In South America, Plasmodium vivax resistance to chloroquine was recently reported in Brazil and Bolivia. The objective of this study was to collect data on chloroquine resistance in French Guiana by associating a retrospective evaluation of therapeutic efficacy with an analysis of recurrent parasitemia from any patients. Patients with P. vivax infection, confirmed by microscopy and a body temperature of ≥37.5°C, were retrospectively identified at Cayenne Hospital between 2009 and 2015.
KEYWORDS: P. vivax, resistance, Amazonia, French Guiana, Guiana Shield, chloroquine, pvcrt-o, pvmdr1
ABSTRACT
In South America, Plasmodium vivax resistance to chloroquine was recently reported in Brazil and Bolivia. The objective of this study was to collect data on chloroquine resistance in French Guiana by associating a retrospective evaluation of therapeutic efficacy with an analysis of recurrent parasitemia from any patients. Patients with P. vivax infection, confirmed by microscopy and a body temperature of ≥37.5°C, were retrospectively identified at Cayenne Hospital between 2009 and 2015. Follow-up and treatment responses were performed according to the World Health Organization protocol. Parasite resistance was confirmed after dosage of a plasma concentration of chloroquine and microsatellite characterization. The pvmdr1 and pvcrt-o genes were analyzed for sequence and gene copy number variation. Among the 172 patients followed for 28 days, 164 presented adequate clinical and parasitological responses. Eight cases of treatment failures were identified (4.7%; n = 8/172), all after 14 days. The therapeutic efficacy of chloroquine was estimated at 95.3% (95% confidence interval [CI], 92.5 to 98.1%; n = 164/172). Among the eight failures, five were characterized: two cases were true P. vivax chloroquine resistance (1.2%; 95% CI, 0 to 2.6%; n = 2/172), and three cases were found with subtherapeutic concentrations of chloroquine. No particular polymorphism in the Plasmodium vivax pvmdr1 and pvcrt-o genes was identified in the resistant parasites. This identified level of resistance of P. vivax to chloroquine in French Guiana does not require a change in therapeutic recommendations. However, primaquine should be administered more frequently to limit the spread of resistance, and there is still a need for a reliable molecular marker to facilitate the monitoring of P. vivax resistance to chloroquine.
INTRODUCTION
In 2017, malaria was still the most prevalent parasitic disease in the world, with 1.4 billion people remaining at risk (1). Representing 40% of malaria cases worldwide, Plasmodium vivax was the second species most responsible for human malaria after Plasmodium falciparum and is the most frequent species outside Africa. The same year in French Guiana, an overseas French territory located on the Guiana Shield in South America, 86% of malaria cases were due to P. vivax, and the remaining were due to P. falciparum, with scarce reports of Plasmodium malariae cases. In this region, the incidence of P. vivax exceeded that of P. falciparum in 2005 (2, 3). Meanwhile the overall number of notified malaria cases decreased from 4,000 in 2009 to 597 in 2017 (4).
Since 1995 in French Guiana, chloroquine (CQ) has no longer been recommended for treatment of P. falciparum and has been replaced by quinine-doxycycline before artemether-lumefantrine (3). However, it is the standard treatment for uncomplicated P. vivax infection. Its posology follows the World Health Organization (WHO) recommendation: an oral dose of 25 mg/kg of body weight distributed over 3 days and 14 days of 30-mg/day primaquine (PQ) to cure dormant hypnozoites (3, 5). Unfortunately, PQ is not systematically administered, mainly because of administrative constraints and difficulties in assessing the glucose-6-phosphate dehydrogenase (G6PD) activity of P. vivax malaria patients in remote areas (3). Without appropriate treatment, these dormant liver forms can cause relapses and participate in transmission (6). As relapses may be caused by the homologous or heterologous genotype, the genetic characterization of parasites is not very useful to characterize failures (7), thus limiting the study of antimalarial drug effectiveness against P. vivax.
