmid maxiprep kit was used (catalogue number NA0310; Sigma) for plasmid DNA extraction. Prior to step 2 of the manufacturer’s protocol, approximately 2.5 mg of recombinant lysostaphin (catalogue number L9043; Sigma) was dissolved in 12 ml of a “resuspension/RNase A” solution. This solution was used to resuspend the cell pellet, and the resulting cell suspension was incubated at 37°C for 10 to 15 min (with vortexing every few minutes) until it cleared, before proceeding with step 3 (alkaline lysis). Following elution and subsequent ethanol-sodium acetate precipitation, this yielded 183 μg of DNA (measured using a Qubit v2 instrument; Thermo Fisher), suspended in 400 μl filtered, deionized H2O.
Sequencing, assembly, and sequence analysis.
For long-read sequencing, approximately 5 μg of purified pWBG731 DNA was used in a single PacBio RSII SMRT cell (carried out by Macrogen, South Korea), multiplexed with five other S. aureus plasmids at similar concentrations. A size-selected ∼5.9-kb library was generated with a concentration of 3.6 ng/μl. PacBio sequencing generated 81,763 sorted, postfilter subreads with a mean length of 2,326 bp, with 43,563 (53%) of reads mapped to the final pWBG731 sequence. Sequence assembly was carried out using the long-read assembler Canu (v1.7) (52). A predicted circular 80,000-bp contig was generated among linear contigs assembled from contaminating genomic DNA. Circlator (53) was used to identify overlapping contig ends and reduced the assembly to 72,160 bp. The pWBG731 sequence start position was manually set to the start codon of the predicted repAN gene adjacent to the smpA-smpX conjugation cluster. Short-read sequencing of pWBG731 was carried out using the Illumina MiSeq system. A paired-end library (2 by 300) was prepared using 1.5 ng of pWBG731 DNA and Nextera XT kit V3 (as described in reference 54). Sequencing generated 2.7 million reads, of which 15% mapped (Bowtie 2 [55]) to the final pWBG731 sequence. Assessment of mapped reads with Pilon (v.1.22) (56) identified and corrected two single-base-pair insertions within poly(A/T) tracts. The final pWBG731 sequence was 72,158 bp, and postassembly mapping statistics provided in Results were produced using Bowtie 2 (55) and Qualimap (v.2.2.1) (57). Whole WBG7565 (pWBG715b) DNA was sequenced using a 2- by 151-bp paired-end library and the Illumina NextSeq system with a 54-fold mapped depth of coverage. De novo assembly was carried out using SPAdes (v3.11.1) (58), which produced 153 contigs of >1 kb and 239 contigs total. BRIG/BLASTN (59) comparisons were carried out with BLASTN and the parameter “–word_size 21.” MLST profiling was carried out using mlst software (44), which makes use of the PubMLST database (45). Plasmid maps were created with Benchling.
Mobilization experiments.
Previously constructed pLI50 plasmids carrying oriT mimics cloned from pWBG762 (23) were introduced into RN4220 by electroporation, and pWBG749e or pWBG731 was subsequently introduced by conjugation, using erythromycin or cadmium together with chloramphenicol to select for exconjugants. The resulting strains carrying both plasmids were used as donors in liquid matings with polyethylene glycol 6000 (PEG 6000) as previously described (23). The RN4220 derivative WBG4515 was used as a recipient, and streptomycin and novobiocin were used to counterselect against donors.
Data availability.
Sequences for pWBG731 and WBG7565 (containing pWBG715b) have been deposited in GenBank under accession numbers MH587574.1 and VLNQ00000000, respectively.
ACKNOWLEDGMENTS
This work was supported by National Health and Medical Research Council (Australia) project grant APP1145697 to N.F. and J.P.R. J.P.R. is the recipient of an Australian Research Council future fellowship (project identifier FT170100235) funded by the Australian Government (http://www.arc.gov.au/grants). K.Y.E. and A.M.D. are recipients of Australian Government Research Training Program (RTP) scholarships administered by the University of Western Australia and Curtin University, respectively.
