The continuous surveillance of polymorphisms in the kelch propeller domain of Plasmodium falciparum from Africa is important for the discovery of the actual markers of artemisinin resistance in the region. The information on the markers is crucial for control strategies involving chemotherapy and chemoprophylaxis for residents and nonimmune travelers to the country.
Keywords: Plasmodium falciparum kelch propeller domain gene (pfk13), single nucleotide polymorphisms (SNPs), antimalarial drug resistance, artemisinin (ART), artemisinin-based combination therapy (ACT), malaria, Ghana
ABSTRACT
The continuous surveillance of polymorphisms in the kelch propeller domain of Plasmodium falciparum from Africa is important for the discovery of the actual markers of artemisinin resistance in the region. The information on the markers is crucial for control strategies involving chemotherapy and chemoprophylaxis for residents and nonimmune travelers to the country. Polymorphisms in the kelch propeller domain of Ghanaian malaria parasites from three different ecological zones at several time periods were assessed. A total of 854 archived samples (2007 to 2016) collected from uncomplicated malaria patients aged ≤9 years old from 10 sentinel sites were used. Eighty-four percent had wild-type sequences (PF3D7_1343700), while many of the mutants had mostly nonsynonymous mutations clustered around codons 404 to 650. Variants with different amino acid changes of the codons associated with artemisinin (ART) resistance validated markers were observed in Ghanaian isolates: frequencies for I543I, I543S, I543V, R561P, R561R, and C580V were 0.12% each and 0.6% for R539I. Mutations reported from African parasites, A578S (0.23%) and Q613L (0.23%), were also observed. Three persisting nonsynonymous (NS) mutations, N599Y (0.005%), K607E (0.004%), and V637G (0.004%), were observed in 3 of the 5 time periods nationally. The presence of variants of the validated markers of artemisinin resistance as well as persisting polymorphisms after 14 years of artemisinin-based combination therapy use argues for continuous surveillance of the markers. The molecular markers of artemisinin resistance and the observed variants will be monitored subsequently as part of ongoing surveillance of antimalarial drug efficacy/resistance studies in the country.
INTRODUCTION
Malaria parasite resistance to antimalarial drugs is of paramount concern in control, elimination, and eradication programs throughout the world. Plasmodium falciparum resistance to artemisinin (ART) and its derivatives is well documented in Southeast Asia (SEA), but comparatively low-level ART resistance has been identified in Africa (1). Ongoing worldwide surveillance is necessary considering the potential tremendous public health impact resistance could have, especially on children under 5 years and pregnant women from Africa as well as nonimmune travelers (2). Artemisinin resistance as defined by the WHO is the slow parasite clearance rate with the use of artemisinin-based combination therapy (ACT) or microscopically detected persistent parasites after 3 days of ACT administration (2). Although the exact mechanism of parasite ART resistance is not clearly understood, genetic factors have been implicated for other antimalarials such as chloroquine (CQ; P. falciparum chloroquine resistance transporter gene pfcrt) and sulfadoxine pyrimethamine (SP; P. falciparum dihydrofolate reductase gene pfdhfr and P. falciparum dihydropteroate synthase gene pfdhps) (3, 4).
ART resistance in the SEA region has been linked to the kelch propeller domain on chromosome 13 (pfk13) (3–6). Single nucleotide polymorphisms (SNPs) and consequent amino acid changes in the gene that occurred as a result of drug pressure are used as molecular markers (7, 8). The pfk13 markers of resistance are nonsynonymous and they include N458Y, R539T, E556D, P574L, R575K, C580Y, S621F, Y493H, R539T, and I543T (3, 9). However, the principal mutations are C580Y, Y493H, R539T, I543T, and N458Y, which were observed in all isolates with the slow clearance phenotypic trait. To date, the principal mutations observed in African isolates are C580Y seen in two samples from Cameroon and Y493H seen in one sample from Ghana out of 1,648 samples from Africa (10). Other mutations that have been observed in ART-resistant parasites from the SEA region and have also been prevalent in African isolates include S522C, P553L, R561H, A675V, and H719N (10, 11). The predominant pfk13 mutation found in African isolates is A578S, which has been observed in all the African countries where the pfk13 mutations have been typed (10, 12–18). Since it is close to the principal mutation C580Y, it is alleged that A578S may be the mutant for ART resistance in African isolates (12); however, there is no phenotypic association with ART resistance (8). Of all the pfk13 mutations observed in African isolates, none has yet been directly linked to ART resistance; thus, further investigations are needed.
In Ghana, the malaria treatment policy was changed in 2005 due to P. falciparum resistance to CQ and SP (19). The ACTs used for uncomplicated malaria treatment are artesunate-amodiaquine (AA), which was started in 2005, and artemether-lumefantrine (AL) as well as dihydroartemisinin piperaquine (DHAP), which were subsequently added in 2008 (20). Recently, reports indicate delayed parasite clearance in isolates from the northern parts of Ghana, and the nationwide cure rates are 96.0% for AL and 99.2% for AA (21). Studies on the molecular markers of ART resistance showed increased P. falciparum multidrug resistance gene (pfmdr1) copy number in 18% of isolates and an increasing trend in the prevalence of haplotype pfmdr1 N86-F184-D1246 (linked to AL resistance) from 2003 to 2010 (22). In addition, the P. falciparum adaptor protein complex 2 gene (pfap2mu) S106N and ubiquitin specific protease 1 gene (pfubp1) E1528D and D1525E mutations, which have been linked to the delayed clearance of parasites to ACT in Kenya and recurrent imported malaria in Britain, were observed in 7.4%, 7.4%, and 4.9%, respectively, of Ghanaian parasites (23–25). This is indicative of the possibility of subtle levels of ART resistance in circulating parasites. As part of ongoing surveillance for molecular markers of ART resistance in Ghana, this study reports the diversity and the prevalence of both novel and known mutations in the pfk13 gene in Ghanaian P. falciparum isolates.