P. vivax multiplication should not occur within 35 days after an adequate CQ treatment. During this time, the mean whole-blood concentrations of CQ and its metabolite desethylchloroquine (dCQ) are normally greater than 100 ng/ml and prevent parasite multiplication. CQ resistance (CQR) is suspected if parasitemia increases during this period (8). Therefore, P. vivax resistance could be identified using plasma concentrations (9). In vitro phenotyping methods are scarce and difficult to implement especially because of the very low synchronicity of parasites belonging to the Chesson South American strain (10). Putative molecular markers of CQR have been identified by homology with those from P. falciparum. Positions 976 and 1076 of the P. vivax multidrug resistance 1 gene (pvmdr1) have been associated with resistance without clear evidence (11, 12). pvmdr1 is considered only a minor determinant for resistance to chloroquine, eventually considered more informative for resistance to mefloquine (13, 14). pvcrt-o, the ortholog gene of pfcrt, the P. falciparum molecular marker for resistance to several antimalarial drugs, has also been described as a putative marker for resistance of P. vivax to chloroquine (15, 16). Gene duplication and the expression level of these genes have also been described as genetic determinants (17).
The first descriptions of well-documented P. vivax resistance in the Americas came from the Republic of Guyana, also part of the Guiana Shield (18). More recently in Oiapoque, Brazil, on the border with French Guiana, 1.1% (n = 1/95) of treatment failures (TFs [i.e., recurrent parasites after treatment]) were reported after supervised treatment with the combination CQ+PQ (19). Manaus, Amazonas, Brazil, is nowadays the hot spot of P. vivax resistance in South America, with 10.1% of TFs (n = 11/109) reported after supervised CQ treatment (20) or 5.2% (n = 7/135) after concomitant administration of CQ+PQ (21).
The present study’s main objective was to bring out new data on P. vivax CQR in French Guiana. This combined study associated the results from a follow-up therapeutic efficacy study of CQ implemented in clinical practices at the Cayenne Hospital with a retrospective analysis of recurrence in patients presenting fever and positive parasitemia within 35 days after the initial infection.
RESULTS AND DISCUSSION
In Cayenne Hospital, the patients infected by P. vivax are mostly young men.
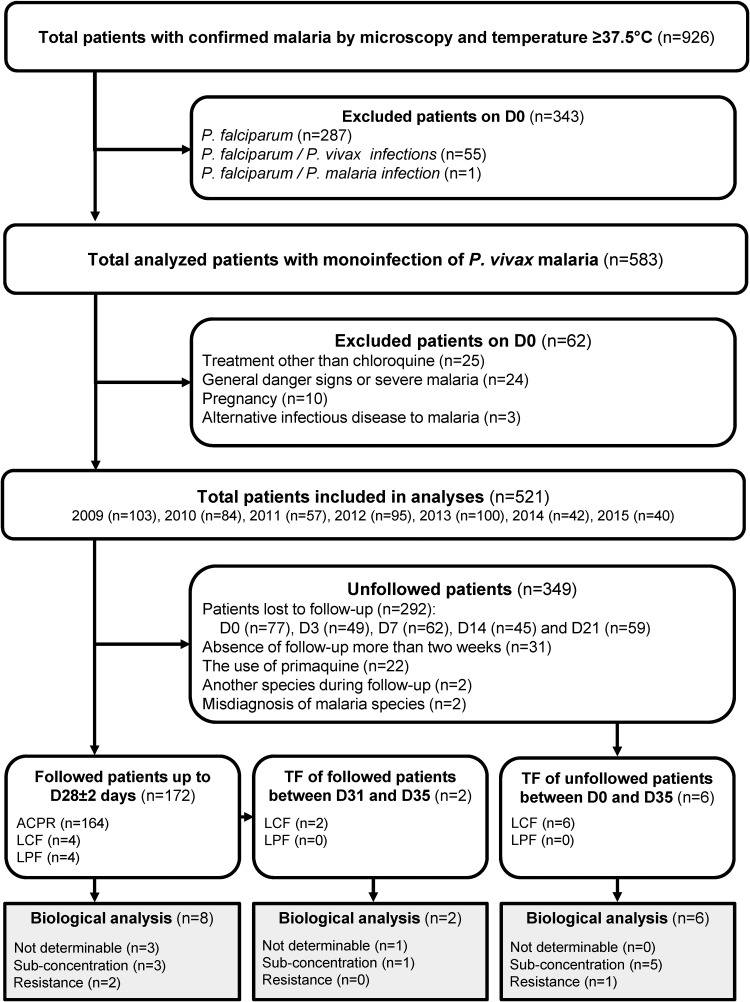
Between 2009 and 2015, 926 patients were screened and diagnosed positive for malaria (Fig. 1). During the clinical examination of the 583 patients with P. vivax infections, the median body temperature was 39.9°C (range, 37.5 to 41.5°C). The median parasitemia was 0.57% (range, 0.01 to 2.00%). During this period, young men were mostly infected by P. vivax (male/female [M/F] sex ratio, 2.05; median age, 31 years [range, 1 to 76 years]). Patients were diagnosed a median of 3 days after their first symptoms. No patient was underweight, and 22.5% (n = 131) of them declared vomiting.
