Mycobacterium tuberculosis spontaneously grows at the air-medium interface, forming pellicle biofilms, which harbor more drug-tolerant persisters than planktonic cultures. The underlying basis for increased persisters in M. tuberculosis biofilms is unknown.
KEYWORDS: tuberculosis, persistence, antibiotics, biofilms
ABSTRACT
Mycobacterium tuberculosis spontaneously grows at the air-medium interface, forming pellicle biofilms, which harbor more drug-tolerant persisters than planktonic cultures. The underlying basis for increased persisters in M. tuberculosis biofilms is unknown. Using a transposon sequencing (Tn-seq) approach, we show here that multiple genes that are necessary for fitness of M. tuberculosis cells within biofilms, but not in planktonic cultures, are also implicated in tolerance of bacilli to a diverse set of stressors and antibiotics. Thus, development of M. tuberculosis biofilms appears to be associated with an enrichment of population, in which challenging growth conditions within biofilm architecture select for cells that maintain intrinsic tolerance to exogenous stresses, including antibiotic exposure. We further observed that the intrinsic drug tolerance of constituent cells of a biofilm determines the frequency of persisters. These findings together allow us to propose that the selection of elite cells during biofilm development promotes the frequency of persisters. Furthermore, probing the possibility that the population enrichment is an outcome of unique environment within biofilms, we demonstrate biofilm-specific induction in the synthesis of isonitrile lipopeptide (INLP). Mutation analysis indicates that INLP is necessary for the architecture development of M. tuberculosis biofilms. In summary, this study offers an insight into persistence of M. tuberculosis biofilms under antibiotic exposure, while identifying INLP as a potential biomarker for further investigation of this phenomenon.
INTRODUCTION
Treatment of tuberculosis (TB), caused by Mycobacterium tuberculosis, entails a multidrug regimen administered for at least 6 months. A lengthy treatment is presumably necessitated by the persistence of a small subpopulation of bacilli, which exhibit phenotypic tolerance to antibiotics (1, 2). Although the mechanism underlying the development of M. tuberculosis persisters during infection is unclear, in vitro studies suggest that these bacilli probably develop through both stochastic and induced mechanisms (3–6).
The persistence of microbes against antibiotics has been closely linked to their ability to grow as sessile, three-dimensionally organized, matrix-encapsulated, multicellular communities called biofilms (7–11). Biofilm-related antibiotic tolerance is rendered particularly relevant by the fact that a majority of chronic microbial infections in humans occur as biofilms (12). Growth and development of biofilms are a multistage process that requires dedicated genetic programs expressed in a spatiotemporal order (13–15). Initial stages involving substratum attachment of planktonic cells and aggregated growth are followed by a maturation stage accompanied by synthesis of an extracellular matrix (13, 14). Physical contacts and chemical communications among cells in developing biofilms, as well as physiological adaptation to self-generated gradients of nutrients and oxygen, stratify the architecture in a way that generates phenotypically heterogeneous cells (15–18). Moreover, molecular mechanisms of stress and antibiotic tolerance in biofilm cells are considered to be linked to those involved in physiological differentiation during architecture development (16, 19).
Environmental and pathogenic mycobacteria, like Mycobacterium avium, Mycobacterium abscessus, Mycobacterium smegmatis, and Mycobacterium tuberculosis spontaneously form biofilms under detergent-free in vitro growth conditions (20–26). Biofilms of M. avium, M. abscessus, Mycobacterium fortuitum, and Mycobacterium ulcerans have also been reported in environmental specimens and in host tissues (27–30). Mutants of mycobacteria that exhibit defective biofilm development, without any defect in planktonic growth, demonstrate the requirement for specialized genes for biofilm development. Moreover, the frequencies of drug-tolerant persisters in M. tuberculosis biofilms are higher than those in planktonic cultures, and their occurrence is tightly linked to the development of a mature three-dimensional architecture (22, 31, 32), similar to that observed in other bacterial species. Previous evidence of GlnR involvement in nitrogen assimilation as well as peroxide resistance during biofilm development of M. smegmatis offers interesting molecular insights into the biofilm-associated stress tolerance (33), although such ideas have been largely unexplored in M. tuberculosis.
Although biofilms of M. tuberculosis during infection remain to be defined, in vitro biofilms represent a suitable model to investigate persistent characteristics of the pathogen. In this study, we report that genes that confer fitness of M. tuberculosis within biofilms are also involved in maintaining tolerance to antibiotics, suggesting that the growth environment in biofilms selects for cells that maintain intrinsic tolerance to exogenous stress, including antibiotics. We further show that intrinsic drug tolerance of the constituent cells within biofilms is a determinant of the persister frequency. Finally, we discover that induced synthesis of isonitrile lipopeptides (INLPs) is specific to M. tuberculosis growth in biofilms, thus demonstrating the unique environment generated in biofilms. These findings begin to explain the basis for biofilm-associated antibiotic tolerance in M. tuberculosis.
RESULTS
Tn-seq of M. tuberculosis biofilms.
To comprehensively determine the genes that confer a fitness advantage to M. tuberculosis in biofilms, we employed a previously developed high-throughput genetic screen, transposon sequencing (Tn-seq) (34). We cultured a transposon insertion mutant library of M. tuberculosis planktonically and in pellicle biofilms (Fig. 1A). Although Sauton’s medium is more suitable for formation of wild-type M. tuberculosis biofilms (22), we chose nutrient-rich Middlebrook 7H9 medium supplemented with oleic acid, albumin, dextrose, and catalase (7H9-OADC) for Tn-seq screening to minimize a potential growth bias introduced due to selective nutrients of Sauton’s medium. Sauton’s medium, however, was used for all subsequent monoculture studies of genes identified from the Tn-seq screen. We also analyzed colonies of the Tn library by Tn-seq (Fig. 1A). While colonies and pellicles represent different growth models in vitro, the two have overlapping gene expression patterns in M. smegmatis (14). Inclusion of both models in the Tn-seq screen allowed similarities and differences in genetic requirements for M. tuberculosis fitness to be determined in these two models of aggregated growth. We recognize that Tn-seq has limitations for screening the colony model of growth, because the procedure involves postexposure outgrowth of individual mutants on agar plates (34). Mutants with an inability to form clonally pure colonies are expected to be eliminated in this screen. Thus, the screen is limited to only those mutants that can form normal colonies in monocultures but display fitness loss under direct competition with the wild type. Furthermore, Tn-seq can also potentially fail to report mutants that are trans-complemented by the wild type in mixed biofilms. Nevertheless, Tn-seq remains a powerful genetic screening approach for fastidious organisms like M. tuberculosis.
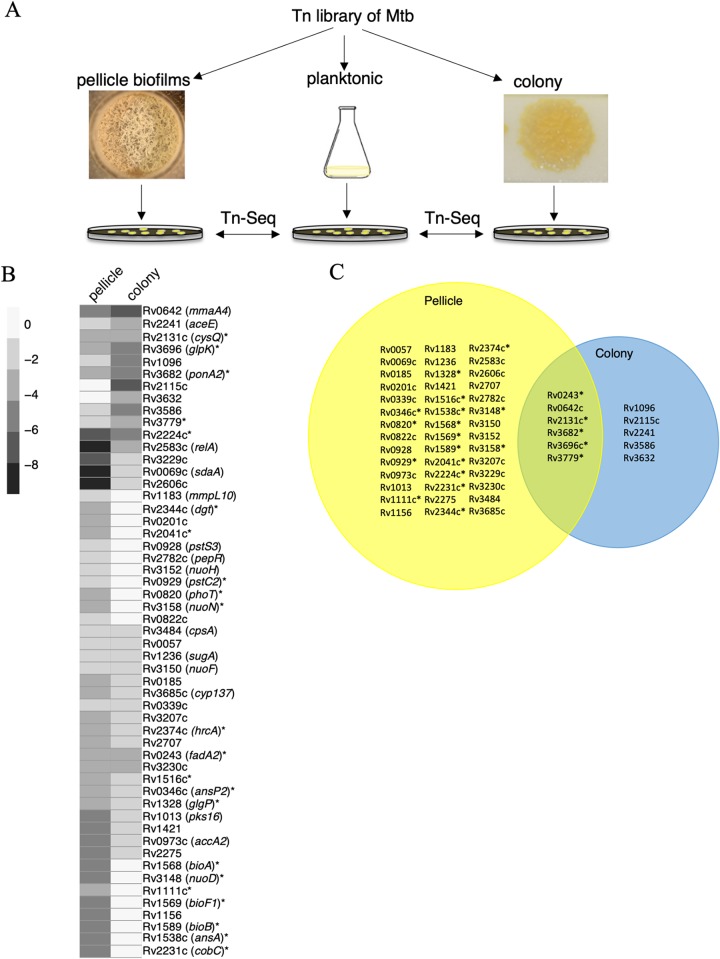
FIG 1.
Genes necessary for M. tuberculosis fitness within biofilms. (A) A schematic outline of Tn-seq-based approach to identify genes required for fitness of M. tuberculosis (Mtb; strain mc27000) in colonies or pellicle biofilms, relative to planktonic cultures. (B and C) Heat map (B) and Venn diagram (C) of 53 genes, mutations in which cause statistically significant underrepresentation of clones either in pellicle biofilms or colonies. While the heat map depicts fold change in the reads, the Venn diagram depicts statistically significant read counts in each of the two growth models. The scale bar for the heat map represents log2 fold change. Fold changes of six genes (Rv0057, -0339, -1236, -2131c, -3150, and -3484), depicted by the heat map as similar in both colonies and pellicles in panel B, are statistically significant only in the pellicle model in panel C. The metadata summary of Tn-seq is provided in Table S1, whereas the complete data set obtained from TRANSIT analysis is in Table S2. Asterisks indicate genes that were followed-up in this study.