We thank Ruth Hall for helpful discussions.
REFERENCES
- 1.D’Agata EMC, Webb GF, Horn MA, Moellering RC, Ruan S. 2009. Modeling the invasion of community-acquired methicillin-resistant Staphylococcus aureus into hospitals. Clin Infect Dis 48:274–284. doi: 10.1086/595844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Seybold U, Kourbatova EV, Johnson JG, Halvosa SJ, Wang YF, King MD, Ray SM, Blumberg HM. 2006. Emergence of community-associated methicillin-resistant Staphylococcus aureus USA300 genotype as a major cause of health care-associated blood stream infections. Clin Infect Dis 42:647–656. doi: 10.1086/499815. [DOI] [PubMed] [Google Scholar]
- 3.Boan P, Tan HL, Pearson J, Coombs G, Heath CH, Robinson JO. 2015. Epidemiological, clinical, outcome and antibiotic susceptibility differences between PVL positive and PVL negative Staphylococcus aureus infections in Western Australia: a case control study. BMC Infect Dis 15:10. doi: 10.1186/s12879-014-0742-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Coombs GW, Daly DA, Pearson JC, Nimmo GR, Collignon PJ, McLaws ML, Robinson JO, Turnidge JD, Australian Group on Antimicrobial Resistance. 2014. Community-onset Staphylococcus aureus Surveillance Programme annual report, 2012. Commun Dis Intell Q Rep 38:E59–E69. [PubMed] [Google Scholar]
- 5.Shearer JE, Wireman J, Hostetler J, Forberger H, Borman J, Gill J, Sanchez S, Mankin A, Lamarre J, Lindsay JA, Bayles K, Nicholson A, O’Brien F, Jensen SO, Firth N, Skurray RA, Summers AO. 2011. Major families of multiresistant plasmids from geographically and epidemiologically diverse staphylococci. G3 (Bethesda) 1:581–591. doi: 10.1534/g3.111.000760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chambers HF, Deleo FR. 2009. Waves of resistance: Staphylococcus aureus in the antibiotic era. Nat Rev Microbiol 7:629–641. doi: 10.1038/nrmicro2200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Townsend DE, Ashdown N, Pearman JW, Annear DI, Grubb WB. 1985. Genetics and epidemiology of methicillin-resistant Staphylococcus aureus isolated in a Western Australian hospital. Med J Aust 142:108–111. [DOI] [PubMed] [Google Scholar]
- 8.Udo EE, Pearman JW, Grubb WB. 1993. Genetic analysis of community isolates of methicillin-resistant Staphylococcus aureus in Western Australia. J Hosp Infect 25:97–108. doi: 10.1016/0195-6701(93)90100-e. [DOI] [PubMed] [Google Scholar]
- 9.O’Brien FG, Coombs GW, Pearman JW, Gracey M, Moss F, Christiansen KJ, Grubb WB. 2009. Population dynamics of methicillin-susceptible and -resistant Staphylococcus aureus in remote communities. J Antimicrob Chemother 64:684–693. doi: 10.1093/jac/dkp285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.van Hal SJ, Steinig EJ, Andersson P, Holden MTG, Harris SR, Nimmo GR, Williamson DA, Heffernan H, Ritchie SR, Kearns AM, Ellington MJ, Dickson E, de Lencastre H, Coombs GW, Bentley SD, Parkhill J, Holt DC, Giffard PM, Tong SYC. 2018. Global scale dissemination of ST93: a divergent Staphylococcus aureus epidemic lineage that has recently emerged from remote Northern Australia. Front Microbiol 9:1453. doi: 10.3389/fmicb.2018.01453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Coombs GW, Pearson JC, O’Brien FG, Murray RJ, Grubb WB, Christiansen KJ. 2006. Methicillin-resistant Staphylococcus aureus clones, Western Australia. Emerg Infect Dis 12:241–247. doi: 