LETTER
Colistin is regarded as one of the last-resort antimicrobials for treatment of Gram-negative bacterial infections (1). Several cases of plasmid-borne colistin resistance genes mcr-1, mcr-3-like, and mcr-4.2 in clinical Enterobacteriaceae isolates, including Escherichia coli, have been reported in Singapore (2–4). However, the mcr-5 gene has not been reported in clinical isolates in Singapore. Previously, we reported the antimicrobial resistance (AMR) genotype and phenotype of E. coli SGEHI2010ENV103 isolates from ready-to-eat food in Singapore and documented the first isolate carrying mcr-5.1 in Singapore (5). In this study, we further analyzed this isolate. Here, we report the first complete nucleotide sequence of a transferable plasmid harboring mcr-5.1 in E. coli isolated from ready-to-eat chicken rice in Singapore. (Chicken rice is a common dish in Singapore which is composed of cooked chicken and seasoned rice, served with sauce and cucumber garnishes.)
This E. coli isolate was obtained through the retail food surveillance program by the National Environmental Agency (NEA). It was isolated from retail chicken rice from a hawker center in Singapore in 2010. The MIC of colistin for this isolate was further determined to be 8 μg/ml using broth dilution assay as described by CLSI (6). Double-disk synergy testing was performed as previously described (5), and no extended-spectrum-β-lactamase (ESBL) phenotype was detected. Conjugation experiments were performed using the filter mating method (7). Sodium azide-resistant E. coli strain J53 was the recipient strain, and transconjugants were selected using 4 μg/ml colistin plus 200 μg/ml sodium azide. PCR confirmed the presence of mcr-5 in transconjugants after 24 h of coculture (8), suggesting that the mcr-5.1 genes were able to be transferred to recipient E. coli strain J53. The transfer frequency determined was 10−6. This suggests that this mcr-5.1-carrying plasmid might have the potential to transfer to other strains. The stability test for the plasmid carrying mcr-5.1 was performed as previously described (9). The plasmid was still stable after 20 successive days of subculture in LB broth (∼200 generations) without colistin selection, demonstrating the stability of the plasmid. The drug MICs of the transconjugant and E. coli J53 were determined using MicroScan Neg MIC panel type 44 (Beckman Coulter, Inc., Brea, CA, USA), in accordance with the manufacturer’s instructions (data not shown). The colistin MIC of transconjugants was further determined by broth dilution assay, and the results showed an 8-fold increase (from 1 to 8 μg/ml).
The total DNA of SGEHI2010ENV103 was extracted and purified using a QIAamp DNA minikit and sequenced using a Pacbio RS II system and a HiSeq 2500 system. The closed plasmid was assembled by Unicycler on the Galaxy platform (https://usegalaxy.org/) using both long-read and short-read data with default parameters (10). It was annotated by Rapid Annotation using Subsystem Technology (RAST) (11) and corrected by the use of BLASTn (12). AMR genes or related site mutations, plasmid type, serotype, and sequence type were determined using the online tools on the Center for Genomic Epidemiology website (https://cge.cbs.dtu.dk/services/), and default settings were used during analysis.
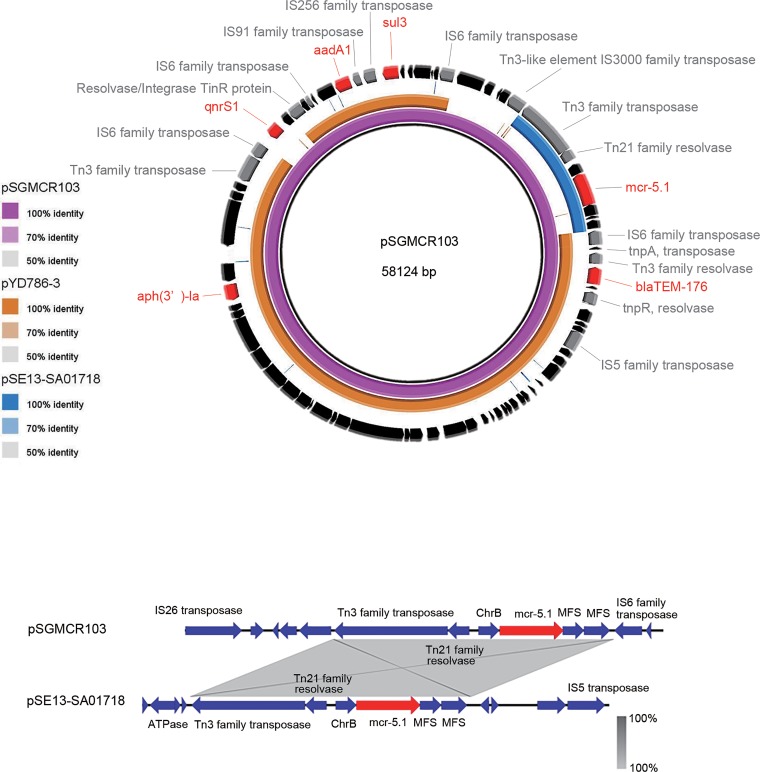
The serotype and sequence type of SGEHI2010ENV103 are determined to be O99:H38 and ST8361, respectively. The sequence of pSGMCR103 revealed a circular plasmid of 58,124 bp in length with 47.7% G+C content. PlasmidFinder showed that it has 98.93% identity with the IncX1 replicon. Members of the IncX group of plasmids are commonly found in Enterobacteriaceae with a narrow host range. They are known to encode fimbriae which enable conjugative transfer (13). Among the five subtypes of the IncX group, only IncX4 was previously reported to be related to mcr genes in E. coli, Salmonella spp., Klebsiella spp., etc. (14, 15). This is the first time that the IncX1 group of plasmids carrying mcr gene has been reported.