RESULTS
The information on the molecular markers of antimalarial drug resistance (a tool for monitoring drug resistance, which gives early warning of the development/emergence of drug resistance) is crucial for control strategies involving chemotherapy and chemoprophylaxis for residents and nonimmune travelers to countries where malaria is endemic. Therefore, polymorphisms in pfk13 of Ghanaian malaria parasites from three different ecological zones at several time periods were assessed. A total of 1,100 samples were sequenced for the pfk13 gene. Of these, 854 isolates with good sequences were used for the analysis. The majority of samples were from the 2015 to 2016 collection, because stored samples from prior years were limited due to usage in other studies. The samples contributed per year from sites in the three ecological zones are shown in Table 1.
TABLE 1.
Ecological zones and year of collection of archived samples at five time periods
| Ecological zones | Yr of collection |
|||||
|---|---|---|---|---|---|---|
| 2007–2008 | 2010–2011 | 2012 | 2013–14 | 2015–16 | Total | |
| Guinea savannah | 26 | 30 | 0 | 42 | 98 | 196 |
| Forest | 73 | 40 | 22 | 86 | 231 | 452 |
| Coastal savannah | 35 | 26 | 26 | 24 | 95 | 206 |
| Total | 134 | 96 | 48 | 152 | 424 | 854 |
Distribution of pfk13 wild type and mutants in the ecological zones.
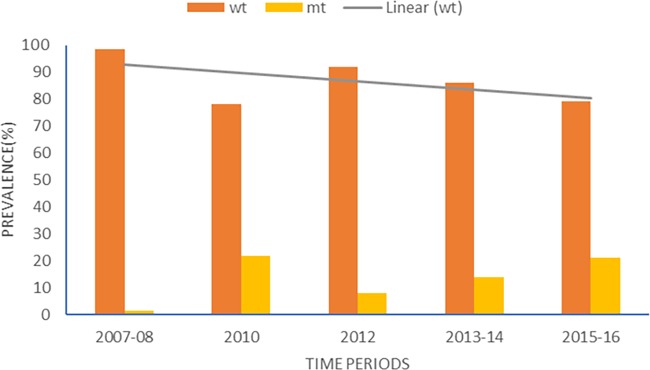
The majority of isolates 84% (716/854) had wild-type sequences like the 3D7 strain (PF3D7_1343700, PLASMODB). The mutants (16%) had SNPs clustered around codons 404 to 650 of the gene that were mostly nonsynonymous (NS) mutations (77.0%). Most of the mutations occurred only once in the samples. Multiple SNPs were observed in some of the samples, which were due to the multiple clones in Ghanaian malaria infections and was obvious from peaks shown in the DNA sequence chromatograms. Seventeen (12.4%) of the 138 samples with mutations had at least two mutations. For the ecological zones, the distributions of wild-type and mutant alleles in the isolates are shown in Fig. 1. Using the pooled data from Ghana, the proportions of wild-type alleles for the 5 time periods showed a significant declining trend (χ2 = 33.9, P = 0.001) over time. Figure 2 shows the distributions of wild-type and mutant alleles for Ghana over the five time periods.
FIG 1.
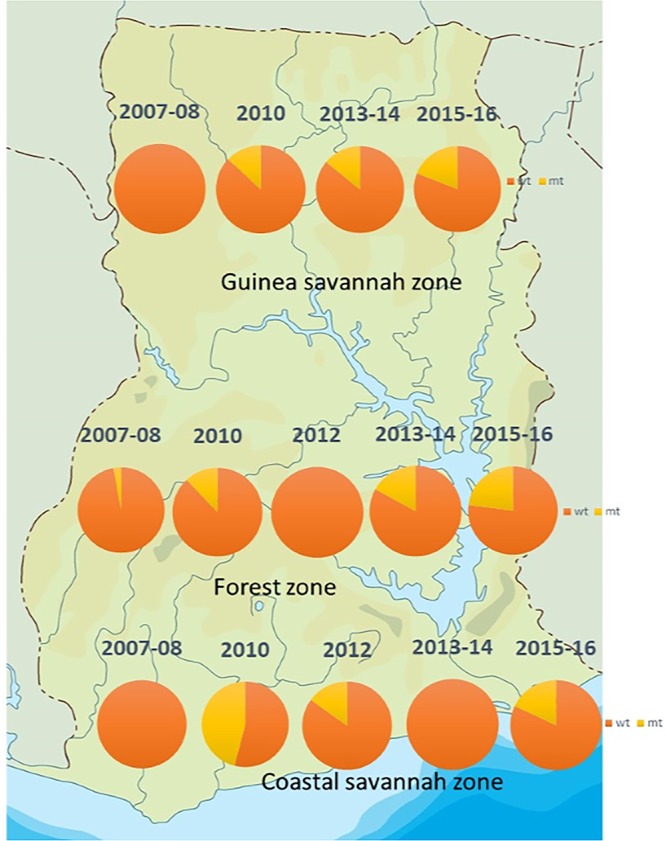
The distribution of pfk13 wild-type and mutant alleles in parasite populations from three ecological zones in Ghana from 2007 to 2016. The orange shade is the proportion for wild-type alleles (wt) and the yellow shade for mutant alleles (mt). (Ghana map adapted with permission from www.netmaps.net.)
FIG 2.
The distribution of wild-type and mutant alleles for the study time periods. The gray line depicts a decreasing trend of pfk13 wild-type alleles with time in Ghanaian parasite populations.
Novel pfk13 mutations in Ghanaian isolates.
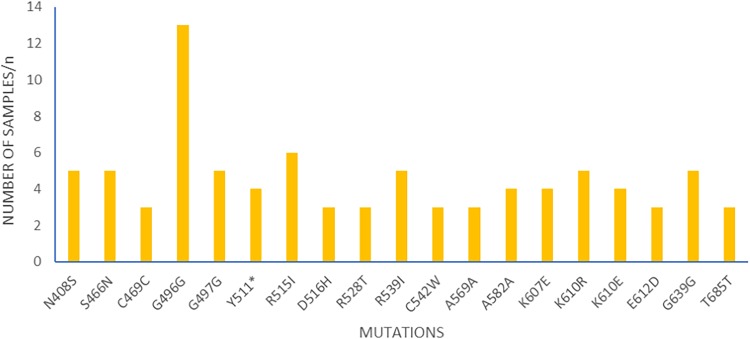
The mutations observed were diverse, and no particular mutation was exceptionally high in frequency in the population over the years. Likewise, no haplotypes were observed, and the majority of the mutations were observed once. Unique mutations with variants of both synonymous (SYN) and NS mutations were also observed. At codon 607 with amino acid lysine (K), variants of NS mutations, K607E (4), K607H (2), K607N (2), K607I (1), K607Q (1), and K607* (* indicates a stop codon) (1) were observed in 11 samples from 2010, 2012, 2013 to 2014, and 2015 to 2016. Another codon with two variants, G496G (13) and G496W (1), was seen in 14 samples from the forest zone in 2015 to 2016. In addition, K610 had three mutant variants, K610R (5), K610E (4), and K610K (1), in 10 samples from 2010, 2012, and 2016. Both SYN and NS mutations which occurred in more than two samples are shown in Fig. 3.