10.3201/eid1202.050454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Byrne ME, Gillespie MT, Skurray RA. 1990. Molecular analysis of a gentamicin resistance transposonlike element on plasmids isolated from North American Staphylococcus aureus strains. Antimicrob Agents Chemother 34:2106–2113. doi: 10.1128/aac.34.11.2106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Archer GL, Johnston JL. 1983. Self-transmissible plasmids in staphylococci that encode resistance to aminoglycosides. Antimicrob Agents Chemother 24:70–77. doi: 10.1128/aac.24.1.70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.O’Brien FG, Ramsay JP, Monecke S, Coombs GW, Robinson OJ, Htet Z, Alshaikh FA, Grubb WB. 2015. Staphylococcus aureus plasmids without mobilization genes are mobilized by a novel conjugative plasmid from community isolates. J Antimicrob Chemother 70:649–652. doi: 10.1093/jac/dku454. [DOI] [PubMed] [Google Scholar]
- 15.Riley TV, Carson CF, Bowman RA, Mulgrave L, Golledge CL, Pearman JW, Grubb WB. 1994. Mupirocin-resistant methicillin-resistant Staphylococcus aureus in Western Australia. Med J Aust 161:397–398. doi: 10.5694/j.1326-5377.1994.tb127503.x. [DOI] [PubMed] [Google Scholar]
- 16.Slattery JT, Udo EE, Pearman JW, Riley T, Grubb WB. 1995. Mupirocin resistance in Staphylococcus aureus in Western Australia, p GP1 In Australian Society for Microbiology Annual Scientific Meeting (in Canberra), vol 16 Reed Business Publishing, Sydney, Australia. [Google Scholar]
- 17.Udo EE, Pearman JW, Grubb WB. 1994. Emergence of high-level mupirocin resistance in methicillin-resistant Staphylococcus aureus in Western Australia. J Hosp Infect 26:157–165. doi: 10.1016/0195-6701(94)90038-8. [DOI] [PubMed] [Google Scholar]
- 18.LaBreck PT, Li Z, Gibbons KP, Merrell DS. 2019. Conjugative and replicative biology of the Staphylococcus aureus antimicrobial resistance plasmid, pC02. Plasmid 102:71–82. doi: 10.1016/j.plasmid.2019.02.006. [DOI] [PubMed] [Google Scholar]
- 19.Townsend DE, Grubb WB, Ashdown N. 1983. Gentamicin resistance in methicillin-resistant Staphylococcus aureus. Pathology 15:169–174. doi: 10.3109/00313028309084707. [DOI] [PubMed] [Google Scholar]
- 20.Odelberg SJ, Weiss RB, Hata A, White R. 1995. Template-switching during DNA synthesis by Thermus aquaticus DNA polymerase I. Nucleic Acids Res 23:2049–2057. doi: 10.1093/nar/23.11.2049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rouch DA, Skurray RA. 1989. IS257 from Staphylococcus aureus: member of an insertion sequence superfamily prevalent among gram-positive and gram-negative bacteria. Gene 76:195–205. doi: 10.1016/0378-1119(89)90160-1. [DOI] [PubMed] [Google Scholar]
- 22.Rossi F, Diaz L, Wollam A, Panesso D, Zhou Y, Rincon S, Narechania A, Xing G, Di Gioia TSR, Doi A, Tran TT, Reyes J, Munita JM, Carvajal LP, Hernandez-Roldan A, Brandão D, van der Heijden IM, Murray BE, Planet PJ, Weinstock GM, Arias CA. 2014. Transferable vancomycin resistance in a community-associated MRSA lineage. N Engl J Med 370:1524–1531. doi: 10.1056/NEJMoa1303359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.O’Brien FG, Yui Eto K, Murphy RJ, Fairhurst HM, Coombs GW, Grubb WB, Ramsay JP. 2015. Origin-of-transfer sequences facilitate mobilisation of non-conjugative antimicrobial-resistance plasmids in Staphylococcus aureus. Nucleic Acids Res 43:7971–7983. doi: 10.1093/nar/gkv755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Weaver KE, Kwong SM, Firth N, Francia MV. 2009. The RepA_N replicons of Gram-positive bacteria: a family of broadly distributed but narrow host range plasmids. Plasmid 61:94–109. doi: 10.1016/j.plasmid.2008.