The plasmid sequence closest to pSGMCR103 in NCBI is that of plasmid pYD786-3 (GenBank accession number KU254580.1) with 77% query coverage and 99% identity, which was carried by one E. coli isolate from a human urine specimen collected in the United States. A comparison of these two plasmids is shown in the top panel of Fig. 1. They share antimicrobial resistance genes aph(3′)-la, aadA1 (aminoglycoside resistance), blaTEM-176 (beta-lactam resistance), and sul3 (sulfonamide resistance). In addition, pSGMCR103 also carries quinolone resistance gene qnrS1 and colistin resistance gene mcr-5.1. Gene mcr-5.1 was harbored on a Tn3 transposon-like element, which is similar to pSE13-SA01718 (accession number KY807921.1) carried by a Salmonella isolate reported before. All three components of Tn3 were found on this plasmid, including beta-lactamase (encoded by gene bla), Tn3 transposase (encoded by gene tnpA), and Tn3 resolvase (encoded by gene tnpR) (Fig. 1, top panel). Its genetic environment is shown in the bottom panel of Fig. 1. Also, other insertion elements such as the IS5, IS6, IS91, and IS256 family (shown in the top panel of Fig. 1) were found on the plasmid, which may indicate the recombinational activity of the plasmid.
FIG 1.
(Top panel) Sequence map of plasmids pSGMCR103, pYD786-3 (GenBank accession number KU254580.1), and pSE13-SA01718 (GenBank accession number KY807921.1). The outermost circle shows the predicted coding sequences of pSGMCR103. The red parts indicate antimicrobial resistance genes; the gray parts indicate the genes encoding mobile element protein. The figure was generated by the use of BRIG (http://brig.sourceforge.net/). (Bottom panel) Genetic environment of the mcr-5.1 gene in comparison to pSE13-SA01718. ME, mobile element; MFS, gene encoding major facilitator superfamily (MFS)-type transporter; ChrB, gene encoding the protein involved in chromate resistance. The figure was drawn by the use of EasyFig 2.2.2 (http://mjsull.github.io/Easyfig/).
To our knowledge, this is the first report of a complete sequence analysis of a plasmid carrying mcr-5 in E. coli in ready-to-eat food, and it is the first report of the association of mcr-5 with the IncX1 plasmid. This has enlarged the range of plasmid types known to harbor mcr-5. The Tn3-like mobile element may enhance the transferability of the mcr-5 gene and make it easier for that gene to coexist with the blaTEM-176 beta-lactamase gene.
Data accessibility.
The sequence of plasmid pSGMCR103 was deposited at GenBank with accession number MK731977. The raw reads of Pacbio sequencing for isolate SGEHI2010ENV103 are available under project identifier PRJNA531074 in the Sequence Read Archive (SRA).
ACKNOWLEDGMENTS
This study was supported by the National Environment Agency (NEA) and Nanyang Technological University Research Initiative.
We declare that we have no conflicts of interest.