FIG 1.
Malaria infections diagnosed at Cayenne Hospital, French Guiana, 2009 to 2015. ACPR, adequate clinical and parasitological response; D0 to D35, days 0 to 35; LCF, late clinical response; LPF, late parasitological response.
Therapeutic efficacy evaluated in one-third of the patients based on the standard hospital protocol for malaria care.
Of the 583 enrolled patients with P. vivax infections, 62 were excluded from the analysis on day 0 (D0) (Fig. 1). Of the 521 remaining patients, 172 patients were followed up to D28, and 292 were lost during follow-up. Fifty-seven patients were excluded during the study (Fig. 1). Among those 172 patients, men were more affected by malaria than women (M/F sex ratio, 2.37) (Table 1). The median age was 32 years (range, 5 to 76 years). Only 13.4% (n = 18) of these 172 patients took a chemoprophylaxis treatment: mainly (94.4%) members of the military using doxycycline. A history of malaria within the last 3 months was reported for 37.6% (n = 56/149) of patients. Within the same period, a history of travelling out of Cayenne was recorded for the majority of patients, of which 71.2% (n = 104/146) declared they had traveled within French Guiana, while 28.8% (n = 42/146) reported not having traveled. Cayenne Hospital is located outside the malaria transmission areas in French Guiana. However, it concentrates people from all over French Guiana, including people contaminated in gold mines (at least n = 32 [data not shown]), such as gold miners or members of the military dedicated to fighting illegal gold mining activities. Thus, the analyzed sample set of this study could be considered representative of the general parasite population circulating in the country.
TABLE 1.
Patient characteristics of uncomplicated P. vivax malaria treated by 3 days of chloroquine administration in French Guiana from 2009 to 2015a
| Parameter | Result for: |
P | ||
|---|---|---|---|---|
| Total | ACPR on D28 |
Recurrent parasitemia on D28 |
||
| Patients, no. (%) | ||||
| Total | 172 | 164 | 8 | |
| By yr | 0.554 | |||
| 2009 | 38 (22.1) | 37 (97.4) | 1 (2.6) | |
| 2010 | 37 (21.5) | 35 (94.6) | 2 (5.4) | |
| 2011 | 24 (14.0) | 24 (100.0) | 0 (0.0) | |
| 2012 | 44 (25.6) | 41 (93.2) | 3 (6.8) | |
| 2013 | 11 (6.4) | 10 (90.9) | 1 (9.1) | |
| 2014 | 8 (4.6) | 7 (87.5) | 1 (12.5) | |
| 2015 | 10 (5.8) | 10 (100.0) | 0 (0.0) | |
| Gender, no. (%) | 0.768 | |||
| M/F ratio | 2.37 | 2.35 | 3.00 | |
| Male | 121 (70.4) | 115 (95.0) | 6 (5.0) | |
| Female | 51 (29.7) | 49 (96.1) | 2 (3.9) | |
| Age, median yr (range) | 33.00 (5–76) | 33.00 (5–76) | 30.50 (17–56) | 0.134 |
| Age group, no. (%) | 0.496 | |||
| Adults | 163 (94.8) | 155 (95.1) | 8 (4.9) | |
| 5–15 yr old | 9 (5.2) | 9 (100.0) | 0 (0.0) | |
| History of malaria, no. (%) | <0.001* | |||
| Yes | 56 (32.5) | 55 (98.2) | 1 (1.8) | |
| No | 93 (54.1) | 91 (97.8) | 2 (2.2) | |
| Unknown | 23 (13.4) | 18 (78.3) | 5 (21.7) | |