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kwong SM, Ramsay JP, Jensen SO, Firth N. 2017. Replication of staphylococcal resistance plasmids. Front Microbiol 8:2279. doi: 10.3389/fmicb.2017.02279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Firth N, Jensen SO, Kwong SM, Skurray RA, Ramsay JP. 2018. Staphylococcal plasmids, transposable and integrative elements. Microbiol Spectr 6:GPP3-0030-2018. doi: 10.1128/microbiolspec.GPP3-0030-2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rice PA. 2015. Serine resolvases. Microbiol Spectr 3:MDNA3-0045-2014. doi: 10.1128/microbiolspec.MDNA3-0045-2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rowland SJ, Dyke KG. 1989. Characterization of the staphylococcal beta-lactamase transposon Tn552. EMBO J 8:2761–2773. doi: 10.1002/j.1460-2075.1989.tb08418.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rowland SJ, Dyke KG. 1990. Tn552, a novel transposable element from Staphylococcus aureus. Mol Microbiol 4:961–975. doi: 10.1111/j.1365-2958.1990.tb00669.x. [DOI] [PubMed] [Google Scholar]
- 30.Rowland SJ, Stark WM, Boocock MR. 2002. Sin recombinase from Staphylococcus aureus: synaptic complex architecture and transposon targeting. Mol Microbiol 44:607–619. doi: 10.1046/j.1365-2958.2002.02897.x. [DOI] [PubMed] [Google Scholar]
- 31.Rouch DA, Messerotti LJ, Loo LS, Jackson CA, Skurray RA. 1989. Trimethoprim resistance transposon Tn4003 from Staphylococcus aureus encodes genes for a dihydrofolate reductase and thymidylate synthetase flanked by three copies of IS257. Mol Microbiol 3:161–175. doi: 10.1111/j.1365-2958.1989.tb01805.x. [DOI] [PubMed] [Google Scholar]
- 32.Berg T, Firth N, Apisiridej S, Hettiaratchi A, Leelaporn A, Skurray RA. 1998. Complete nucleotide sequence of pSK41: evolution of staphylococcal conjugative multiresistance plasmids. J Bacteriol 180:4350–4359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Leelaporn A, Firth N, Byrne ME, Roper E, Skurray RA. 1994. Possible role of insertion sequence IS257 in dissemination and expression of high- and low-level trimethoprim resistance in staphylococci. Antimicrob Agents Chemother 38:2238–2244. doi: 10.1128/aac.38.10.2238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Firth N, Skurray RA. 1998. Mobile elements in the evolution and spread of multiple-drug resistance in staphylococci. Drug Resist Updat 1:49–58. doi: 10.1016/S1368-7646(98)80214-8. [DOI] [PubMed] [Google Scholar]
- 35.Bjorland J, Steinum T, Sunde M, Waage S, Sviland S, Oppegaard H, Heir E. 2006. Deletion of pT181-like sequence in an smr-encoding mosaic plasmid harboured by a persistent bovine Staphylococcus warneri strain. J Antimicrob Chemother 57:46–51. doi: 10.1093/jac/dki407. [DOI] [PubMed] [Google Scholar]
- 36.Alam MM, Kobayashi N, Uehara N, Watanabe N. 2003. Analysis on distribution and genomic diversity of high-level antiseptic resistance genes qacA and qacB in human clinical isolates of Staphylococcus aureus. Microb Drug Resist 9:109–121. doi: 10.1089/107662903765826697. [DOI] [PubMed] [Google Scholar]