FIG 3.
The pfk13 synonymous and nonsynonymous mutations that occurred in more than two samples; n is the number of samples with the mutations.
Other mutations were observed in specific ecological zones. The mutations seen from all zones were L422F, N408S, N599Y, S466N, and Y511*. Those seen in samples from forest and Guinea savannah zones were K607E, R539I, C542W, and R515I, while A627S seen in Guinea and coastal savannah samples. Those seen only in the forest zone were E606D, G496G, and L407R. The frequencies of the novel mutations and the time periods for the ecological zones are shown in Table 2. There were unique mutations to specific sites: Wa, I461I (2 samples); Hohoe, R528T (3 samples) and D421N (2 samples); Cape Coast, Q613P (2 samples); Begoro, A578S (2 samples); Navrongo, I590T (2 samples); and Bekwai, D584D (2 samples). The persisting NS mutations observed at three time periods include N599Y (2010, 2012, and 2013 to 2014), K607E (2010, 2013 to 2014, and 2015 to 2016), and V637G (2010, 2013 to 2014, and 2015 to 2016), and the zones in which they were observed are shown in Table 2.
TABLE 2.
Frequencies of pfk13 mutations unique to or shared by the ecological zones at different time periods
| Mutation | Frequency (%) |
||||||||
|---|---|---|---|---|---|---|---|---|---|
| Guinea savannah |
Forest |
Coastal savannah |
|||||||
| 2010 (n = 30) | 2013–2014 (n = 42) | 2015–2016 (n = 98) | 2010 (n = 40) | 2013–2014 (n = 86) | 2015–2016 (n = 231) | 2010 (n = 26) | 2012 (n = 26) | 2015–2016 (n = 95) | |
| L407R | —a | — | — | — | 3.5 | — | — | — | — |
| N408S | — | — | 1.0 | — | — | 0.9 | — | — | 1.1 |
| L422F | — | — | 2.0 | — | — | 0.9 | — | — | |
| S466N | — | — | 1.0 | 2.5 | — | 0.9 | 3.8 | — | — |
| G496G | — | — | — | — | — | 5.6 | — | — | — |
| Y511* | — | — | 2.0 | — | — | 0.9 | — | — | — |
| R515I | — | — | 1.0 | — | — | 2.2 | — | — | — |
| R539I | — | — | 2.0 | 2.5 | — | 0.9 | — | — | — |
| C542W | 6.7 | — | — | — | — | 0.4 | — | — | — |
| N599Y | 3.3 | — | 1.0 | — | 1.2 | — | — | 3.8 | — |
| E606D | — | — | — | — | — | 0.9 | — | — | — |
| K607E | — | 2.4 | 1.0 | 2.5 | — | 0.4 | — | — | — |
| A627S | — | — | 1.0 | — | — | — | — | — | 1.1 |
| V637G | — | — | — | 2.5 | 1.2 | — | — | — | 1.1 |
—, no data available.
ART resistance validated and candidate pfk13 mutations in Ghanaian isolates.
The codons bearing the mutations linked to artemisinin resistance in the SEA region were observed with different amino acid substitutions in P. falciparum strains from Ghana. Some of the mutations were found at specific sentinel sites and time periods. They include C580V, detected in one sample from Cape Coast, R539I, detected in 5 isolates from Hohoe (1), Begoro (1), Sunyani (1), and Wa (2), variants of codon 543, I543I, I543S, and I543V, in single isolates from Navrongo, Sunyani, and Hohoe, respectively, R561P from Hohoe, and R561R from Sunyani. The M476I mutation (found in the Tanzanian ART resistant isolate, African descent) was seen in 1 sample from Wa, and a variant mutant, M476V, was detected in Begoro.
The effect of amino acid changes in proteins using the method by Brick and Pizzi (26), CCF53_62 matrices, indicates that the smaller the stabilization score, the more negative and drastic the change is on the proteins. Referring to the CCF53_62 matrices, the change from C to Y in C580Y gives a stabilization score of −2, and the change from C to V in C580V gives a stabilization score of −1. This implies that both amino acid changes have effects on the protein structure and function. However, C580Y will have a more drastic change due to its more negative stabilization score. The change from R to T in R539T gives a stabilization sore of −1, and the change from R to I in R539I observed in Ghanaian isolates gives a stabilization score of −3, which is much lower, implying that the change from R to I may have a graver effect on the protein structure and function. Similarly, the change from I to T in I543T gives a score of −1, whereas the change from I to S at the same codon is −2.
Previously reported African pfK13 mutations observed in Ghanaian isolates.
This study detected mutations that were previously described from African malaria parasites which are likely candidates for markers of drug resistance. The A578S seen in both SEA and African isolates was seen in 2 isolates from Begoro, while the additional variants A578P and A578A were seen in isolates from Wa and Sunyani, respectively. Variants of most of the reported mutations from African countries were observed except for C469C in 3 isolates from Begoro, Wa, and Navrongo, M608L and N629Y in samples from Cape Coast, and C532C from Begoro. The validated mutations as well as other candidate mutations for ART resistance in Africa reported in published articles and the variants observed in Ghanaian isolates are shown in Table 3.
TABLE 3.