Using massively parallel sequencing, we compared the frequency of transposon junction site per gene in colonies and biofilms relative to planktonic cultures. Reads corresponding to junction sequences of transposon from each growth condition were obtained through sequential filtering and trimming using the TRANSIT pipeline (35). Of the total 74,605 potential transposon insertion (TA) sites, the mapped TA sites in samples ranged from 29.5% to 55.7% coverage (see Table S1 in the supplemental material). However, no correlation was observed between growth conditions and density of mapped reads (Table S1). Only one sample had a library density below the recommended threshold of 35% coverage (35). One of the replicates in the colony model had the least (29.5%) library density and also returned significantly less reads than the other sequencing libraries (Table S1). Removing this replicate from TRANSIT did not affect the comparison of Tn hits between colonies and planktonic cultures; Pearson correlation coefficients between the sequencing replicates for each growth condition indicated statistically significant consistency (see Fig. S1 in the supplemental material).
We identified 53 candidate genes that were significantly (q-value of <0.05) underrepresented in either pellicles or colonies or both relative to planktonic cultures (Fig. 1B and C; see Table S2 in the supplemental material). Of these, 48 mutants were underrepresented exclusively in pellicle biofilms, while 11 were underrepresented in colony biofilms. The differences in fitness patterns of the mutants between pellicles and colonies is consistent with a distinct growth environment in these cultures. The list of 53 genes included mmaA4, pks16, and relA (Fig. 1B and C), which were previously implicated in formation of pellicle biofilms (22, 31, 36). Consistent with the altered morphology of ΔmmaA4 colonies, and its deficiency in formation of pellicle biofilms (31), Tn mutants of mmaA4 were underrepresented in both colonies and pellicles (Fig. 1B and C).
Monocultures of mutants distinguish absolute from relative fitness deficiency.
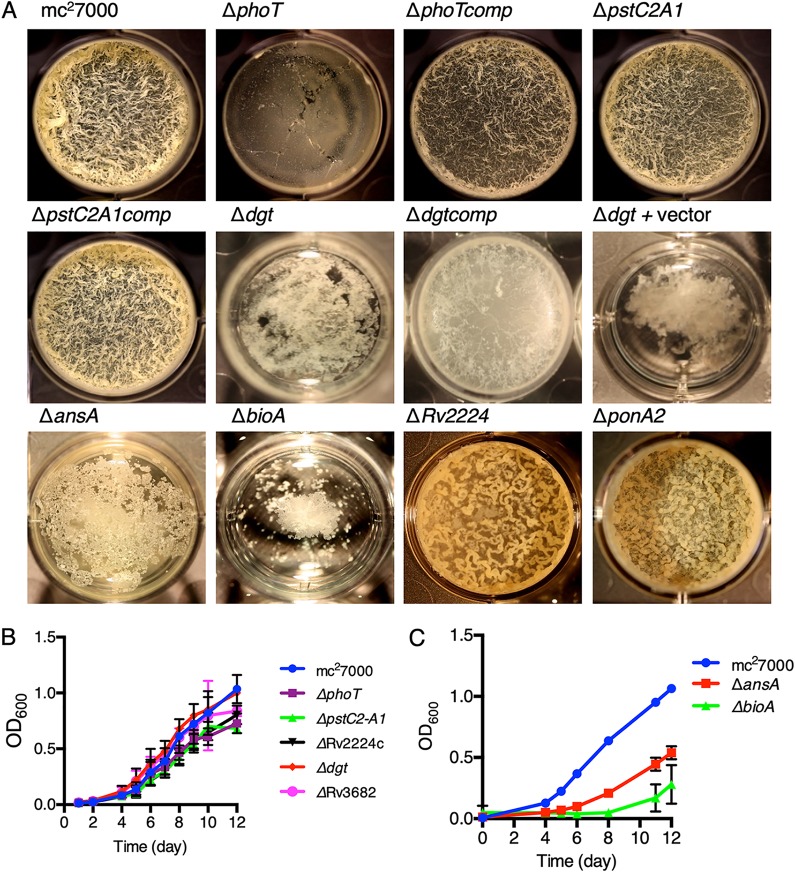
Excluding the previously reported mma4, relA, and pks16 genes, we selected 22 candidates (indicated in Fig. 1B) for investigation of their role in biofilm formation. The selection was based on published significance of these genes in survival during host infection and pathogenesis of M. tuberculosis (37, 38). Additional genes like fadA2, glgP, and Rv1516c were also included in our investigation, because we speculated these to have important role in biofilm formation. Isogenic deletions in selected genes were constructed, and their phenotypes in monoculture pellicle biofilms were compared with their planktonic growth. We note that mutation in pstS3C2 (Rv0928–0929) could not be accomplished, so we used a previously constructed ΔpstC2A1 (Rv0929–0930) mutant. We reasoned that the mutant would behave similarly given that Tn insertion in any of the three genes caused underrepresentation in biofilms; insertion in pstA1 was underrepresented but did not meet the statistical threshold of the TRANSIT pipeline (Table S2). The comparison of the isogenic mutants in planktonic and biofilm cultures revealed three categories of mutants. In the first and the largest category, 16 mutants were indistinguishable from wild type in their ability to form biofilms, suggesting that mutations in these genes exhibit fitness deficiency only when they are in direct competition with the wild type in biofilms. In the second category, two mutants (ΔansA and ΔbioA) were severely growth retarded in both planktonic and biofilm cultures (Fig. 2A to C). This suggests that growth in Sauton’s medium, which contains l-asparagine as the primary nitrogen source and lacks supplemental biotin, requires the activity of l-asparaginase (ansA) and biotin synthase (bioA) in cells. The activities of ansA and bioA may have greater consequence in biofilm formation than in planktonic growth because mutations in these genes cause a biofilm-specific fitness disadvantage in the Tn-seq screen, which was performed in biotin- and nitrogen-rich 7H9-OADC medium. The last category of four mutants formed either morphologically altered (ΔRv2224c and ΔponA2), delayed (ΔphoT), or severely deficient (Δdgt) pellicle biofilms, but remained growth competent in planktonic cultures (Fig. 2A and B). The ΔphoT mutant only formed normal biofilms upon extended incubation (see Fig. S2 in the supplemental material). The Δdgt phenotype was particularly striking because the mutant accumulated biomass at the bottom of the container, but no significant growth was observed at the air-medium interface (Fig. 2A; see Fig. S3 in the supplemental material). Plasmid-borne expression of dgt under a constitutive hsp60 promoter partially complemented Δdgt (Fig. 2A), suggesting that additional chromosomal elements are perhaps necessary. Slow development of ΔphoT biofilms was also rescued by plasmid-borne expression of phoT by the hsp60 promoter (Fig. 2A).
FIG 2.
Genes required for development of pellicle biofilms of M. tuberculosis. (A) A top down view of pellicles of mc27000 (wild type) and a subset of isogenic mutants of genes identified from the Tn-seq screen (Fig. 1C). Complemented strains of the ΔphoT (ΔphoTcomp), ΔpstC2A1 (ΔpstC2A1comp), and Δdgt (Δdgtcomp) mutants carrying a plasmid expressing the corresponding genes under the control of either its own promoter (for pstC2A1) or the hsp60 promoter are also shown. The Δdgt strain carrying the empty vector pMH94 was the control for the Δdgtcomp strain. Pellicles were grown at the air-medium interface in 12-well tissue culture plates in Sauton’s medium for 5 weeks at 37°C. While the biomass of the Δdgt strain remained at the bottom of the container, growth of the ΔphoT strain was delayed, and the strain formed normal pellicles upon further incubation (also see Fig. S2 and S3). (B and C) Planktonic growth of mc27000 and the mutant strains described in panel A. All strains were cultured in detergent-free Sauton’s medium. Cultures were shaken once daily, and their optical densities at 600 nm (OD600) were measured after dispersion of 1-ml aliquots in Tween 80. Data represent mean ± standard deviation (SD) (n = 3).
The low abundance of pstC2A1 and phoT Tn mutants revealed by the Tn-seq screen of biofilms suggests the importance of phosphorus (Pi) homeostasis in fitness of M. tuberculosis cells within biofilms. In bacteria, phosphate-specific transporter (Pst) is a multisubunit complex of PstSCAB, which imports Pi under limiting conditions and transduces adaptive changes in gene expression through a phospho-sensing mechanism (39). PstS is the periplasmic component of the complex and directly binds phosphate (Pi) to present it to the transmembrane channel formed by PstC and PstA, which is complexed with cytosolic PstB that powers the release of Pi in the cytosol through ATP hydrolysis (40). The M. tuberculosis genome encode multiple paralogues of PstS (Rv0928, Rv0932c, and Rv0934), PstC (Rv0929 and Rv0935), PstA (Rv0930 and Rv0936), and PstB (Rv0933 and Rv0820 or phoT), although Rv0928-30-encoded PstS3, PstC2, and PstA1 as well as Rv0933-encoded PstB are considered to constitute the active Pi-transporter complex (41, 42). It was therefore surprising to observe biofilm deficiency in ΔphoT, but not in ΔpstC2A1, in the monoculture assay (Fig. 2A), while both mutants exhibited phenotype in the Tn-seq screen (Table S2). A transcriptome sequencing (RNA-seq) analysis of the two mutants in comparison with the wild type offers a possible explanation. The transcriptomic profiles of the ΔpstC2A1 and ΔphoT mutants exhibited extensive similarity in the patterns of gene expression relative to the wild type (see Fig. S4 and Table S3 in the supplemental material), consistent with the idea that both the loci indeed function in Pi homeostasis. RegX3, a transcription regulator that responds to Pi depletion through its cognate sensor SenX3 (43, 44), and the downstream target operon pstS3C2A1 were upregulated in both the mutants (Table S3). Expression levels of PhoT remained constitutively low in all the strains, but transcripts corresponding to its paralogue (Rv0933) were constitutively higher than those of PstS3C2A1 (Table S3). Therefore, the upregulation of the pstS3C2A1 operon and high constitutive levels of PstB (Rv0933) in the ΔphoT mutant would likely increase the levels of functional PstS3C2A1B complex, thereby increasing the abundance of active Pi transporters and causing dysregulation of Pi homeostasis. This scenario is inconceivable in the ΔpstC2A1 mutant, which may perhaps result in less severe dysregulation of Pi homeostasis than the ΔphoT mutant. However, other deficiencies related to globally dysregulated gene expression in both the ΔphoT and ΔpstC2A1 mutants could possibly underlie their retarded growth when they are in direct competition with the wild type in mixed-culture (Tn library) biofilms.