- 37.Ubeda C, Tormo MA, Cucarella C, Trotonda P, Foster TJ, Lasa I, Penades JR. 2003. Sip, an integrase protein with excision, circularization and integration activities, defines a new family of mobile Staphylococcus aureus pathogenicity islands. Mol Microbiol 49:193–210. doi: 10.1046/j.1365-2958.2003.03577.x. [DOI] [PubMed] [Google Scholar]
- 38.Wessler SR, Bureau TE, White SE. 1995. LTR-retrotransposons and MITEs: important players in the evolution of plant genomes. Curr Opin Genet Dev 5:814–821. doi: 10.1016/0959-437X(95)80016-X. [DOI] [PubMed] [Google Scholar]
- 39.Fattash I, Rooke R, Wong A, Hui C, Luu T, Bhardwaj P, Yang G. 2013. Miniature inverted-repeat transposable elements: discovery, distribution, and activity. Genome 56:475–486. doi: 10.1139/gen-2012-0174. [DOI] [PubMed] [Google Scholar]
- 40.Siguier P, Gourbeyre E, Chandler M. 2014. Bacterial insertion sequences: their genomic impact and diversity. FEMS Microbiol Rev 38:865–891. doi: 10.1111/1574-6976.12067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Perez-Roth E, Armas-Gonzalez E, Alcoba-Florez J, Mendez-Alvarez S. 2011. PCR-based amplification of heterogeneous IS257-ileS2 junctions for molecular monitoring of high-level mupirocin resistance in staphylococci. J Antimicrob Chemother 66:471–475. doi: 10.1093/jac/dkq493. [DOI] [PubMed] [Google Scholar]
- 42.LaBreck PT, Rice GK, Paskey AC, Elassal EM, Cer RZ, Law NN, Schlett CD, Bennett JW, Millar EV, Ellis MW, Hamilton T, Bishop-Lilly KA, Merrell DS. 2018. Conjugative transfer of a novel staphylococcal plasmid encoding the biocide resistance gene, qacA. Front Microbiol 9:2664. doi: 10.3389/fmicb.2018.02664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Li Z, Hiasa H, Kumar U, DiGate RJ. 1997. The traE gene of plasmid RP4 encodes a homologue of Escherichia coli DNA topoisomerase III. J Biol Chem 272:19582–19587. doi: 10.1074/jbc.272.31.19582. [DOI] [PubMed] [Google Scholar]
- 44.Seemann T. 2019. mlst. GitHub https://github.com/tseemann/mlst.
- 45.Jolley KA, Maiden MC. 2010. BIGSdb: scalable analysis of bacterial genome variation at the population level. BMC Bioinformatics 11:595. doi: 10.1186/1471-2105-11-595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Becker K, van Alen S, Idelevich EA, Schleimer N, Seggewiß J, Mellmann A, Kaspar U, Peters G. 2018. Plasmid-encoded transferable mecB-mediated methicillin resistance in Staphylococcus aureus. Emerg Infect Dis 24:242–248. doi: 10.3201/eid2402.171074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ramsay JP, Firth N. 2017. Diverse mobilization strategies facilitate transfer of non-conjugative mobile genetic elements. Curr Opin Microbiol 38:1–9. doi: 10.1016/j.mib.2017.03.003. [DOI] [PubMed] [Google Scholar]
- 48.Ramsay JP, Kwong SM, Murphy RJT, Yui Eto K, Price KJ, Nguyen QT, O’Brien FG, Grubb WB, Coombs GW, Firth N. 2016. An updated view of plasmid conjugation and mobilization in Staphylococcus. Mob Genet Elements 6:e1208317. doi: 10.1080/2159256X.2016.1208317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Pollet RM, Ingle JD, Hymes JP, Eakes TC, Eto KY, Kwong SM, Ramsay JP, Firth N, Redinbo MR. 2016. Processing of nonconjugative resistance plasmids by conjugation nicking enzyme of staphylococci. J Bacteriol 198:888–897. doi: 10.1128/JB.00832-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Stegger M, Price LB, Larsen AR, Gillece JD, Waters AE, Skov R, Andersen PS. 2012. Genome sequence of Staphylococcus aureus strain 