Variants of ART resistance validated and candidate mutations in Ghanaian isolates
| Region | Mutation type | References | Previously reported mutations | Mutations in Ghanaian isolates (n)a | Study sitesb |
|---|---|---|---|---|---|
| Southeast Asia | Validated ART resistance mutations | 3, 8, 11 | N458Y | — | |
| M476I | M476I (1), M476V (1) | Wa, Begoro | |||
| Y493H | — | ||||
| R539T | R539I (5) | Begoro, Wa, Sunyani, Hohoe | |||
| I543T | I543I (1), I543S (1), I543V (1) | Navrongo, Sunyani, Hohoe | |||
| R561H | R561P (1), R561R (1) | Sunyani, Hohoe | |||
| C580Y | C580V (1) | Cape Coast | |||
| Sub-Saharan Africa | Shared mutations in African parasites | 10, 12–14, 16–18, 28, 29, 37–39 | C469C | C469C (3), C469R (1) | Wa, Cape Coast |
| S522C | S522R (1) | Wa | |||
| C532S | C532S (1) | Begoro | |||
| V534I, V534L | V534V (2) | Cape Coast, Sunyani | |||
| P553L | P553P (1) | Accra | |||
| A578S | A578S (2), A578A (1), A578P (1) | Begoro, Sunyani, Wa | |||
| N585K | N585H (1), N585N (1) | Hohoe, Navrongo | |||
| V589I | V589G (1) | Cape Coast | |||
| M608L | M608L (1) | Cape Coast | |||
| E612D | E612D (3) | Cape Coast, Sunyani | |||
| Q613L | Q613L (2), Q613P (1), Q613R (1) | Navrongo, Begoro, Cape Coast | |||
| N629Y | N629Y (1) | Cape Coast |
n is the number of isolates with the mutation. Mutations in boldface font are the same mutations as published. —, not detected.
Study sites in boldface font are where the published mutations were detected.
DISCUSSION
The pfk13 mutations linked to ART resistance in the SEA region are being used as a paradigm for ongoing surveillance to detect emerging malaria parasite resistance to ACTs in countries in sub-Saharan Africa (sSA) where malaria is endemic. With the use of ACTs in Ghana for the past 14 years, there is the growing concern that drug-resistant parasites are evolving and are under selective pressure. This study identified pfk13 polymorphisms in Ghanaian P. falciparum populations from uncomplicated malaria cases reporting at health centers over 9 years (2007 to 2016). Analysis of the pfk13 genetic diversity in parasites from across 3 ecological zones in Ghana uncovered both known and novel mutations, with the majority being nonsynonymous. Although none of the validated mutations for ART resistance were observed, variants with different amino acid substitutions of the codons conferring resistance were observed (R539I instead of T). In addition, some candidate mutations from African isolates were also observed but at low frequencies, such as A578S.
The observed diverse mutations in the Ghanaian isolates corroborate what was observed by Kamau and others in their baseline report of pfk13 polymorphisms in sub-Saharan Africa (12). Although the period of sample collection from the sites was not disclosed, it is noteworthy that the majority of mutations observed in that study were from Ghanaian parasites (Cape Coast and Navrongo). The paper also mentioned the polyclonality of infections observed in samples from Ghana and Kenya, which was apparent by the presence of minor peaks in the DNA sequence chromatograms (12). A similar observation as seen in Ghanaian infections was made in some of the samples analyzed in this study due to the multiplicity of infection (MOI). Depending on the region, the MOIs ranged from 1 to 4 genotypes per infection (27), as such, some samples had more than one pfk13 mutation. This observation is in contrast to the often seen monoclonal infections in the SEA region, where one mutation per sample was observed for the parasites (3). Ocan and others have also indicated that there is a higher diversity of pfk13 mutations in the African region than in the SEA region from a systematic review of the prevalence of the mutations in the two geographic malaria regions (28).
After 2 to 3 years of ACT implementation (2007 to 2008), the wild-type population for pfk13 was approximately 99%, and subsequently, there has been an overall significant decline in the ecological zones. The Guinea savannah (GS) and the coastal savannah (CS) zones had no mutants in the first time period (2007 to 2008), but the mutants were seen from the second time period onwards. This implies that there was an independent emergence of spontaneous mutations with time as observed in Senegal, where no mutations were detected in 2010 but started coming up in the subsequent years (29, 30). There was also an observation of no mutations in the 2012 forest and 2013 to 2014 CS samples, which may be due to the samples available for use (22 and 24, respectively). It is possible that most of the samples with mutants may have been missed due to usage in other studies. However, the overall analysis showed a decline the pfk13 wild-type population with time, suggesting the evolution of the pfk13 C-terminal region with the introduction of ACTs due to the increase in mutations over time. However, there are no prior data to ACT use to confirm this assertion. Three mutations were also observed at three consistent time periods nationally but were seen at 2 time periods for the zones. This is indicative of a possible selection of the mutations with drug pressure, but the frequencies decreased with time. These mutations will be tracked in the subsequent years to investigate the spread in the population.
The observation of mutations at the codons of the ART resistance molecular markers but with different amino acid substitutions is of great interest. The variants of the validated mutations seen include R539I, I543S, R561P, and C580V. The CCF53_62 matrix analysis by Brick and Pizzi (26) which determines the effect of amino acid changes in proteins using bioinformatics indicates that the smaller the stabilization score of the amino acid, the more negative and drastic the change in the protein. From the analysis, R539I, I543S, R561P, and C580V had lower stabilization scores than R539T, I543T, R561H, and C580Y, respectively, which implies an effect on the protein structure and function. The functional roles of the substituted amino acids need to be investigated to reveal any disruption or modification of the domain scaffold and the phenotypic effect on parasite artemisinin resistance. It is worth noting that since the molecular markers of CQ and SP for the SEA region and sSA regions were of different haplotypes, there is a likelihood of a similar scenario with pfk13 mutations.
Differences in the number and distribution of the polymorphisms from the different geographic locations depict the probable role of transmission dynamics as well as and human migratory patterns. High transmission intensity enhances genetic recombination in parasite populations and consequently spontaneous mutations that could be heritable. Although there were mutations that were shared among populations from the different ecological zones, others were zone specific. This implies that the possibility of intracountry gene flow as a result parasite transport with human migratory patterns cannot be overlooked, which has been observed in The Gambia (Alfred Amambua-Ngwa, personal communication). The observation of previously reported mutations in circulating African parasites corroborates an assertion that mutations from the African parasites probably independently evolved locally; however, the possibility of gene flow from the SEA region cannot be overlooked, although there is no evidence of that yet (10).