Relationship between fitness in biofilms and persistence against antibiotics.
Multiple genes of diverse functional categories (like mma4, Rv2224c, ponA2, and pstC2A1) identified in our Tn-seq screen have also been implicated in maintaining intrinsic tolerance to a panel of stressors and antibiotics, including but not limited to acidic pH, lysozyme, SDS, isoniazid, and rifampin (RIF) (31, 45). A reasonable interpretation of this observation is that the restrictive conditions formed in biofilms may favor the growth of cells that are tolerant to both endogenous and exogenous stresses. Thus, more cells with intrinsic antibiotic tolerance are expected to occur in biofilm population than in planktonic cultures. Based on published evidence that intrinsic defense mechanisms against antibiotic-induced damage promote persister frequency in a bacterial population (46), we hypothesized that enrichment of drug-tolerant cells in M. tuberculosis biofilms contribute to high persister frequency (Fig. 3A). An implication of our hypothesis is that monoculture biofilms of mutant cells that are intrinsically more sensitive to antibiotics than the wild type would also harbor fewer persisters than the wild type. To test this, we exploited the ΔpstC2A1 strain, which exhibits hypersensitivity to RIF. Importantly, the mutant forms biofilms indistinguishable from the wild type in a monoculture assay (Fig. 2A), thereby allowing a direct assessment of the effect of intrinsic property of the constituent cells without any compounding effect from an altered architecture, which is generally observed with other antibiotic-sensitive mutants identified in this study (e.g., ponA2 and mma4). Matured biofilms of the ΔpstC2A1 mutant and wild-type strains were exposed to 50 μg/ml of RIF, which is 100 times the MIC, for 7 days, and the persister frequency was scored as described previously (22). Relative to the wild type, the mutant biofilms contained significantly (∼20-fold) fewer RIF-tolerant persisters. The persister frequency was substantially restored upon complementation (Fig. 3B and C). A mutation in pstA1 also renders M. tuberculosis hypersensitive to RIF and produces fewer persisters in planktonic cultures (44). This indicates that the intrinsic drug sensitivity of cells constituting a population is a key determinant of the persister frequency, regardless of the growth condition. This is consistent with our model that a growth condition, such as that produced in biofilms, that restricts drug-sensitive clones will also promote the occurrence of persister cells.
FIG 3.
Intrinsic antibiotic tolerance of constituent cells determines persister frequency in M. tuberculosis biofilms. (A) A schematic representation of biofilm (bf)-associated drug tolerance in M. tuberculosis. Enrichment of tolerant (red) over sensitive (green) cells in biofilms increases the frequency of persisters. Thus, monoculture biofilms of a drug-sensitive mutant are expected to harbor fewer persisters than the wild type. (B and C) Frequency of rifampin-tolerant persisters in biofilms of mc27000, a RIF-sensitive ΔpstC2A1 mutant, and the ΔpstC2A1comp complemented strain. Five-week pellicles were exposed to 50 μg/ml RIF for 1 week and then homogenized with Tween 80, and dilutions were plated on 7H11-OADC agar plates to enumerate viable cells. Cells exposed to DMSO (a solvent for rifampin) were used as the untreated control (Ctrl). Data represent three biologically independent experiments. Average percentages of survival calculated from the panel B plot are shown in panel C (n = 3; P < 0.01; unpaired t test).
Unique growth environment in biofilms is marked by INLPS induction.
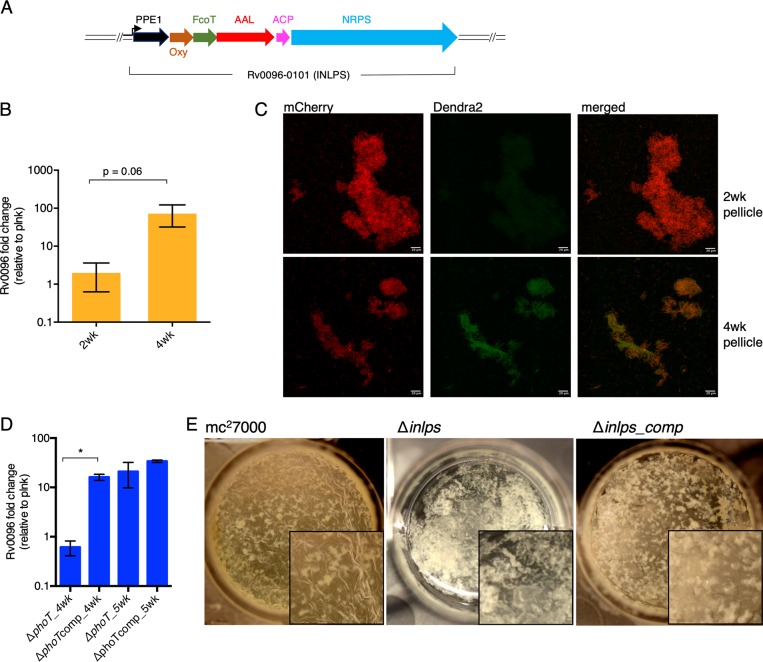
We hypothesize that the enrichment of stress/antibiotic-tolerant cells in biofilms likely originates from their complex microenvironment that emerges from the uniquely stratified architecture. To demonstrate a distinctive growth environment of M. tuberculosis biofilms, we sought to identify a gene expression pattern specific to M. tuberculosis growth in biofilms. We compared transcriptomic analysis of planktonic and biofilm cultures (see Table S4 in the supplemental material). Coinduction in biofilms of a 6-gene operon (Rv0096–0101), which is involved in the synthesis of a putative isonitrile lipopeptide (INLP) and thus called INLP synthase (INLPS) complex (Fig. 4A) (47, 48), was immediately apparent (Table S4). We confirmed the induced expression of Rv0096 in 4-week biofilms by reverse transcription-PCR (RT-PCR), which was not evident during an earlier (2-week) stage (Fig. 4B). Expression of a Dendra2 reporter gene fused to the inlps promoter (Pinlps) was only detected at the 4-week stage. In contrast, the reporter expression was not observed at an earlier (2-week) stage of biofilm development, in planktonic culture, or in colonies grown on an agar surface (Fig. 4C; see Fig. S5 in the supplemental material). Reporter expression was further confirmed in 4-week biofilms of the virulent strain of M. tuberculosis (Erdman) (Fig. S5). Thus, INLPS induction appears to occur specifically during later developmental stages of pellicle biofilms, but not in any other forms of growth. This pellicle-specific INLPS induction further highlights the distinction between pellicle biofilms and colony growth, as suggested by the Tn-seq results. Expression of the INLPS gene cluster is negatively regulated by RegX3 (49), suggesting that the retarded biofilms of the ΔphoT mutant could also be contributed by a suboptimal induction of the cluster. While Rv0096 induction was not apparent in 4-week biofilms of the ΔphoT mutant, the expression was fully restored upon further incubation (Fig. 4D). This implies that factors other than RegX3, possibly responding to signals generated during maturation of biofilms, likely regulate INLPS. The correlation between INLPS induction and timing of biofilm maturation further suggested an important role of INLP during the maturation stages. A severe defect in biofilm formation of the Δinlps mutant was confirmed (Fig. 4E), without any growth defect in planktonic culture (see Fig. S6 in the supplemental material). The mutant phenotype was partially restored upon complementation with a cosmid bearing inlps and its upstream promoter region (Fig. 4E).
FIG 4.
Induced expression of INLPS is required for biofilm development. (A) Schematic representation of inlps gene cluster comprising of six ORFs organized in an operon. Except for Rv0096, which has the conserved Pro-Pro-Glu (PPE) motif, likely functions of other genes have been assigned based on previous studies (47, 48). Oxy, oxidase; FcoT, thioesterase; AAL, acyl ACP ligase; ACP, acyl carrier protein; NRPS, nonribosomal peptide synthase. The arrow indicates the putative promoter. (B) RT-PCR-based expression analysis of Rv0096 in mc27000 biofilms at the 2- and 4-week stages, relative to planktonic (plnk) culture. (C) Microscopic analysis of Dendra2 expression from the promoter of inlps at the 2- and 4-week stages of mc27000 biofilms. Cells constitutively expressing mCherry from the hsp60 promoter were smeared in 80% glycerol on a glass slide, and images were captured with 20× objective on a confocal microscope, analyzed by FIJI software, and shown as maximum projection. The scale bar corresponds to 20 μm. (D) An RT-PCR analysis of Rv0096 expression in the ΔphoT mutant and a complemented (ΔphoTcomp) strain at the 4- and 5-week stages of pellicle biofilm development. SigA transcripts were used as an endogenous control for normalization of CT values in RT-PCR experiments described in panels B and D. Data in these panels represent average from at least two biologically independent experiments. *, P < 0.05 (paired t test). (E) A top-down view of 4-week pellicle biofilms of mc27000, its isogenic Δinlps mutant, and a complemented strain with cosmid-borne inlps. While the parent wild-type strain and the complemented strain had visible texture development at the air-medium interface, the biomass of the mutant was predominantly at the bottom of the well.
Chemical evidence of INLP accumulation in biofilms.