| Prophylaxis, no. (%) | 0.121 | |||
| Yes | 18 (10.5) | 18 (100.0) | 0 (0.0) | |
| No | 116 (67.4) | 112 (96.6) | 4 (3.4) | |
| Unknown | 38 (22.1) | 34 (89.5) | 4 (10.5) | |
| Days before consultation, median no. (range) |
3 (0–15) | 3 (0–15) | 1 (0–7) | 0.428 |
| Body wt, median kg (range) |
72 (49–158) | 72 (49–158) | 68 (58–83) | 0.493 |
| Body temp, median °C (range) |
39.0 (37.7–40.9) | 39.0 (37.7–40.9) | 38.9 (37.5–39.6) | 0.672 |
| Vomiting, no. (%) | 0.671 | |||
| Yes | 59 (34.3) | 57 (96.6) | 2 (3.4) | |
| No | 106 (61.6) | 100 (94.3) | 6 (5.7) | |
| Unknown | 7 (4.1) | 7 (100.0) | 0 (0.0) | |
| Parasitemia, median % infected blood cells (range) |
0.15 (0.01–2.00) | 0.15 (0.01–2.00) | 0.13 (0.02–0.76) | 0.218 |
| Hospitalization, no. (%) | 0.946 | |||
| Yes | 23 (13.4) | 22 (95.7) | 1 (4.3) | |
| No | 98 (57.0) | 93 (94.9) | 5 (5.1) | |
| Unknown | 51 (29.6) | 49 (96.1) | 2 (3.9) | |
ACPR, adequate clinical and parasitological response; D28, day 28. *, P < 0.05 (significant difference).
High but incomplete (95.3%) CQ therapeutic efficacy against P. vivax in French Guiana between 2009 and 2015.
In French Guiana, between 2009 and 2015, the D28 follow-up estimated a therapeutic efficacy of CQ at 95.3% (95% confidence interval [CI], 92.5 to 98.1; n = 164/172) to treat uncomplicated P. vivax monoinfection. This study included a large number of patients, compared to the WHO recommendation (n = 172 versus 73). Therefore, these results were associated with a confidence level of 95% and a margin error of 2.8%. A cross-analysis was done within samples received at the National Reference Center (around 50% of the total number declared each year in the country) to track potential recurrent parasitemia in patients enrolled in the protocol but followed outside the Cayenne Hospital. This allowed us to identify one additional recurrent parasitemia. This confirms the robustness of the results presented from this Cayenne Hospital follow-up. However, recurrent cases could be underestimated in miners as they rarely complete the follow-up regardless of the medical recommendations so they can rapidly travel back into the deep forest (22).
With an endpoint at D28, eight patients experienced recurrent parasitemia (4 with late clinical failure [LCF] and 4 with late parasitological failure [LPF])—all after D14. No difference between years of infection was observed (P = 0.5542 [Table 1]). Therapeutic efficacy was stable for a 7-year period, suggesting that drug pressure on the parasite population did not participate in a rapid spread of resistance through the parasite population. The only significant difference between the adequate clinical and parasitological response (ACPR) group and the group experiencing recurrent parasitemia was history of malaria (P = 0.0022). Therefore, in the absence of systematic PQ prescription, this observation could suggest a large part of recurrence was due to relapses.