The data show that after 14 years of the introduction of ACT use in Ghana, there has been a steady increase in the variability of mutations in the pfk13 gene as well as the number of mutants from 2007 to 2016. The pfk13 polymorphisms observed were both SYN and NS mutations which include known mutations from SEA and Africa malaria regions, but in most cases, with a different in amino acid substitution. Novel mutations were also detected in Ghanaian isolates, and three of them were observed consistently at three time periods nationally. In the advent of the unknown markers for ACT resistance in Africa, the observed mutations will be tracked in isolates from ongoing surveillance of antimalarial drug resistance in Ghana for the possible validation of the resistance-conferring mutations in Africa.
MATERIALS AND METHODS
Study sites.
A collaboration between the National Malaria Control Program (NMCP) and the Noguchi Memorial Institute for Medical Research (NMIMR) established 10 sentinel sites in the ten regions of Ghana for the surveillance of antimalarial drug efficacy in the country (Fig. 4). The sites include Navrongo (10.8940°N, 1.0921°W), Wa (10.0601°N, 2.5099°W), and Yendi (9.4450°N, 0.0093°W) in the Guinea savannah ecological zone with seasonal malaria transmission, Begoro (6.3916°N, 0.3795°W), Bekwai (6.4532°N, 1.5838°W), Hohoe (7.1519°N, 0.4738°E), Sunyani (7.3349°N, 2.3123°W), and Tarkwa (5.3018°N, 1.9930°W) in the forest ecological zone with perennial malaria transmission, and Accra (5.6037°N, 0.1870°W) and Cape Coast (5.1315°N, 1.2795°W) in the coastal savannah zone with perennial malaria transmission.
FIG 4.
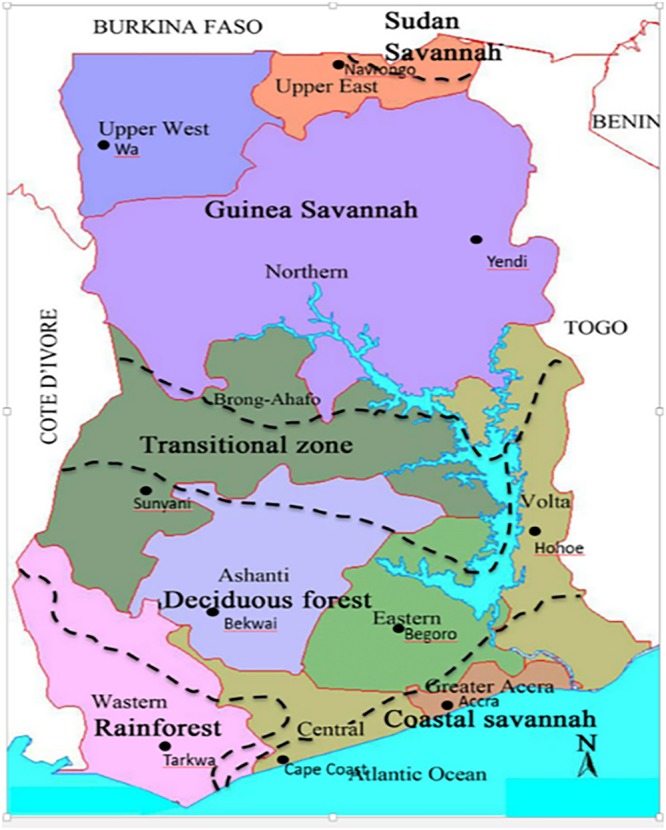
Map showing the 10 sentinel sites for surveillance of antimalarial drug resistance in Ghana. (Map is from https://binged.it/31uEjfO, and the URL of the original copyright holder [newafrica.com] is no longer active.)
Study samples.
Archived filter paper blood blots collected from uncomplicated malaria patients aged ≤9 years were used. The samples were originally collected during the wet transmission seasons of five time periods (2007 to 2008, 2010 to 2011, 2012, 2013 to 2014, and 2015 to 2016) as determined by the availability of funds. The information on sample collection over the years was previously reported (21, 22, 31–36). The samples were collected before (day 0) and after treatment (with AL or AA) on days 2, 3, 7, 14, 21, and 28 per protocol with microscopically and PCR-detected infection. For this work, only day 0 samples were used. The samples (100 μl blood) were collected on Whatman 3 filter paper (Little Chalfont, UK), stored in plastic bags containing silica gels, and kept at room temperature until use. Mixed infections were not looked at, because one of the criteria for recruitment in our treatment efficacy study is to have solely P. falciparum parasites not in combination with the other species.
Molecular analysis for the detection of pfk13 SNPs in isolates.
Malaria parasite DNA was extracted from 1,100 filter paper blood blots by using a QIAamp DNA minikit (Qiagen, Hilden, Germany) according to the manufacturer’s protocol. Nested PCR (nPCR) was performed to amplify the region of pfk13 gene using published protocols (9) with minimal modification. PCR products were sequenced using Sanger sequencing at Macrogen, Netherlands. For quality control purposes, 20 filter paper blood blots of known parasite sequences were sent to Venkatachalam Udhayakumar’s laboratory at the Malaria Centre, Centers for Disease Control and Prevention (CDC), Atlanta, GA, for analysis and confirmation of results.
pfk13 sequence analysis.
Sequences were analyzed with the BLAST program (http://blast.ncbi.nlm.nih.gov/) to determine the authenticity of the sequences. Multiple sequences were aligned with MAFFT (EMBL.EBI, Hinxton, Cambridge, UK) using the 3D7 wild-type pfk13 sequence (PF3D7_1343700) as a reference. Consensus sequence editing and single nucleotide polymorphism (SNP) detection was carried out using the CLC Main Workbench 7.9.1 (Qiagen, Aarhus, Denmark) and the Benchling website (San Francisco, CA, USA). Frequencies of mutations were determined for each sample by individual counts. The chi-square test was used to determine the prevalence trends of the wild types over time. The CCF53_62 matrix analysis by Brick and Pizzi (26) was used to determine the effect of amino acid changes in proteins.
Ethics statement.
The study protocol was approved by the NMIMR and Naval Medical Research Center’s Institutional Review Boards in compliance with all applicable federal regulations governing the protection of human subjects.