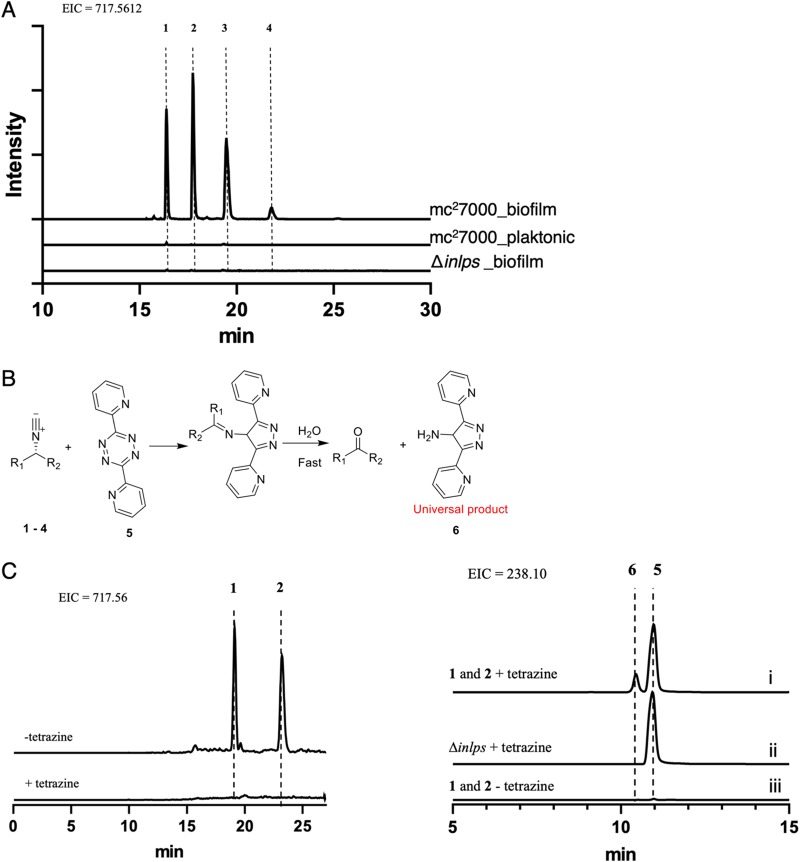
We next sought to determine if expression of inlps correlates with new metabolite accumulation in M. tuberculosis biofilms. INLP was originally identified upon overexpression of Rv0097–0101 and its homologues in Escherichia coli (47), but the structure of the INLP native to M. tuberculosis is unknown. We therefore took an unbiased approach and compared lipid profiles of crude organic extract from planktonic and biofilm cultures of wild-type M. tuberculosis. The biofilm culture of the Δinlps mutant was also included for comparison. The comparative profiling by liquid chromatography–high-resolution mass spectrometry (LC-HRMS) analysis detected an increased abundance of more than 200 small molecules in wild-type biofilm extracts relative to other extracts (data not shown), complicating the identification of metabolic products of the inlps gene cluster. Further careful analysis of these metabolites identified four compounds with identical masses (m/z 717.5612, proposed molecular formulas C40H73N6O5+), which are likely synthesized by the inlps cluster (Fig. 5, compounds 1 to 4; see Fig. S7 in the supplemental material). Our analysis was based on three criteria. First, these four compounds were detected exclusively in wild-type biofilm extracts and were not detected in either planktonic wild-type or Δinlps cultures (Fig. 5A). Identical fragmentation patterns by liquid chromatography-tandem mass spectrometry (LC-MS/MS) analysis of these metabolites were observed (Fig. S7), indicating that these compounds are isomers. Second, click reactions with tetrazine confirmed the presence of unique isonitrile functionality in the compounds (50) (Fig. 5B and C). Tetrazine treatment of partially purified compounds 1 and 2 caused disappearance of their mass signals with concomitant appearance of the predicted reaction product. Third, weak but noticeable incorporation of [13C]glycine was observed in these four metabolites (see Fig. S8 in the supplemental material). Glycine is a direct precursor utilized by INLPS (47, 50). Therefore, its incorporation in compounds 1 to 4 is in agreement with the characterized biosynthetic mechanism for INLP synthesis. We note that the structure of these molecules remains to be determined, largely due to the difficulty in purification of these molecules in sufficient quantity necessary for nuclear magnetic resonance (NMR) analysis. Nonetheless, our data provide substantial evidence that compounds 1 to 4 are isonitrile lipopeptides, and their accumulation in biofilms is a direct consequence of induced expression of inlps.
FIG 5.
Accumulation of INLP in biofilms of M. tuberculosis. (A) Extracted ion chromatograms (EICs) show the presence of four isomers of INLP (compounds 1 to 4; m/z = 717.5612) exclusively in the wild-type biofilm extract, but neither in planktonic wild-type nor Δinlps mutant cultures. A 10-ppm mass error tolerance was used for each trace. (B) Scheme of click reaction of INLP with tetrazine showing [4 + 1] cyclo-addition between compounds 1 and 4 and tetrazine. The product rapidly hydrolyzes in a trace amount of water to generate a primary amine 6. (C) Extracted ion chromatograms show disappearance of partially purified compounds 1 and 2 after reaction with compound 5 and concomitant appearance of the predicted reaction product 6. Control reactions without compound 5 and compound 5-treated Δinlps mutant extract are also shown.
Signal-inducing INLP originates in biofilm architecture.
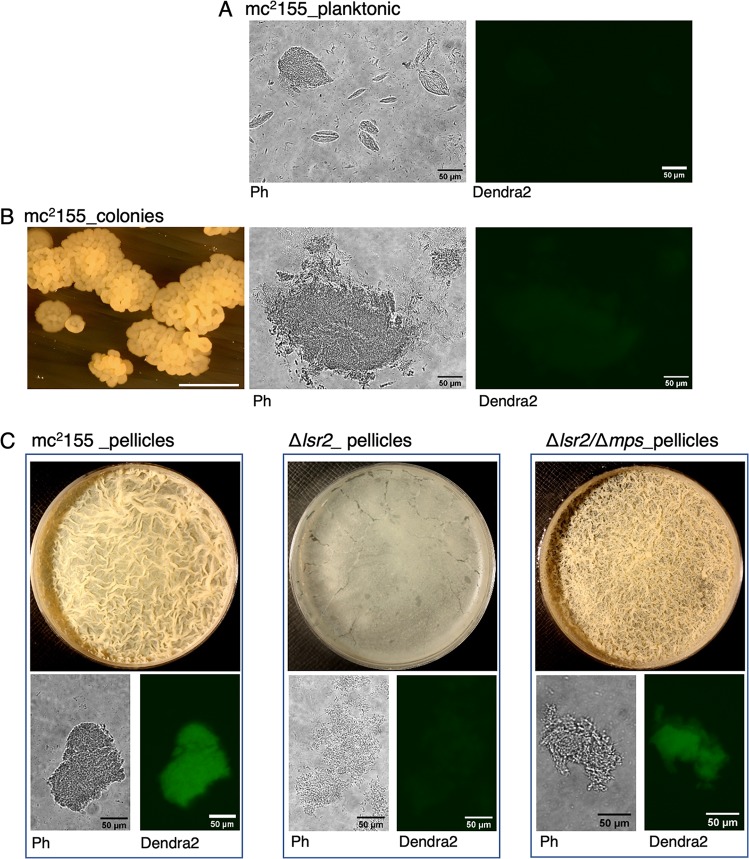
We next sought to determine the origin of the signal that regulates the inlps gene cluster. No changes were detected in RegX3-Senx3 transcripts during biofilm development (Table S3), thereby ruling out the possibility that inlps upregulation is signaled by changes in Pi homeostasis. Moreover, the level of inlps induction in a regX3 mutant (49) is about an order of magnitude less than those observed in biofilms (Table S4). We thus hypothesized that the signal regulating inlps induction is dependent on the specific nature of the growth environment in mature pellicle biofilms, which are not created by other growth conditions, including during colony development on agar surfaces and in high-density detergent-free shaken cultures. To address this hypothesis, we used a previously described, hyperaggregating suppressor strain of M. smegmatis that reports gene regulation responsive to cell surface properties and/or biofilm microenvironments (14). The strain carries a suppressor mutation in the glycopeptidolipid biosynthesis gene (mps) in a Δlsr2 background that rescues the deficiencies in cell-cell adhesion and biofilm formation of the parent Δlsr2 strain (14). Importantly, the suppressor mutation does not repress the expression of genes that are induced upon lsr2 deletion, implying that these genes are directly repressed by Lsr2 (14). Thus, the lsr2-independent genes responsive to the suppressor mutation are likely responding to signals sensing changes in environments that occur during aggregated growth such as pellicle development. We therefore reasoned that if M. smegmatis encodes regulators that recognize the inlps promoter and regulate its expression in a manner that responds to cellular growth in biofilms, then the regulation will be diminished in lsr2 mutation, but restored in the Δlsr2/Δmps suppressor. The expression pattern of a Pinlps-Dendra2 reporter in M. smegmatis was similar to those observed in M. tuberculosis: induced expression was observed only during later stages (4 days or later) of pellicle development, and the induction was not apparent in planktonic cultures or colonies (Fig. 6A to C). Induced expression of Dendra2 reporter was not observed in impaired biofilms of the Δlsr2 mutant, but was substantially restored in the Δlsr2/Δmps suppressor (Fig. 6C). Together, the findings in M. smegmatis suggest that signals inducing inlps expression likely originate from the unique environment formed within biofilms. The Pinlps-Dendra2 reporter therefore can potentially serve as a marker for biofilm development.
FIG 6.
Expression pattern of INLPS promoter of M. tuberculosis in M. smegmatis. (A to C) Reporter (Pinlps-Dendra2) expression in recombinant M. smegmatis (mc2155) either in shaken detergent-free Sauton’s medium incubated overnight (planktonic) (A), as 3-day colonies on Sauton’s medium agar plate (B), or as 4-day pellicle biofilms in static Sauton’s medium (C). Reporter strains of a biofilm-defective Δlsr2 mutant and its Δlsr2/Δmps isogenic suppressor were also examined for Pinlps-Dendra2 expression in biofilm culture. Cells from each culture were smeared on a slide in 50% glycerol and imaged under wide-field fluorescent-light microscope at ×20 magnification under phase contrast (Ph) or a blue filter (Dendra2). Morphologies of pellicles and colonies, from which cells were collected, are also shown. Cells in detergent-free shaken culture predominantly contain microscopic clumps (A). The white scale bar in the colony image of panel B represents 10 mm; the scale bar in all microscopic images represents 50 μm.