P. vivax resistance occurs at the minimum prevalence of 1.2%.
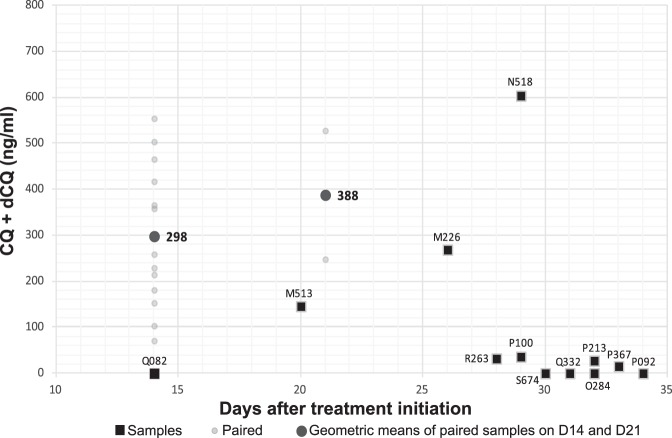
Two out of five analyzable failures on the D28 follow-up had a drug level that normally kills or at least suppresses parasite multiplication (M513, 146 ng/ml on D20; N518, 604 ng/ml on D29 [Table 2]). After comparison of plasma dosages of CQ with those of samples associated with ACPR around the same day of follow-up, these concentrations were compatible with an efficient antimalarial activity on sensitive parasites (Fig. 2). These results demonstrated that parasite resistance to CQ was present in at least 1.2% (95% CI, 0 to 2.6%; n = 2/172) of the patients in French Guiana. The six microsatellite markers showed homologous genetic background of parasites at D0 and the day of treatment failure in these cases (DF).
TABLE 2.
Characterization of P. vivax samples associated with a recurrent parasitemia after chloroquine treatment, French Guiana, 2009 to 2015a
| ID | BT (°C) | Parasitemia (%) | Treatment | Treatment response on D28 |
CQ+dCQ (ng/ml) |
Pvmdr1
copy no. |
Pvmdr1
sequence |
Pvcrt-o
copy no. |
Insertion of pvcrt-o K10 |
Sizes of microsatellite loci 13.239, 3.27, 5.504, 11.162, MS9, and MS8f |
|---|---|---|---|---|---|---|---|---|---|---|
| Recurrence in patients followed D28 ± 2 days | ||||||||||
| Not determinable reasonsb | ||||||||||
| P063 | ||||||||||
| D0 | 39.0 | 0.1000 | CQ | ND | A/WT | 0.94 ± 0.19 | Yes | 192–196, 109–117, 206–213–272, 182–186, 112–120, 199–207–211 | ||
| D22 | 36.1 | 0.1000 | No | LPF | NR | ND | A | ND | ND | 192–196, 109, ND, 182–186, 112–120, 199–207 |
| N183 | ||||||||||
| D0 | 39.0 | 0.1800 | CQ | NR | NR | NR | NR | NR | ||
| D30 | 38.7 | 0.3000 | CQ | LCF | NR | NR | NR | NR | NR | NR |
| NR | ||||||||||
| D0 | 38.9 | 0.1000 | CQ | NR | NR | NR | NR | NR | ||
| D22 | 39.0 | 1.2000 | CQ | LCF | NR | NR | NR | NR | NR | NR |
| Subtherapeutic chloroquine concentrations observedc | ||||||||||
| P100 | ||||||||||
| D0 | 39.6 | 0.1600 | CQ | 2 | A | 0.76 ± 0.02 | No | 192, 133, 199, 262–266–270, 116, 203 | ||
| D29 | 36.8 | 0.0010 | No | LPF | 36 + <10 | 2 | A | 0.61 ± 0.01 | No | Identical to D0 |
| Q082 | ||||||||||
| D0 | 38.1 | 0.0300 | CQ | 1 | A | 0.79 ± 0.05 | Yes | 191–196, 109, 59, 182, 112–116–120, 207 | ||
| D14 | 37.0 | 0.0300 | No | LPF | <10 + <10 | 1 | A | 0.73 ± 0.01 | Yes | Identical to D0 |
| R263 | ||||||||||
| D0 | 36.0 | 0.0020 | CQ | 1 | A | 0.69 ± 0.07 | Yes | 192, 109, 213, 186, 116-120-124, 199 | ||
| D28 | 39.0 | 0.1400 | CQ | LCF | 17 + 15 | 1 | A | 0.97 ± 0.01 | Yes | Identical to D0 |
| Chloroquine therapeutic failure associated with parasite resistanced | ||||||||||
| M513 | ||||||||||
| D0 | 38.9 | 0.2500 | CQ | 1 | A | 1.01 ± 0.08 | Yes | 200, 129, 213, 186, 108, 207 | ||
| D2 | 36.7 | 0.0005 | ||||||||
| D6 | 36.5 | 0.0000 | ||||||||