ACKNOWLEDGMENTS
The study was funded by the Armed Forces Health Surveillance Branch (AFHSB) and its Global Emerging Infections Surveillance and Response section (GEIS). The field work for the collection of the samples was partly funded by GEIS and the Global Fund to Fight AIDS, Tuberculosis and Malaria (GFATM)/National Malaria Control Programme (NMCP; Ghana). The funders played no role in the design of the study, sample collection, analysis, and interpretation of results as well as manuscript preparation and submission.
We thank Venkatachalam Udhayakumar and Naomi Lucchi of the Malaria Centre, CDC, Atlanta, GA, for the quality control analysis at their laboratory. We also thank the participants for the various studies under the Surveillance of Antimalarial Drug Resistance in Ghana. We declare no competing interests. The views expressed in this article are those of the authors and do not necessary reflect the official policy or position of the Department of the Navy, Department of Defense, nor the U.S. Government.
Andrew Letizia is a military Service member and Anne Fox is an employee of the U.S. government. This work was prepared as part of their official duties. Title 17, U.S.C., §105 provides that copyright protection under this title is not available for any work of the U.S. Government. Title 17, U.S.C., §101 defines a U.S. Government work as a work prepared by a military service member or employee of the U.S. Government as part of that person’s official duties.
REFERENCES
- 1.WHO. 2018. Malaria fact sheet 2018. http://www.who.int/news-room/fact-sheets/detail/malaria. Accessed 22 May 2018.
- 2.WHO. 2017. Artemisinin and artemisinin-based combination therapy resistance 2017. http://apps.who.int/iris/bitstream/10665/255213/1/WHO-HTM-GMP-2017.9-eng.pdf. Accessed 16 February 2018.
- 3.Ariey F, Witkowski B, Amaratunga C, Beghain J, Langlois A-C, Khim N, Kim S, Duru V, Bouchier C, Ma L, Lim P, Leang R, Duong S, Sreng S, Suon S, Chuor CM, Bout DM, Ménard S, Rogers WO, Genton B, Fandeur T, Miotto O, Ringwald P, Le Bras J, Berry A, Barale J-C, Fairhurst RM, Benoit-Vical F, Mercereau-Puijalon O, Ménard D. 2014. A molecular marker of artemisinin-resistant Plasmodium falciparum malaria. Nature 505:50–55. doi: 10.1038/nature12876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cheeseman IH, Miller BA, Nair S, Nkhoma S, Tan A, Tan JC, Al Saai S, Phyo AP, Moo CL, Lwin KM, McGready R, Ashley E, Imwong M, Stepniewska K, Yi P, Dondorp AM, Mayxay M, Newton PN, White NJ, Nosten F, Ferdig MT, Anderson TJ. 2012. A major genome region underlying artemisinin resistance in malaria. Science 336:79–82. doi: 10.1126/science.1215966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Anderson TJ, Nair S, Nkhoma S, Williams JT, Imwong M, Yi P, Socheat D, Das D, Chotivanich K, Day NP, White NJ, Dondorp AM. 2010. High heritability of malaria parasite clearance rate indicates a genetic basis for artemisinin resistance in western Cambodia. J Infect Dis 201:1326–1330. doi: 10.1086/651562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Phyo AP, Nkhoma S, Stepniewska K, Ashley EA, Nair S, McGready R, Ler Moo C, Al-Saai S, Dondorp AM, Lwin KM, Singhasivanon P, Day NP, White NJ, Anderson TJ, Nosten F. 2012. Emergence of artemisinin-resistant malaria on the western border of Thailand: a longitudinal study. Lancet 379:1960–1966. doi: 10.1016/S0140-6736(12)60484-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ashley EA, Dhorda M, Fairhurst RM, Amaratunga C, Lim P, Suon S, Sreng S, Anderson JM, Mao S, Sam B, Sopha C, Chuor CM, Nguon C, Sovannaroth S, Pukrittayakamee S, Jittamala P, Chotivanich K, Chutasmit K, Suchatsoonthorn C, Runcharoen R, Hien TT, Thuy-Nhien NT, Thanh NV, Phu NH, Htut Y, Han KT, Aye KH, Mokuolu OA, Olaosebikan RR, Folaranmi OO, Mayxay M, Khanthavong M, Hongvanthong B, Newton PN, Onyamboko MA, Fanello CI, Tshefu AK, Mishra N, Valecha N, Phyo AP, Nosten F, Yi P, Tripura R, Borrmann S, Bashraheil M, Peshu J, Faiz MA, Ghose A, Hossain MA, Samad R, et al. . 2014. Spread of artemisinin resistance in Plasmodium falciparum malaria. N Engl J Med 371:411–423. doi: 10.1056/NEJMoa1314981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Menard D, Khim N, Beghain J, Adegnika AA, Shafiul-Alam M, Amodu O, Rahim-Awab G, Barnadas C, Berry A, Boum Y, Bustos MD, Cao J, Chen JH, Collet L, Cui L, Thakur GD, Dieye A, Djallé D, Dorkenoo MA, Eboumbou-Moukoko CE, Espino FE, Fandeur T, Ferreira-da-Cruz MF, Fola AA, Fuehrer HP, Hassan AM, Herrera S, Hongvanthong B, Houzé S, Ibrahim ML, Jahirul-Karim M, Jiang L, Kano S, Ali-Khan W, Khanthavong M, Kremsner PG, Lacerda M, Leang R, Leelawong M, Li M, Lin K, Mazarati JB, Ménard S, Morlais I, Muhindo-Mavoko H, Musset L, Na-Bangchang K, Nambozi M, Niaré K, Noedl H, et al. . 2016. A worldwide map of Plasmodium falciparum K13-propeller polymorphisms. N Engl J Med 374:2453–2464. doi: 10.1056/NEJMoa1513137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Talundzic E, Okoth SA, Congpuong K, Plucinski MM, Morton L, Goldman IF, Kachur PS, Wongsrichanalai C, Satimai W, Barnwell JW, Udhayakumar V. 2015. Selection and spread of artemisinin-resistant alleles in Thailand prior to the global artemisinin resistance containment campaign. PLoS Pathog 11:e1004789. doi: 10.1371/journal.ppat.1004789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.MalariaGen Plasmodium falciparum Community Project. 