DISCUSSION
In this study, we propose a model for elevated level of drug tolerance and high frequency of persisters of M. tuberculosis in biofilms. It is apparent from the Tn-seq analysis that significantly more genes are required for fitness of M. tuberculosis cells in biofilms than in other growth conditions. Cells growing within the complex biofilm architecture must be able to adapt to limiting nutrients and oxygen, as well as mechanical stresses. It is therefore not surprising that genes involved in the stringent response pathway (relA), cell wall integrity (mma4, ponA2, mmpL10, fadA2, pks16, etc.) or nutrient homeostasis (sugA, Rv2606c, sdaA, pstC2, pstA1, and phoT) are more crucial for M. tuberculosis fitness in biofilms than in planktonic culture. However, the correlation of a subset of these genes with antibiotic tolerance—e.g., mma4 (31), ponA2 (45), pstA1 (44), and Rv2224c (45)—suggests that a greater fraction of biofilm cells can survive for a longer exposure to antibiotics than planktonic cells. The extended survival of the biofilm population under antibiotic exposure would allow greater opportunity for the occurrence of persister cells, which emerge through mechanisms related to intrinsic defense against antibiotic-induced toxicity (46). In support of this hypothesis, we detected a lower frequency of persisters in morphologically normal biofilms of the ΔpstC2A1 mutant, providing a reasonable platform to systematically evaluate the contribution of other identified genes in this study in drug tolerance of M. tuberculosis biofilms. We note that rifampin sensitivity was also observed in the ΔphoT and Δdgt mutants (J.P.R. and A.O., unpublished results). Moreover, the normal morphology of ΔpstC2A1 mutant biofilms suggests that the physical barrier presumably formed by the three-dimensional architecture of biofilms has a limited contribution to the development of RIF-tolerant persisters; instead the intrinsic sensitivity of resident cells to the drug plays a significant role. Thus, a primary role of the biofilm architecture is to create growth environments that foster enrichment of cells maintaining tolerance to antibiotics.
A stringent genetic requirement for M. tuberculosis fitness in biofilms implies that conditions within biofilms are uniquely challenging. The unique environment of M. tuberculosis biofilms is clearly supported by the biofilm-specific induction of INLP. Further investigations on INLP function and its regulation will likely identify the conditions and signals that induce inlps expression within biofilms. SigC appears to be a strong candidate regulating the expression of INLPS in M. tuberculosis because it binds directly to the region upstream of Rv0096 (51), and overexpression of SigC induces the expression of Rv0096 (52). However, M. smegmatis, which also exhibits biofilm-specific upregulation of INLPS (Fig. 6), does not encode a SigC homolog, suggesting the likelihood of a functional equivalent of SigC in this species. Regardless, the fast-growing species offers a suitable model to identify the regulator and the signal inducing INLPS. INLP-like isonitrile compounds in Streptomyces thioluteus appear to be involved in copper transport (53), suggesting that INLP could be involved in metal transport in M. tuberculosis cells during biofilm development. Low intracellular zinc and nickel in a Δinlps mutant of Mycobacterium marinum (47) support INLP-dependent metal import in mycobacteria. Moreover, zinc is an important nutrient for M. tuberculosis biofilm formation (22). However, other functions of INLP cannot be ruled out, particularly because an intrinsic defect such as zinc import fails to explain why an inlps mutant could be trans-complemented by wild-type—as implied by the lack of inlps mutants in our Tn-seq screen.
Although direct evidence of M. tuberculosis biofilms during infection remains to be obtained, several genes required for fitness in biofilms are also important in M. tuberculosis survival during the early acute phase of infection—characterized by survival in macrophages (e.g., ponA2 [45], Rv2224c [45, 54], mma4 [55], and pstC2A1 [56])—and in the chronic phase characterized by survival beyond 8 weeks of infection in a mouse model (e.g., relA [57]). This fits with an argument that in vivo biofilms of M. tuberculosis would likely represent a persistent community of bacilli that can successfully survive both innate and adaptive host resistance mechanisms. INLP has been implicated in M. tuberculosis survival during both acute and chronic phases of infection (58, 59), although it is unclear if the expression level of INLP varies during infection. It is tempting to speculate that a basal level of INLP expression has a different consequence for M. tuberculosis pathogenesis than an induced level. An INLP-like reporter could potentially address these possibilities and define in vivo biofilms of M. tuberculosis during infection.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
Unless otherwise indicated, an attenuated strain of M. tuberculosis, mc27000 (22), was used as the parent wild type in the study. For planktonic cultures of mc27000 or its recombinant strains, cells were grown at 37°C in Middlebrook 7H9 medium (Difco) supplemented with 10% (vol/vol) albumin, dextrose, catalase and oleic acid (OADC) (Difco), 0.05% (vol/vol) Tween 80 (Sigma), and 100 μg/ml pantothenic acid (Sigma). For plate cultures, Middlebrook 7H11 agar supplemented with 10% OADC, and 100 μg/ml pantothenic acid, or Sauton’s medium with 1% agarose and 100 μg/ml pantothenic acid, were used. As necessary, zeocin, kanamycin, and hygromycin were added at concentrations of 25, 20, and 50 μg/ml, respectively, while selecting the recombinant strains. For pellicle biofilms, logarithmic-phase planktonic cultures of the tested strains were washed twice and then resuspended in detergent-free 7H9-OADC or Sauton’s medium and diluted 1:100 into the corresponding detergent-free medium. Pellicles were grown at 37°C with 4 ml per well in 12-well polystyrene tissue culture plates wrapped in Parafilm for 5 weeks. If necessary, 12-well plates were replaced with 50-ml polystyrene conical tubes (Fisher Scientific), while all other conditions were maintained the same. For colonies grown for the Tn-seq, 1:10 dilutions of washed and resuspended cultures were spotted onto 13-mm Whatman polycarbonate membranes (Millipore Sigma, catalog no. WHA110407), which were then dried for 1 h to allow cells to attach. Inoculated membranes were placed on sterilized stacks of cardstock soaked in a pool of either detergent-free 7H9-OADC medium or Sauton’s medium inside a 100- by 35-mm polystyrene culture plate and then grown at 37°C for the indicated period of time. Medium was replenished as needed. For Mycobacterium smegmatis, the mc2155 strain (wild type) was grown at 37°C in Middlebrook 7H9 medium supplemented with 10% (vol/vol) albumin, dextrose, and catalase (ADC) (Difco) and 0.05% (vol/vol) Tween 80 or in detergent-free Sauton’s medium. Agar plates of these media were used to obtain colonies. For selection of M. smegmatis, 150 μg/ml of hygromycin or 25 μg/ml of zeocin was used as necessary. M. smegmatis pellicle biofilms were grown in Sauton’s medium, as previously described (14), by inoculating 10 μl from a saturated planktonic culture into 10 ml of detergent-free Sauton’s medium in 60-mm polystyrene culture plates and incubating at 37°C for 4 days. For molecular cloning, Escherichia coli GC5 cells were grown at 37°C in Luria broth or on LB agar under antibiotic selection conditions: 100 μg/ml of carbenicillin, 50 μg/ml of kanamycin, 150 μg/ml of hygromycin, or 25 μg/ml of zeocin.
Tn-seq screen and analysis.
A transposon insertion mutant library of mc27000 consisting of approximately 100,000 isolated mutant colonies was constructed using ΦMycoMarT7 bacteriophage carrying the Himar-1 transposon. The library was grown in 7H9-OADC either as a planktonic suspension to mid-logarithmic phase, in pellicle biofilms for 5 weeks, or as colonies on polycarbonate membrane for 18 days. Cells from each growth model, including planktonic suspension, were harvested and resuspended in phosphate-buffered saline (PBS) with 0.25% (vol/vol) Tween 80, vortexed for 30 s to disperse biomass, and sonicated in a Branson bath sonicator for 10 min. Ten-fold dilutions of bacteria from each sample were then plated out on ten 100-mm 7H11-OADC plates per replicate and incubated for 21 days. Colonies were harvested with a cell scraper, and genomic DNA was extracted for further processing as described previously (34). Briefly, 2 to 5 μg of genomic DNA was sheared with a Covaris M220 Focused-ultrasonicator (Covaris) into 400- to 600-bp fragments, which were resolved by gel electrophoresis. The resolved fragments were gel extracted (Qiagen catalog no. 28606), and end blunted using the Epicentre end repair kit (catalog no. ER0720). The fragments were then adenylated at the 3′ sequence end, and custom adapter oligonucleotides were ligated using the Epicentre FastLink DNA ligation kit (catalog no. LK6201H). Two-step heminested PCR amplification of DNA fragments with a mixture of four staggered primers with homology to the end region of the transposon insertion resulted in a library of PCR amplicons at the junction site of transposon insertion and genomic DNA. The primers used for heminested PCR and DNA sequencing are listed in Table S5 in the supplemental material. PCR with a set of short primers was performed with the following settings: 95°C for 5 min, followed by 20 cycles of 95°C for 30 s, 58°C for 30 s, and 72°C for 45 s, then 72°C for 5 min. This was followed by heminested PCR with a staggered primer set at 95°C for 5 min: 10 cycles of 95°C for 30 s, 58°C for 30 s, and 72°C for 45 s, then 72°C for 5 min. The library, prepared from a pool of three biologically independent sources of genomic DNA for every growth model, was sequenced at the transposon junction site using the Illumina Hi-Seq 2500 platform (Tn-seq). Two biologically independent sets of libraries were sequenced for each growth model. The sequencing results were analyzed by the TRANSIT (35) pipeline, using terminal total reads (TTR) as the normalization method, to identify open reading frames (ORFs) with significant underrepresentation in either pellicle or colony biofilms, relative to planktonic cultures.
Plasmids and mutant constructions.