| D13 | 37.6 | 0.0000 | ||||||||
| D20 | 36.5 | 0.0000 | 93 + 53 | |||||||
| D26 | 36.7 | 0.0005 | No | LPF | NR | 1 | A | ND | ND | Identical to D0 |
| N518 | ||||||||||
| D0 | 38.8 | 0.7600 | CQ | 1 | A | 1.09 ± 0.04 | No | 192, 109, 213, 186, 116–120, 207 | ||
| D1 | NR | 0.0000 | ||||||||
| D3 | NR | 0.0000 | ||||||||
| D13 | NR | 0.0000 | ||||||||
| D29 | 39.5 | 0.2000 | CQ | LCF | 224 + 379 | 1 | A | 0.78 ± 0.03 | No | Identical to D0 |
| Extended or no follow-upe |
||||||||||
| Not determinable reasonsb | ||||||||||
| NR | ||||||||||
| D0 | 39.2 | 1.0000 | CQ | NR | NR | NR | NR | NR | ||
| D35 | 40.0 | 0.6000 | CQ | ACPR | NR | NR | NR | NR | NR | NR |
| Subtherapeutic chloroquine concentrations observedc | ||||||||||
| O284 | ||||||||||
| D0 | 39.2 | 0.5000 | CQ | 1 | A/WT | 1.48 ± 0.04 | No | 192, 109–129-149, 213, 186, 116–120-124, 207 | ||
| D32 | 38.6 | 0.4500 | CQ | Lost | <10 + <10 | 1 | A/WT | 0.96 ± 0.15 | No | Identical to D0 |
| P092 | ||||||||||
| D0 | 39.0 | 0.4600 | CQ | 1 | A | 0.83 ± 0.09 | No | 188, 109, 213, 186, 116–120–124, 199 | ||
| D34 | 39.7 | 0.1700 | CQ | Lost | <10 + <10 | 2 | A | 1.33 ± 0.21 | Yes | 192, 109, 213, 186, 116-120-124, 199 |
| P213 | ||||||||||
| D0 | 39.0 | 0.1000 | CQ | 2 | A | 0.80 ± 0.06 | No | 196, 121, 262, 186, 116–120–124, 215-219 | ||
| D32 | 39.0 | 0.0700 | CQ | ACPR | 17 + 10 | 2 | A | 0.70 ± 0.08 | No | Identical to D0 |
| P367 | ||||||||||
| D0 | 38.3 | 0.1000 | CQ | 1 | A | 0.75 ± 0.03 | Yes | 196, 109, 213, 182, 116-120, 207 | ||
| D33 | 38.6 | 0.1100 | CQ | Lost | 15 + <10 | 1 | A | 1.19 ± 0.15 | Yes | Identical to D0 |
| Q332 | ||||||||||
| D0 | 37.6 | 0.3500 | CQ | 1 | A | 1.11 ± 0.03 | No | 192–196, 97–145, 206–213, 182–186, 112–116–120, 207 | ||
| D31 | 40.0 | 0.0500 | CQ | Lost | <10 + <10 | 1 | A | 1.00 ± 0.04 | No | Identical to D0 |
| S674 | ||||||||||
| D0 | 39.5 | 0.1800 | CQ | 1 | A | 1.02 ± 0.02 | No | 192, 129, 59-206, 182, 124–128–132, 199 | ||
| D30 | 38.1 | 0.0200 | CQ | Lost | <10 + <10 | 1 | A | 0.74 ± 0.13 | No | Identical to D0 |
| Chloroquine therapeutic failure associated with parasite resistanced | ||||||||||
| M226 | ||||||||||
| D0 | 38.6 | 0.2000 | CQ | 2 | A | 0.89 ± 0.02 | Yes | 196, 109, 213, 186, 120–124, 219 | ||
| D7 | 0.0000 | |||||||||
| D26 | 38.6 | 0.0500 | CQ | Lost | 65 + 204 | 2 | A | 0.64 ± 0.06 | Yes | Identical to D0 |
A, Guy-A; ACPR, adequate clinical and parasitological response; BT, body temperature; D0 to D35, days 0 to 35; dCQ, desethylchloroquine; ID, identification; LCF, late clinical response; Lost, lost to follow-up; LPF, late parasitological response; ND, not determinable; No, no modification of the treatment; NR, not received; pvmdr1, Plasmodium vivax multidrug resistance 1 gene; WT, wild type. Pvcrt-o gene copy numbers have been determined based on two technical replicates.