2016. Genomic epidemiology of artemisinin resistant malaria. Elife 5:e08714. doi: 10.7554/eLife.08714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Miotto O, Amato R, Ashley EA, MacInnis B, Almagro-Garcia J, Amaratunga C, Lim P, Mead D, Oyola SO, Dhorda M, Imwong M, Woodrow C, Manske M, Stalker J, Drury E, Campino S, Amenga-Etego L, Thanh TN, Tran HT, Ringwald P, Bethell D, Nosten F, Phyo AP, Pukrittayakamee S, Chotivanich K, Chuor CM, Nguon C, Suon S, Sreng S, Newton PN, Mayxay M, Khanthavong M, Hongvanthong B, Htut Y, Han KT, Kyaw MP, Faiz MA, Fanello CI, Onyamboko M, Mokuolu OA, Jacob CG, Takala-Harrison S, Plowe CV, Day NP, Dondorp AM, Spencer CC, McVean G, Fairhurst RM, White NJ, Kwiatkowski DP. 2015. Genetic architecture of artemisinin-resistant Plasmodium falciparum. Nat Genet 47:226–234. doi: 10.1038/ng.3189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kamau E, Campino S, Amenga-Etego L, Drury E, Ishengoma D, Johnson K, Mumba D, Kekre M, Yavo W, Mead D, Bouyou-Akotet M, Apinjoh T, Golassa L, Randrianarivelojosia M, Andagalu B, Maiga-Ascofare O, Amambua-Ngwa A, Tindana P, Ghansah A, MacInnis B, Kwiatkowski D, Djimde AA. 2015. K13-propeller polymorphisms in Plasmodium falciparum parasites from sub-Saharan Africa. J Infect Dis 211:1352–1355. doi: 10.1093/infdis/jiu608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ouattara A, Kone A, Adams M, Fofana B, Maiga AW, Hampton S, Coulibaly D, Thera MA, Diallo N, Dara A, Sagara I, Gil JP, Bjorkman A, Takala-Harrison S, Doumbo OK, Plowe CV, Djimde AA. 2015. Polymorphisms in the K13-propeller gene in artemisinin-susceptible Plasmodium falciparum parasites from Bougoula-Hameau and Bandiagara, Mali. Am J Trop Med Hyg 92:1202–1206. doi: 10.4269/ajtmh.14-0605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Taylor SM, Parobek CM, DeConti DK, Kayentao K, Coulibaly SO, Greenwood BM, Tagbor H, Williams J, Bojang K, Njie F, Desai M, Kariuki S, Gutman J, Mathanga DP, Martensson A, Ngasala B, Conrad MD, Rosenthal PJ, Tshefu AK, Moormann AM, Vulule JM, Doumbo OK, Ter Kuile FO, Meshnick SR, Bailey JA, Juliano JJ. 2015. Absence of putative artemisinin resistance mutations among Plasmodium falciparum in Sub-Saharan Africa: a molecular epidemiologic study. J Infect Dis 211:680–688. doi: 10.1093/infdis/jiu467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Conrad MD, Bigira V, Kapisi J, Muhindo M, Kamya MR, Havlir DV, Dorsey G, Rosenthal PJ. 2014. Polymorphisms in K13 and falcipain-2 associated with artemisinin resistance are not prevalent in Plasmodium falciparum isolated from Ugandan children. PLoS One 9:e105690. doi: 10.1371/journal.pone.0105690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Isozumi R, Uemura H, Kimata I, Ichinose Y, Logedi J, Omar AH, Kaneko A. 2015. Novel mutations in K13 propeller gene of artemisinin-resistant Plasmodium falciparum. Emerg Infect Dis 21:490–492. doi: 10.3201/eid2103.140898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.de Laurent ZR, Chebon LJ, Ingasia LA, Akala HM, Andagalu B, Ochola-Oyier LI, Kamau E. 2018. Polymorphisms in the K13 gene in Plasmodium falciparum from different malaria transmission areas of Kenya. Am J Trop Med Hyg 98:1360–1366. doi: 10.4269/ajtmh.17-0505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ishengoma DS, Mandara CI, Francis F, Talundzic E, Lucchi NW, Ngasala B, Kabanywanyi AM, Mahende MK, Kamugisha E, Kavishe RA, Muro F, Mohamed A, Mandike R, Mkude S, Chacky F, Paxton L, Greer G, Kitojo CA, Njau R, Martin T, Venkatesan M, Warsame M, Halsey ES, Udhayakumar V. 2019. Efficacy and safety of artemether-lumefantrine for the treatment of uncomplicated malaria and prevalence of Pfk13 and Pfmdr1 polymorphisms after a decade of using artemisinin-based combination therapy in mainland Tanzania. Malar J 18:88. doi: 10.1186/s12936-019-2730-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Koram KA, Abuaku B, Duah N, Quashie N. 2005. Comparative efficacy of antimalarial drugs including ACTs in the treatment of uncomplicated malaria among children under 5 years in Ghana. Acta Trop 95:194–203. doi: 10.1016/j.actatropica.2005.06.018. [DOI] [PubMed] [Google Scholar]
- 20.MOH. 2009. Anti-malaria drug policy for Ghana. Ministry of Health, Ghana. [Google Scholar]
- 21.Abuaku B, Duah NO, Quaye L, Matrevi S, Quashie N, Gyasi A, Owusu-Antwi F, Malm K, Koram K. 2019. Therapeutic efficacy of artesunate-amodiaquine and artemether-lumefantrine combinations for uncomplicated malaria in 10 sentinel sites across Ghana: 2015–2017. Malar J 18:206. doi: 10.1186/s12936-019-2848-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Duah NO, Matrevi SA, de Souza DK, Binnah DD, Tamakloe MM, Opoku VS, Onwona CO, Narh CA, Quashie NB, Abuaku B, Duplessis C, Kronmann KC, Koram KA. 2013. Increased pfmdr1 gene copy number and the decline in pfcrt and pfmdr1 resistance alleles in Ghanaian Plasmodium falciparum isolates after the change of anti-malarial drug treatment policy. Malar J 12:377. doi: 10.1186/1475-2875-12-377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sutherland CJ, Lansdell P, Sanders M, Muwanguzi J, van Schalkwyk DA, Kaur H, Nolder D, Tucker J, Bennett HM, Otto TD, Berriman M, Patel TA, Lynn R, Gkrania-Klotsas E, Chiodini PL. 2017. pfk13-independent treatment failure in four imported cases of Plasmodium falciparum malaria treated with artemether-lumefantrine in the United