The strains and plasmids used in this study are listed in Tables S6 and S7, respectively, in the supplemental material. A modified recombineering method, as described previously (14), was used for construction of isogenic deletions in the indicated genes. For allelic exchange substrates (AESs) of a target gene, 250-bp PCR amplicons corresponding to upstream and downstream of the gene were joined to the either side of a loxP-flanked zeocin-resistance cassette by “sewing PCR.” Oligonucleotides used for AES generation are listed in Table S5. The AESs were electroporated into a recombineering strain of mc27000 carrying the plasmid pJV53-SacB (14), and plated on 7H11-OADC agar with 25 μg/ml of zeocin and 20 μg/ml kanamycin. The genotype of Zeor colonies was confirmed by PCR. The recombineering plasmid, pJV53-SacB, was removed by selecting the mutant strains on 7H11-OADC agar with 15% (wt/vol) sucrose and 25 μg/ml zeocin. The sucrose-resistant colonies were screened for kanamycin sensitivity. A second PCR was performed to confirm gene deletion gene using primers homologous to upstream and downstream ends of AESs and to the zeocin-resistance cassette. As necessary, mutants were complemented by the corresponding gene cloned along with a 500-bp upstream promoter region, or fused to hsp60 promoter, on an integrative plasmid (pMH94). For a reporter strain of a mutant, an integrative plasmid expressing mCherry from the constitutive hsp60 promoter was electroporated into the mutant, and transformants were selected on 7H11-OADC plates with 25 μg/ml zeocin and 20 μg/ml kanamycin. For construction of the INLP expression reporter, the putative promoter and regulator elements of INLP harbored in the 500 bp upstream of the gene cluster were amplified using the primers listed in Table S5 and cloned upstream of Dendra2 at XbaI and NdeI restriction sites in plasmid pYL026. The resulting plasmid, pJR36, was electroporated into mc2155, mc27000, and M. tuberculosis (Erdman) strains for analysis of the reporter expression.
Microscopic imaging and analysis.
For imaging on a wide-field fluorescence microscope, Nikon Eclipse TE2000-E microscope with either a 20× (NA, 0.75) objective was used. Images were taken with a 600-nm exposure using Pro-Image 7.0 software under transmitted light (phase contrast) or with X-Cite 120 fluorescence illumination system with a blue GFP(R)-BP filter (excitation, 460 to 500 nm; BA, 510 to 560 nm) for Dendra2 fluorescence or a yellow Y-2E filter (excitation, 540 to 580 nm; BA, 600 to 660 nm) for mCherry. Confocal scanning laser microscopy (CSLM) imaging was done with a Leica SP5 confocal microscope with a 488-nm laser for Dendra2 and a 593-nm laser for mCherry, under the settings described above. All postacquisition analysis was done using FIJI image analysis software. All settings on the microscopes as well as in FIJI were maintained across compared samples during image acquisition and analysis.
Analysis of persisters in pellicle biofilms.
Biofilms of the indicated strains were grown in pellicles as described above and then exposed to 50 μg/ml of RIF (or an equal volume of dimethyl sulfoxide [DMSO] as a control) for 7 days. After exposure, pellicles were collected and washed 3 times with 1× PBS with 0.25% Tween 80 to remove residual antibiotic and then left on a shaker at 4°C overnight in the presence of sterile 10-mm glass beads. Serial dilutions were plated on 7H11 agar to enumerate the viable colonies.
Gene expression analysis by RNA-seq and RT-PCR.
Using a Qiagen RNeasy kit (catalog no. 74104), total RNA from desired cultures of the indicated strains of M. tuberculosis were isolated from 20-ml cultures obtained from the mid-exponential growth phase in 7H9-OADC medium. For biofilm transcriptomics, mc27000 was grown planktonically to mid-exponential phase in detergent-free Sauton’s medium or into pellicles for 5 weeks in detergent-free Sautons’s medium as described above. The pellicle biomass portion was scooped out of the liquid medium using Parafilm. RNA was extracted from the pellicles using the RNAeasy kit (Qiagen, Inc.). Contaminating genomic DNA from the RNA preparations was removed with the Thermo Fisher Scientific Turbo DNA-free kit (catalog no. AM1907). The rRNA from a total of 5 μg RNA was removed with the Illumina RiboZero kit (now discontinued). Strand-specific cDNA libraries were prepared from 100 ng mRNA of each sample using the Illumina Scriptseq v2 Complete kit (catalog no. SSV21106). Libraries were sequenced on the Illumina NextSeq500 platform at the Wadsworth Center, and results were analyzed by Rockhopper (60) at the default settings using the M. tuberculosis H37Rv reference genome (NC_000962). All oligonucleotides used for reverse transcription-quantitative PCR (RT-qPCR) are listed in Table S5. For RT-qPCR, DNA-free RNA was extracted from desired bacterial cultures as described above for RNA-seq. Using the Fisher Maxima first strand cDNA synthesis kit (catalog no. K1641), cDNA was generated from 200 ng of total RNA from each specified sample. RT-qPCR mixtures were prepared using 2 μl of cDNA reaction mixture, 0.5 μl from 10 μM stock for each gene-specific primer per reaction, and Applied Biosystems SYBR green master mix (catalog no. A25742) as per the manufacturer’s instruction. qPCR was performed on an Applied Biosystems 7500 Fast real-time PCR system (catalog no. 4351106) using the following cycle conditions: 95°C for 10 min, followed by 40 cycles of 95°C for 20 s, and then 60°C for 1 min. Primers corresponding to SigA transcripts were used as an endogenous control. Reaction mixtures without the cDNA were used as the no-template negative control. Expression of a target gene (tgene) in pellicles relative to planktonic was calculated as 2−[ΔCT(pellicle_tgene − pellicle_sigA) − ΔCT(plnk_tgene − plnk_sigA)], where “CT” is the cycle threshold.
INLP analysis.
Pellicle biofilms or planktonic cultures were grown in detergent-free Sauton’s medium prior to crude lipid extraction. A 1 mM concentration of [2-13C]Gly (Sigma) was included in selective biofilm cultures for isotope incorporation. Cells from planktonic cultures were transferred to glass tubes, and an equal volume of chloroform-methanol (2:1) was added to each culture, followed by intermittent vortexing for 30 s every 15 min for 2 h. The mixture was then centrifuged at ∼900 × g for 10 min on a Thermo Scientific Sorvall Legend XTR centrifuge (catalog no. 75004521). The lower, organic phase was separated by Pasteur pipette and dried under N2 gas. Pellicle biomass was separated from the liquid portion of the cultures using Parafilm. The biomass was washed and resuspended in 5 ml PBS. An equal volume of chloroform-methanol (2:1) was added to the suspension, and the lipid in organic phase was collected and dried as described above. Dried crude extracts were dissolved in methanol to a final concentration of 0.1 mg/ml. Samples were centrifuged at 12,000 rpm for 5 min, and the supernatant was used for LC-MS analysis.
Liquid chromatography-mass spectrometry (Orbitrap).
Samples of extracted metabolites were analyzed using a liquid chromatography system (1200 series, Agilent, Santa Clara, CA) that was connected in-line with an LTQ-Orbitrap-XL mass spectrometer equipped with an electrospray ionization source (Thermo Fisher Scientific, Waltham, MA). The liquid chromatograph was equipped with a reverse-phase analytical column (length, 150 mm; inner diameter, 1.0 mm; particle size, 5 μm [Viva C18; Restek, Bellefonte, PA]). Acetonitrile, formic acid (Optima grade, 99.5+%; Fisher, Pittsburgh, PA), and water purified to a resistivity of 18.2 MΩ·cm (at 25°C) using a Milli-Q gradient ultrapure water purification system (Millipore, Billerica, MA) were used to prepare mobile-phase solvents. Solvent A was 99.9% water–0.1% formic acid, and solvent B was 99.9% acetonitrile–0.1% formic acid (vol/vol). The elution program consisted of isocratic conditions at 5% B for 2 min, a linear gradient to 98% B over 25 min, and isocratic conditions at 98% B for 10 min, at a flow rate of 150 μl/min. Full-scan, high-resolution mass spectra were acquired over the m/z range of 70 to 1,000 using the Orbitrap mass analyzer, in the positive-ion mode and profile format, with a mass resolution setting of 100,000 (measured at full width at half-maximum peak height [FWHM] at m/z = 400). For tandem mass spectrometry (MS/MS or MS2) analysis, precursor ions were fragmented using higher-energy collisional dissociation (HCD) under the following conditions: MS/MS spectra acquired using the Orbitrap mass analyzer, in centroid format, with a mass resolution setting of 7,500 (at m/z = 400, FWHM), isolation width of 5 m/z units, normalized collision energy of 38%, default charge state of 1+, activation time of 30 ms, and first m/z value of 100. Mass spectrometry data acquisition and analysis were performed using Xcalibur software (version 2.0.7; Thermo). Comparative metabolomics analysis was performed using MS-DIAL (61).
Liquid chromatography-mass spectrometry (QTOF).
LC-MS analysis was performed using an Agilent Technologies 6520 Accurate-Mass quadrupole time of flight (Q-TOF) LC-MS instrument and an Agilent Eclipse Plus C18 column (4.6 by 100 mm). A linear gradient of 2 to 98% acetonitrile (vol/vol) over 30 min in H2O with 0.1% formic acid (vol/vol) at a flow rate of 0.5 ml/min was used. HRMS/MS analysis was conducted using targeted MS/MS with collision energy of 20 V.
Click chemistry analysis.