Reasons for recurrence not determinable because no drug concentration was available.
Recurrence linked to subtherapeutic concentration of drug: the CQ+dCQ concentration was <100 ng/ml.
Treatment failure (i.e., parasite resistance): the CQ+dCQ concentration was >100 ng/ml, with the same parasite genotype observed on D0 and DF.
Recurrence observed in patients with extended follow-up (D31 to D35) or patients without follow-up (D1 to D35).
Values separated with a hyphen mean that this microsatellite has a multiclonal stucture represented by the different observed sizes at the studied locus.
FIG 2.
Chloroquine plasma concentrations (including chloroquine [CQ] plus desethylchloroquine [dCQ]) per day in 16 patients experiencing treatment failure (black squares) and 15 patients with adequate treatment responses (small gray circles) in French Guiana from 2009 to 2015. Mean of adequate treatement response concentrations are also represented (large gray circles).
Until 1995, chloroquine was also recommended to treat P. falciparum (3). However, resistance of P. vivax has not evolved as quickly as for P. falciparum. French Guiana is the second country of the Guiana Shield reporting cases of CQR to P. vivax after the Republic of Guyana. In South America, resistance was reported in Manaus, Brazil (20), in more than 10% of cases after CQ treatment. Other studies also reported CQR in Amazonia but after CQ+PQ treatment: 5.2% in Manaus, Brazil (CQ+PQ) (21), 1.1% in Oiapoque, Brazil (CQ+PQ) (19), and 6.5% in Bolivia (CQ+PQ) (23). However, comparison with these results is impossible because of the potentialization of CQ action by PQ when the drugs are coadministered (24). In France, coadministration is rare because PQ is never given without a preliminary evaluation of the G6PD activity of the patient by quantitative laboratory methods.
Beside the eight treatment failures observed during the D28 follow-up, eight cases of treatment failures were also identified from the D35 extended follow-up or patients who were not followed but had returned to the hospital because of fever (Fig. 1). Biological analyses were conducted to identify resistance in seven of these samples because for one case, plasma and/or DNA was missing. In this context, one new case of P. vivax resistance was identified with a CQ+dCQ plasma concentration ([CQ]) of 269 ng/ml (Fig. 2). The other six were associated with very low or undetectable chloroquine concentration despite the fact that until day 35, the chloroquine concentration should normally be above 100 ng/ml (8). Therefore, these recurrent cases of parasitemia were probably due to relapses. In fact, the Chesson strain circulating in South America generates relapses around D28 (25). With a limited and variable [CQ] in blood after D28, these results underlined the importance of ending the follow-up at D28 during a chloroquine efficacy study in case of infection by a Chesson strain, before the occurrence of natural relapses.
A relevant molecular marker is required to easily monitor P. vivax resistance to chloroquine.