Kingdom. Antimicrob Agents Chemother 61:e02382-16. doi: 10.1128/AAC.02382-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Henriques G, Hallett RL, Beshir KB, Gadalla NB, Johnson RE, Burrow R, van Schalkwyk DA, Sawa P, Omar SA, Clark TG, Bousema T, Sutherland CJ. 2014. Directional selection at the pfmdr1, pfcrt, pfubp1, and pfap2mu loci of Plasmodium falciparum in Kenyan children treated with ACT. J Infect Dis 210:2001–2008. doi: 10.1093/infdis/jiu358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Adams T, Ennuson NAA, Quashie NB, Futagbi G, Matrevi S, Hagan OCK, Abuaku B, Koram KA, Duah NO. 2018. Prevalence of Plasmodium falciparum delayed clearance associated polymorphisms in adaptor protein complex 2 mu subunit (pfap2mu) and ubiquitin specific protease 1 (pfubp1) genes in Ghanaiaian isolates. Parasit Vectors 11:175. doi: 10.1186/s13071-018-2762-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Brick K, Pizzi E. 2008. A novel series of compositionally biased substitution matrices for comparing Plasmodium proteins. BMC Bioinformatics 9:236. doi: 10.1186/1471-2105-9-236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Duah NO, Matrevi SA, Quashie NB, Abuaku B, Koram KA. 2016. Genetic diversity of Plasmodium falciparum isolates from uncomplicated malaria cases in Ghana over a decade. Parasit Vectors 9:416. doi: 10.1186/s13071-016-1692-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ocan M, Akena D, Nsobya S, Kamya MR, Senono R, Kinengyere AA, Obuku E. 2019. K13-propeller gene polymorphisms in Plasmodium falciparum parasite population in malaria affected countries: a systematic review of prevalence and risk factors. Malar J 18:60. doi: 10.1186/s12936-019-2701-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Boussaroque A, Fall B, Madamet M, Camara C, Benoit N, Fall M, Nakoulima A, Dionne P, Fall KB, Diatta B, Dieme Y, Wade B, Pradines B. 2016. Emergence of mutations in the K13 propeller gene of Plasmodium falciparum isolates from Dakar, Senegal, in 2013–2014. Antimicrob Agents Chemother 60:624–627. doi: 10.1128/AAC.01346-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Torrentino-Madamet M, Fall B, Benoit N, Camara C, Amalvict R, Fall M, Dionne P, Ba Fall K, Nakoulima A, Diatta B, Diemé Y, Ménard D, Wade B, Pradines B. 2014. Limited polymorphisms in k13 gene in Plasmodium falciparum isolates from Dakar, Senegal in 2012–2013. Malar J 13:472. doi: 10.1186/1475-2875-13-472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Abuaku B, Duah N, Quaye L, Quashie N, Malm K, Bart-Plange C, Koram K. 2016. Therapeutic efficacy of artesunate-amodiaquine and artemether-lumefantrine combinations in the treatment of uncomplicated malaria in two ecological zones in Ghana. Malar J 15:6. doi: 10.1186/s12936-015-1080-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Abuaku B, Duah N, Quaye L, Quashie N, Koram K. 2012. Therapeutic efficacy of artemether-lumefantrine combination in the treatment of uncomplicated malaria among children under five years of age in three ecological zones in Ghana. Malar J 11:388. doi: 10.1186/1475-2875-11-388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Duah NO, Quashie NB, Abuaku BK, Sebeny PJ, Kronmann KC, Koram KA. 2012. Surveillance of molecular markers of Plasmodium falciparum resistance to sulphadoxine-pyrimethamine 5 years after the change of malaria treatment policy in Ghana. Am J Trop Med Hyg 87:996–1003. doi: 10.4269/ajtmh.2012.12-0202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Koram K, Quaye L, Abuaku B. 2008. Efficacy of amodiaquine/artesunate combination therapy for uncomplicated malaria in children under five years in Ghana. Ghana Med J 42:55–60. [PMC free article] [PubMed] [Google Scholar]
- 35.Quashie NB, Duah NO, Abuaku B, Koram KA. 2007. The in-vitro susceptibilities of Ghanaian Plasmodium falciparum to antimalarial drugs. Ann Trop Med Parasitol 101:391–398. doi: 10.1179/136485907X176553. [DOI] [PubMed] [Google Scholar]
- 36.Quashie NB, Duah NO, Abuaku B, Quaye L, Ayanful-Torgby R, Akwoviah GA, Kweku M, Johnson JD, Lucchi NW, Udhayakumar V, Duplessis C, Kronmann KC, Koram KA. 2013. A SYBR green 1-based in vitro test of susceptibility of Ghanaian Plasmodium falciparum clinical isolates to a panel of anti-malarial drugs. Malar J 12:450. doi: 10.1186/1475-2875-12-450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Heuchert A, Abduselam N, Zeynudin A, Eshetu T, Loscher T, Wieser A, Pritsch M, Berens-Riha N. 2015. Molecular markers of anti-malarial drug resistance in southwest Ethiopia over time: regional surveillance from 2006 to 2013. Malar J 14:208. doi: 10.1186/s12936-015-0723-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ocan M, Bwanga F, Okeng A, Katabazi F, Kigozi E, Kyobe S, Ogwal-Okeng J, Obua C. 2016. Prevalence of K13-propeller gene polymorphisms among Plasmodium falciparum parasites isolated from adult symptomatic patients in northern Uganda. BMC Infect Dis 16:428. doi: 10.1186/s12879-016-1777-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dama S, Niangaly H, Ouattara A, Sagara I, Sissoko S, Traore OB, Bamadio A, Dara N, Djimde M, Alhousseini ML, Goita S, Maiga H, Dara A, Doumbo OK, Djimde AA. 2017. Reduced ex vivo susceptibility of Plasmodium falciparum after oral artemether-lumefantrine treatment in Mali. Malar J 16:59. doi: 10.1186/s12936-017-1700-8. [DOI] [PMC free article] [PubMed] [Google Scholar]