Compounds 1 and 2 were partially purified from the wild-type culture extract using high-performance liquid chromatography (HPLC). This was conducted using an Agilent 1200 high-performance liquid chromatograph with a Waters Atlantis T3 OBD column (10 by 250 mm) using a linear gradient of 5 to 95% CH3CN (vol/vol) over 30 min in H2O without formic acid at a flow rate of 3.5 ml/min. Fractions were screened using LC-MS as described above. Fractions containing compounds 1 and 2 were combined, dried, and redissolved in methanol, followed by addition of 100 μM 3,6-di-2-pyridyl-1,2,4,5-tetrazine and incubated at room temperature for 4.5 h before LC-MS analysis.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by grants to A.K.O. (NIH: AI132422, NIH: AI144474) and W.Z. (NIH: DP2AT009148, the Alfred P. Sloan Foundation, and the Chan Zuckerberg Biohub Investigator Program). Support from the Wadsworth Center core facilities Fluorescent and Light Microscopy Imaging and Analysis and Applied Genomics Technology is acknowledged. Support to the QB3/Chemistry Mass Spectrometry Facility, University of California, Berkeley, by NIH grant (1S10OD020062) is acknowledged.
We acknowledge Yong Yang and Jennifer Gundrum for technical help. We are also thankful to William R. Jacobs, Jr., for the gift of the inlps cosmid, Christopher M. Sassetti for help with Tn-seq experiments, and Anthony T. Iavarone for assistance with LC-MS (Orbitrap) analysis.
J.P.R. and A.K.O. designed and performed experiments, as well as analyzed data on all experiments except chemical analysis of INLP. W.C., N.A.Z., and W.Z. designed and performed experiments and analyzed data on INLP analysis. J.P.R., W.C., N.A.Z., and W.Z. wrote the manuscript.
The authors declare no financial conflict of interest.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/AAC.01213-19.
REFERENCES
- 1.Jindani A, Aber VR, Edwards EA, Mitchison DA. 1980. The early bactericidal activity of drugs in patients with pulmonary tuberculosis. Am Rev Respir Dis 121:939–949. [DOI] [PubMed] [Google Scholar]
- 2.Wallis RS, Patil S, Cheon SH, Edmonds K, Phillips M, Perkins MD, Joloba M, Namale A, Johnson JL, Teixeira L, Dietze R, Siddiqi S, Mugerwa RD, Eisenach K, Ellner JJ. 1999. Drug tolerance in Mycobacterium tuberculosis. Antimicrob Agents Chemother 43:2600–2606. doi: 10.1128/AAC.43.11.2600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wakamoto Y, Dhar N, Chait R, Schneider K, Signorino-Gelo F, Leibler S, McKinney JD. 2013. Dynamic persistence of antibiotic-stressed mycobacteria. Science 339:91–95. doi: 10.1126/science.1229858. [DOI] [PubMed] [Google Scholar]
- 4.Torrey HL, Keren I, Via LE, Lee JS, Lewis K. 2016. High persister mutants in Mycobacterium tuberculosis. PLoS One 11:e0155127. doi: 10.1371/journal.pone.0155127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Aldridge BB, Fernandez-Suarez M, Heller D, Ambravaneswaran V, Irimia D, Toner M, Fortune SM. 2012. Asymmetry and aging of mycobacterial cells lead to variable growth and antibiotic susceptibility. Science 335:100–104. doi: 10.1126/science.1216166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jain P, Weinrick BC, Kalivoda EJ, Yang H, Munsamy V, Vilcheze C, Weisbrod TR, Larsen MH, O’Donnell MR, Pym A, Jacobs WR Jr.. 2016. Dual-reporter mycobacteriophages (Phi2DRMs) reveal preexisting Mycobacterium tuberculosis persistent cells in human sputum. mBio 7:e01023-16. doi: 10.1128/mBio.01023-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Stoodley P, Sauer K, Davies DG, Costerton JW. 2002. Biofilms as complex differentiated communities. Annu Rev Microbiol 56:187–209. doi: 10.1146/annurev.micro.56.012302.160705. [DOI] [PubMed] [Google Scholar]
- 8.López D, Vlamakis H, Kolter R. 2010. Biofilms. Cold Spring Harb Perspect Biol 2:a000398. doi: 10.1101/cshperspect.a000398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Flemming H-C, Wingender J, Szewzyk U, Steinberg P, Rice SA, Kjelleberg S. 2016. Biofilms: an emergent form of bacterial life. Nat Rev Microbiol 14:563. doi: 10.1038/nrmicro.2016.94. [DOI] [PubMed] [Google Scholar]
- 10.Islam MS, Richards JP, Ojha AK. 2012. Targeting drug tolerance in mycobacteria: a perspective from mycobacterial biofilms. Expert Rev Anti Infect Ther 10:1055–1066. doi: 10.1586/eri.12.88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fux CA, Costerton JW, Stewart PS, Stoodley P. 2005. Survival strategies of infectious biofilms. Trends Microbiol 13:34–40. doi: 10.1016/j.tim.2004.11.010. [DOI] [PubMed] [Google Scholar]
- 12.Davies D. 2003. Understanding biofilm resistance to antibacterial agents. Nat Rev Drug Discov 2:114. doi: 10.1038/nrd1008. [DOI] [PubMed] [Google Scholar]
- 13.Petrova OE, Sauer K. 2009. A novel signaling network essential for regulating Pseudomonas aeruginosa biofilm development. PLoS Pathog 5:e1000668. doi: 10.1371/journal.ppat.1000668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yang Y, Thomas J, Li Y, Vilchèze C, Derbyshire KM, Jacobs WR, Ojha AK. 2017. Defining a temporal order of genetic requirements for development of mycobacterial biofilms. Mol Microbiol 105:794–809. doi: 10.1111/mmi.13734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Vlamakis H, Aguilar C, Losick R, Kolter R. 2008. Control of cell fate by the formation of an architecturally complex bacterial community. Genes Dev 22:945–953. doi: 10.1101/gad.1645008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Serra DO, Hengge R. 2014. Stress responses go three dimensional—the spatial order of physiological differentiation in bacterial macrocolony biofilms. Environ Microbiol 16:1455–1471. doi: 10.1111/1462-2920.12483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rani SA, Pitts B, Beyenal H, Veluchamy RA, Lewandowski Z, Davison WM, Buckingham-Meyer K, Stewart PS. 2007. Spatial patterns of DNA replication, protein synthesis, and oxygen concentration within bacterial biofilms reveal diverse physiological states. J Bacteriol 189:4223–4233. doi: 10.1128/JB.00107-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.An D, Parsek MR. 2007. The promise and peril of transcriptional profiling in biofilm communities. Curr Opin Microbiol 10:292–296. doi: 10.1016/j.mib.2007.05.011. [DOI] [PubMed] [Google Scholar]
- 19.Mah TF, O'Toole GA. 2001. Mechanisms of biofilm resistance to antimicrobial agents. Trends Microbiol 9:34–39. doi: 10.1016/S0966-842X(00)01913-2. [DOI] [PubMed] [Google Scholar]
- 20.Yamazaki Y, Danelishvili L, Wu M, Hidaka E, Katsuyama T, Stang B, Petrofsky M, Bildfell R, Bermudez LE. 2006. The ability to form biofilm influences Mycobacterium avium invasion and translocation of bronchial epithelial cells. Cell Microbiol 8:806–814. doi: 10.1111/j.1462-5822.2005.00667.x. [DOI] [PubMed] [Google Scholar]
- 21.Hall-Stoodley L, Brun OS, Polshyna G, Barker LP. 2006. Mycobacterium marinum biofilm formation reveals cording morphology. FEMS Microbiol Lett 257:43–49. doi: 10.1111/j.1574-6968.2006.00143.x. [DOI] [PubMed] [Google Scholar]
- 22.Ojha AK, Baughn AD, Sambandan D, Hsu T, Trivelli X, Guerardel Y, Alahari A, Kremer L, Jacobs WR Jr, Hatfull GF. 2008. Growth of Mycobacterium tuberculosis biofilms containing free mycolic acids and harbouring drug-tolerant bacteria. Mol Microbiol 69:164–174. doi: 10.1111/j.1365-2958.2008.06274.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Howard ST, Rhoades E, Recht J, Pang X, Alsup A, Kolter R, Lyons CR, Byrd TF. 2006. Spontaneous reversion of Mycobacterium abscessus from a smooth to a rough morphotype is associated with reduced expression of glycopeptidolipid and reacquisition of an invasive phenotype. Microbiology 152:1581–1590. doi: 10.1099/mic.0.28625-0. [DOI] [PubMed] [Google Scholar]
- 24.Hall‐Stoodley L, Keevil CW, Lappin-Scott HM. 1998. Mycobacterium fortuitum and Mycobacterium chelonae biofilm formation under high and low nutrient conditions. J Appl Microbiol 85:60S–69S. doi: 10.1111/j.1365-2672.1998.tb05284.x. [DOI] [PubMed] [Google Scholar]
- 25.Recht J, Martinez A, Torello S, Kolter R. 2000. Genetic analysis of sliding motility in Mycobacterium smegmatis. J Bacteriol 182:4348–4351. doi: 10.1128/jb.182.15.4348-4351.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ojha A, Anand M, Bhatt A, Kremer L, Jacobs WR Jr, Hatfull GF. 2005. GroEL1: a dedicated chaperone involved in mycolic acid biosynthesis during biofilm formation in mycobacteria. Cell 123:861–873. doi: 10.1016/j.cell.2005.09.012. [DOI] [PubMed] [Google Scholar]
- 27.Falkinham JO III, Norton CD, LeChevallier MW. 2001. Factors influencing numbers of Mycobacterium avium, Mycobacterium intracellulare, and other mycobacteria in drinking water distribution systems. Appl Environ Microbiol 67:1225–1231. doi: 10.1128/AEM.67.3.1225-1231.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schulze-Robbecke R, Janning B, Fischeder R. 1992. Occurrence of mycobacteria in biofilm samples. Tuber Lung Dis 73:141–144. doi: 10.1016/0962-8479(92)90147-C. [DOI] [PubMed] [Google Scholar]
- 29.Bosio S, Leekha S, Gamb SI, Wright AJ, Terrell CL, Miller DV. 2012. Mycobacterium fortuitum prosthetic valve endocarditis: a case for the pathogenetic role of biofilms. Cardiovasc Pathol 21:361–364. doi: 10.1016/j.carpath.2011.11.001. [DOI] [PubMed] [Google Scholar]