REFERENCES
- 1.Sun J, Zhang H, Liu Y-H, Feng Y. 2018. Towards understanding MCR-like colistin resistance. Trends Microbiol 26:794–808. doi: 10.1016/j.tim.2018.02.006. [DOI] [PubMed] [Google Scholar]
- 2.Teo J-M, Ong R-H, Xia E, Koh T-H, Khor C-C, Lee S-Y, Lim T-P, Kwa A-H. 2016. mcr-1 in multidrug-resistant blaKPC-2-producing clinical Enterobacteriaceae isolates in Singapore. Antimicrob Agents Chemother 60:6435–6437. doi: 10.1128/AAC.00804-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Teo JW, Kalisvar M, Venkatachalam I, Ng OT, Lin RT, Octavia S. 2017. mcr-3 and mcr-4 variants in carbapenemase-producing clinical Enterobacteriaceae do not confer phenotypic polymyxin resistance. J Clin Microbiol 56:e01562-17. doi: 10.1128/JCM.01562-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Teo JW, Chew KL, Lin RT. 2016. Transmissible colistin resistance encoded by mcr-1 detected in clinical Enterobacteriaceae isolates in Singapore. Emerg Microbes Infect 5:1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Guo S, Tay MY, Aung KT, Seow KL, Ng LC, Purbojati RW, Drautz-Moses DI, Schuster SC, Schlundt J. 2019. Phenotypic and genotypic characterization of antimicrobial resistant Escherichia coli isolated from ready-to-eat food in Singapore using disk diffusion, broth microdilution and whole genome sequencing methods. Food Control 99:89–97. doi: 10.1016/j.foodcont.2018.12.043. [DOI] [Google Scholar]
- 6.Clinical and Laboratory Standards Institute (CLSI). 2017. M100. Performance standards for antimicrobial susceptibility testing, 27th ed Clinical and Laboratory Standards Institute, Wayne, Pa. [Google Scholar]
- 7.Zhao F, Feng Y, Lü X, McNally A, Zong Z. 2017. Remarkable diversity of Escherichia coli carrying mcr-1 from hospital sewage with the identification of two new mcr-1 variants. Front Microbiol 8:2094. doi: 10.3389/fmicb.2017.02094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rebelo AR, Bortolaia V, Kjeldgaard JS, Pedersen SK, Leekitcharoenphon P, Hansen IM, Guerra B, Malorny B, Borowiak M, Hammerl JA, Battisti A, Franco A, Alba P, Perrin-Guyomard A, Granier SA, De Frutos Escobar C, Malhotra-Kumar S, Villa L, Carattoli A, Hendriksen RS. 2018. Multiplex PCR for detection of plasmid-mediated colistin resistance determinants, mcr-1, mcr-2, mcr-3, mcr-4 and mcr-5 for surveillance purposes. Eurosurveillance 23:e00672-17. doi: 10.2807/1560-7917.ES.2018.23.6.17-00672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ma S, Sun C, Hulth A, Li J, Nilsson LE, Zhou Y, Börjesson S, Bi Z, Bi Z, Sun Q, Wang Y. 2018. Mobile colistin resistance gene mcr-5 in porcine Aeromonas hydrophila. J Antimicrob Chemother 73:1777–1780. doi: 10.1093/jac/dky110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wick RR, Judd LM, Gorrie CL, Holt KE. 2017. Unicycler: resolving bacterial genome assemblies from short and long sequencing reads. PLoS Comput Biol 13:e1005595. doi: 10.1371/journal.pcbi.1005595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Aziz RK, Bartels D, Best AA, DeJongh M, Disz T, Edwards RA, Formsma K, Gerdes S, Glass EM, Kubal M, Meyer F, Olsen GJ, Olson R, Osterman AL, Overbeek RA, McNeil LK, Paarmann D, Paczian T, Parrello B, Pusch GD, Reich C, Stevens R, Vassieva O, Vonstein V, Wilke A, Zagnitko O. 2008. The RAST server: rapid annotations using subsystems technology. BMC Genomics 9:75. doi: 10.1186/1471-2164-9-75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Madden T. 2013. The BLAST sequence analysis tool. In The NCBI handbook, 2nd ed U.S. National Center for Biotechnology Information, Bethesda, MD: https://www.ncbi.nlm.nih.gov/books/NBK153387/. [Google Scholar]
- 13.Johnson TJ, Bielak EM, Fortini D, Hansen LH, Hasman H, Debroy C, Nolan LK, Carattoli A. 2012. Expansion of the IncX plasmid family for improved identification and typing of novel plasmids in drug-resistant Enterobacteriaceae. Plasmid 68:43–50. doi: 10.1016/j.plasmid.2012.03.001. [DOI] [PubMed] [Google Scholar]
- 14.Alba P, Leekitcharoenphon P, Franco A, Feltrin F, Ianzano A, Caprioli A, Stravino F, Hendriksen RS, Bortolaia V, Battisti A. 2018. Molecular epidemiology of mcr-encoded colistin resistance in Enterobacteriaceae from food-producing animals in italy revealed through the EU Harmonized Antimicrobial Resistance Monitoring. Front Microbiol 9:1217. doi: 10.3389/fmicb.2018.01217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fernandes MR, McCulloch JA, Vianello MA, Moura Q, Pérez-Chaparro PJ, Esposito F, Sartori L, Dropa M, Matté MH, Lira DPA, Mamizuka EM, Lincopan N. 2016. First report of the globally disseminated IncX4 plasmid carrying the mcr-1 gene in a colistin-resistant Escherichia coli sequence type 101 isolate from a human infection in Brazil. Antimicrob Agents Chemother 60:6415–6417. doi: 10.1128/AAC.01325-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The sequence of plasmid pSGMCR103 was deposited at GenBank with accession number MK731977. The raw reads of Pacbio sequencing for isolate SGEHI2010ENV103 are available under project identifier PRJNA531074 in the Sequence Read Archive (SRA).