11819-97, an ST80-IV European community-acquired methicillin-resistant isolate. J Bacteriol 194:1625–1626. doi: 10.1128/JB.06653-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Stegger M, Wirth T, Andersen PS, Skov RL, De Grassi A, Simoes PM, Tristan A, Petersen A, Aziz M, Kiil K, Cirkovic I, Udo EE, del Campo R, Vuopio-Varkila J, Ahmad N, Tokajian S, Peters G, Schaumburg F, Olsson-Liljequist B, Givskov M, Driebe EE, Vigh HE, Shittu A, Ramdani-Bougessa N, Rasigade JP, Price LB, Vandenesch F, Larsen AR, Laurent F. 2014. Origin and evolution of European community-acquired methicillin-resistant Staphylococcus aureus. mBio 5:e01044-14. doi: 10.1128/mBio.01044-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Koren S, Walenz BP, Berlin K, Miller JR, Bergman NH, Phillippy AM. 2017. Canu: scalable and accurate long-read assembly via adaptive k-mer weighting and repeat separation. Genome Res 27:722–736. doi: 10.1101/gr.215087.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hunt M, Silva ND, Otto TD, Parkhill J, Keane JA, Harris SR. 2015. Circlator: automated circularization of genome assemblies using long sequencing reads. Genome Biol 16:294. doi: 10.1186/s13059-015-0849-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Murphy RJT, Lee YT, Pang S, Bastholm TR, Crow JE, Davis AM, Coombs GW, O’Dea MA, Abraham S, Ramsay JP. 2018. Complete genome sequence of a Staphylococcus aureus sequence type 612 isolate from an Australian horse. Microbiol Resour Announc 7:e00869-18. doi: 10.1128/MRA.00869-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Langmead B, Salzberg SL. 2012. Fast gapped-read alignment with Bowtie 2. Nat Methods 9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Walker BJ, Abeel T, Shea T, Priest M, Abouelliel A, Sakthikumar S, Cuomo CA, Zeng Q, Wortman J, Young SK, Earl AM. 2014. Pilon: an integrated tool for comprehensive microbial variant detection and genome assembly improvement. PLoS One 9:e112963. doi: 10.1371/journal.pone.0112963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Okonechnikov K, Conesa A, Garcia-Alcalde F. 2016. Qualimap 2: advanced multi-sample quality control for high-throughput sequencing data. Bioinformatics 32:292–294. doi: 10.1093/bioinformatics/btv566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Nurk S, Bankevich A, Antipov D, Gurevich AA, Korobeynikov A, Lapidus A, Prjibelski AD, Pyshkin A, Sirotkin A, Sirotkin Y, Stepanauskas R, Clingenpeel SR, Woyke T, McLean JS, Lasken R, Tesler G, Alekseyev MA, Pevzner PA. 2013. Assembling single-cell genomes and mini-metagenomes from chimeric MDA products. J Comput Biol 20:714–737. doi: 10.1089/cmb.2013.0084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Alikhan NF, Petty NK, Ben Zakour NL, Beatson SA. 2011. BLAST Ring Image Generator (BRIG): simple prokaryote genome comparisons. BMC Genomics 12:402. doi: 10.1186/1471-2164-12-402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Magis C, Taly JF, Bussotti G, Chang JM, Di Tommaso P, Erb I, Espinosa-Carrasco J, Notredame C. 2014. T-Coffee: tree-based consistency objective function for alignment evaluation. Methods Mol Biol 1079:117–129. doi: 10.1007/978-1-62703-646-7_7. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Sequences for pWBG731 and WBG7565 (containing pWBG715b) have been deposited in GenBank under accession numbers MH587574.1 and VLNQ00000000, respectively.