- 30.Marsollier L, Brodin P, Jackson M, Korduláková J, Tafelmeyer P, Carbonnelle E, Aubry J, Milon G, Legras P, André J-P, Leroy C, Cottin J, Guillou MLJ, Reysset G, Cole ST. 2007. Impact of Mycobacterium ulcerans biofilm on transmissibility to ecological niches and Buruli ulcer pathogenesis. PLoS Pathog 3:e62. doi: 10.1371/journal.ppat.0030062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sambandan D, Dao DN, Weinrick BC, Vilcheze C, Gurcha SS, Ojha A, Kremer L, Besra GS, Hatfull GF, Jacobs WR Jr.. 2013. Keto-mycolic acid-dependent pellicle formation confers tolerance to drug-sensitive Mycobacterium tuberculosis. mBio 4:e00222-13. doi: 10.1128/mBio.00222-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ojha AK, Jacobs WR Jr, Hatfull GF. 2015. Genetic dissection of mycobacterial biofilms. Methods Mol Biol 1285:215–226. doi: 10.1007/978-1-4939-2450-9_12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yang Y, Richards JP, Gundrum J, Ojha AK. 2018. GlnR activation induces peroxide resistance in mycobacterial biofilms. Front Microbiol 9:1428. doi: 10.3389/fmicb.2018.01428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Griffin JE, Gawronski JD, Dejesus MA, Ioerger TR, Akerley BJ, Sassetti CM. 2011. High-resolution phenotypic profiling defines genes essential for mycobacterial growth and cholesterol catabolism. PLoS Pathog 7:e1002251. doi: 10.1371/journal.ppat.1002251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.DeJesus MA, Ambadipudi C, Baker R, Sassetti C, Ioerger TR. 2015. TRANSIT—a software tool for Himar1 TnSeq analysis. PLoS Comput Biol 11:e1004401. doi: 10.1371/journal.pcbi.1004401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Weiss LA, Stallings CL. 2013. Essential roles for Mycobacterium tuberculosis Rel beyond the production of (p)ppGpp. J Bacteriol 195:5629–5638. doi: 10.1128/JB.00759-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rengarajan J, Bloom BR, Rubin EJ. 2005. Genome-wide requirements for Mycobacterium tuberculosis adaptation and survival in macrophages. Proc Natl Acad Sci U S A 102:8327–8332. doi: 10.1073/pnas.0503272102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sassetti CM, Rubin EJ. 2003. Genetic requirements for mycobacterial survival during infection. Proc Natl Acad Sci U S A 100:12989–12994. doi: 10.1073/pnas.2134250100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hsieh YJ, Wanner BL. 2010. Global regulation by the seven-component Pi signaling system. Curr Opin Microbiol 13:198–203. doi: 10.1016/j.mib.2010.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Saier MH Jr, Reddy VS, Tsu BV, Ahmed MS, Li C, Moreno-Hagelsieb G. 2016. The Transporter Classification Database (TCDB): recent advances. Nucleic Acids Res 44:D372–D379. doi: 10.1093/nar/gkv1103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sarin J, Aggarwal S, Chaba R, Varshney GC, Chakraborti PK. 2001. B-subunit of phosphate-specific transporter from Mycobacterium tuberculosis is a thermostable ATPase. J Biol Chem 276:44590–44597. doi: 10.1074/jbc.M105401200. [DOI] [PubMed] [Google Scholar]
- 42.Tischler AD, Leistikow RL, Ramakrishnan P, Voskuil MI, McKinney JD. 2016. Mycobacterium tuberculosis phosphate uptake system component PstA2 is not required for gene regulation or virulence. PLoS One 11:e0161467. doi: 10.1371/journal.pone.0161467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Glover RT, Kriakov J, Garforth SJ, Baughn AD, Jacobs WR Jr.. 2007. The two-component regulatory system senX3-regX3 regulates phosphate-dependent gene expression in Mycobacterium smegmatis. J Bacteriol 189:5495–5503. doi: 10.1128/JB.00190-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Namugenyi SB, Aagesen AM, Elliott SR, Tischler AD. 2017. Mycobacterium tuberculosis PhoY proteins promote persister formation by mediating Pst/SenX3-RegX3 phosphate sensing. mBio 8:e00494-17. doi: 10.1128/mBio.00494-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Vandal OH, Roberts JA, Odaira T, Schnappinger D, Nathan CF, Ehrt S. 2009. Acid-susceptible mutants of Mycobacterium tuberculosis share hypersusceptibility to cell wall and oxidative stress and to the host environment. J Bacteriol 191:625–631. doi: 10.1128/JB.00932-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wilmaerts D, Windels EM, Verstraeten N, Michiels J. 2019. General mechanisms leading to persister formation and awakening. Trends Genet 35:401–411. doi: 10.1016/j.tig.2019.03.007. [DOI] [PubMed] [Google Scholar]
- 47.Harris NC, Sato M, Herman NA, Twigg F, Cai W, Liu J, Zhu X, Downey J, Khalaf R, Martin J, Koshino H, Zhang W. 2017. Biosynthesis of isonitrile lipopeptides by conserved nonribosomal peptide synthetase gene clusters in actinobacteria. Proc Natl Acad Sci U S A 114:7025–7030. doi: 10.1073/pnas.1705016114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wang F, Langley R, Gulten G, Wang L, Sacchettini JC. 2007. Identification of a type III thioesterase reveals the function of an operon crucial for Mtb virulence. Chem Biol 14:543–551. doi: 10.1016/j.chembiol.2007.04.005. [DOI] [PubMed] [Google Scholar]
- 49.Parish T, Smith DA, Roberts G, Betts J, Stoker NG. 2003. The senX3-regX3 two-component regulatory system of Mycobacterium tuberculosis is required for virulence. Microbiology 149:1423–1435. doi: 10.1099/mic.0.26245-0. [DOI] [PubMed] [Google Scholar]
- 50.Harris NC, Born DA, Cai W, Huang Y, Martin J, Khalaf R, Drennan CL, Zhang W. 2018. Isonitrile formation by a non-heme iron(II)-dependent oxidase/decarboxylase. Angew Chem Int Ed Engl 57:9707–9710. doi: 10.1002/anie.201804307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Minch KJ, Rustad TR, Peterson EJ, Winkler J, Reiss DJ, Ma S, Hickey M, Brabant W, Morrison B, Turkarslan S, Mawhinney C, Galagan JE, Price ND, Baliga NS, Sherman DR. 2015. The DNA-binding network of Mycobacterium tuberculosis. Nat Commun 6:5829. doi: 10.1038/ncomms6829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Rustad TR, Minch KJ, Ma S, Winkler JK, Hobbs S, Hickey M, Brabant W, Turkarslan S, Price ND, Baliga NS, Sherman DR. 2014. Mapping and manipulating the Mycobacterium tuberculosis transcriptome using a transcription factor overexpression-derived regulatory network. Genome Biol 15:502. doi: 10.1186/PREACCEPT-1701638048134699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wang L, Zhu M, Zhang Q, Zhang X, Yang P, Liu Z, Deng Y, Zhu Y, Huang X, Han L, Li S, He J. 2017. Diisonitrile natural product SF2768 functions as a chalkophore that mediates copper acquisition in Streptomyces thioluteus. ACS Chem Biol 12:3067–3075. doi: 10.1021/acschembio.7b00897. [DOI] [PubMed] [Google Scholar]
- 54.Rengarajan J, Murphy E, Park A, Krone CL, Hett EC, Bloom BR, Glimcher LH, Rubin EJ. 2008. Mycobacterium tuberculosis Rv2224c modulates innate immune responses. Proc Natl Acad Sci U S A 105:264–269. doi: 10.1073/pnas.0710601105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Dubnau E, Chan J, Raynaud C, Mohan VP, Laneelle MA, Yu K, Quemard A, Smith I, Daffe M. 2000. Oxygenated mycolic acids are necessary for virulence of Mycobacterium tuberculosis in mice. Mol Microbiol 36:630–637. doi: 10.1046/j.1365-2958.2000.01882.x. [DOI] [PubMed] [Google Scholar]
- 56.Tischler AD, Leistikow RL, Kirksey MA, Voskuil MI, McKinney JD. 2013. Mycobacterium tuberculosis requires phosphate-responsive gene regulation to resist host immunity. Infect Immun 81:317–328. doi: 10.1128/IAI.01136-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Dahl JL, Kraus CN, Boshoff HI, Doan B, Foley K, Avarbock D, Kaplan G, Mizrahi V, Rubin H, Barry CE III, 2003. The role of RelMtb-mediated adaptation to stationary phase in long-term persistence of Mycobacterium tuberculosis in mice. Proc Natl Acad Sci U S A 100:10026–10031. doi: 10.1073/pnas.1631248100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Dhar N, McKinney JD. 2010. Mycobacterium tuberculosis persistence mutants identified by screening in isoniazid-treated mice. Proc Natl Acad Sci U S A 107:12275–12280. doi: 10.1073/pnas.1003219107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Bhatt K, Machado H, Osorio NS, Sousa J, Cardoso F, Magalhaes C, Chen B, Chen M, Kim J, Singh A, Ferreira CM, Castro AG, Torrado E, Jacobs WR Jr, Bhatt A, Saraiva M. 2018. A nonribosomal peptide synthase gene driving virulence in Mycobacterium tuberculosis. mSphere 3:e00352-18. doi: 10.1128/mSphere.00352-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.McClure R, Balasubramanian D, Sun Y, Bobrovskyy M, Sumby P, Genco CA, Vanderpool CK, Tjaden B. 2013. Computational analysis of bacterial RNA-Seq data. Nucleic Acids Res 41:e140. doi: 10.1093/nar/gkt444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Tsugawa H, Cajka T, Kind T, Ma Y, Higgins B, Ikeda K, Kanazawa M, VanderGheynst J, Fiehn O, Arita M. 2015. MS-DIAL: data-independent MS/MS deconvolution for comprehensive metabolome analysis. Nat Methods 12:523–526. doi: 10.1038/nmeth.3393. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.