The genotype and copy number of the pvmdr1 and pvcrt-o genes were analyzed in the 13 available pairs of samples (D0-DF) associated with a treatment failure whatever the reason. The pvmdr1 mutation T958M previously described as prevalent in French Guiana was identified in 86.5% of the samples (26). No difference within D0-DF pairs of samples was observed. The mutations Y976F and F1076L were absent even in the chloroquine-resistant parasites associated with the three chloroquine treatment failures (M226, M513, and N518). The pvmdr1 copy number was 2 in seven samples (29.2%), without any associated with the in vivo phenotype. The general percentage of multicopy samples was significantly higher than what was observed in the same period in the general parasite population of French Guiana (12.8%; n = 43/335; P < 0.05) (data not shown). The pvcrt-o part of the gene including an extra amino acid at position 10 (K10) has been sequenced in D0-DF pairs as well as in a sample set of 28 samples in order to compare to the genetic profile specific of French Guiana (27). No difference within D0-DF pairs of samples was observed for the pvcrt-o K10 insertion (Table 2). This polymorphism was observed in 57.1% of samples (95% CI, 38.8 to 75.5%; n = 16/28 [Fig. 3A]). The pvcrt-o gene was monocopy in the sample set, whether associated or not with therapeutic failure (Fig. 3B).
FIG 3.
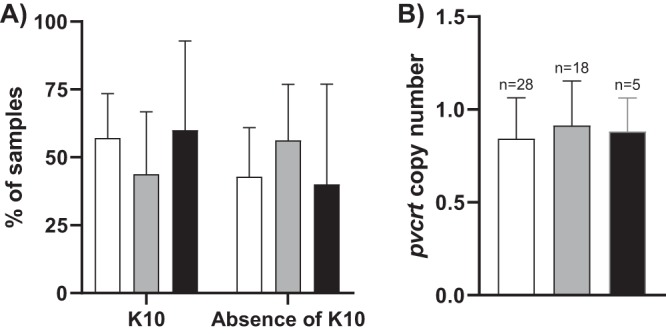
Genotyping of the pvcrt-o gene. (A) Prevalence of the insertion of K10. (B) Gene copy number. Results are presented according to the therapeutic response to chloroquine for each group of P. vivax isolates: adequate clinical and therapeutic response in white (n = 28), failure in the absence of resistance in gray (n = 9 D0-DF pairs), and failure associated with parasite resistance in black (M226 and N518 D0-DF pairs and M513 D0).
The pvmdr1 and pvcrt-o genotypes (sequence and copy number) were not associated with CQR in French Guiana. As previously described, these markers are probably not markers of chloroquine resistance or even minor determinants for resistance (13). However, their expression levels have been described as being associated with P. vivax resistance in the Amazon region (17). These have not been analyzed in this study because of the absence of RNA collection.
P. vivax resistance to CQ exists in French Guiana and needs to be controlled using primaquine.
P. vivax CQR exists in French Guiana but at a low prevalence. Therefore, these results do not justify a change in treatment regimen and regional recommendations. However, a better implementation of the coadministration of CQ and PQ is crucial to avoid the spread of these resistant parasites. To do so, the use of rapid quantitative screening methods for G6PD deficiency and the systematic recording of patient’s G6PD status should be considered.
MATERIALS AND METHODS
Study site, patients, and treatment monitoring.
All cases of malaria confirmed by microscopy and an axillary temperature of ≥37.5°C or subjects with a history of fever during the past 24 h between March 2009 and October 2015 were retrospectively included in this study. Cases were excluded from the analysis under the following circumstances: (i) cases of severe P. vivax malaria according to the WHO definitions regarding P. falciparum, (ii) the presence of concomitant infectious disease or comorbid conditions, (iii) pregnant women, and (iv) patients not treated by CQ (Nivaquine) at a total dose of 25-mg base/kg body wt (10-mg base/kg body wt on day 0 [D0], 10 mg/kg body wt on D1, and 5 mg/kg body wt on D2). Treatment administration was not supervised.
Patients treated by antimalarial drugs followed the common clinical practices of Cayenne Hospital. Patients were invited to come back for clinical and biological examinations on D3, D7 ± 1, D14 ± 1, D21 ± 2, and D28 ± 2. Hospitalized patients were also followed on D1 and D2. Patients who did not experience TF during the standard 28-day follow-up had an extended follow-up until D35. Additionally, patients were informed to come back to the hospital in case of symptom resurgence without waiting for the next scheduled visit.
Any patients who did not attend the visit on D28 were classified as lost to follow-up. However, patients who attended the D28 visit but were not followed for more than 2 weeks during this period were withdrawn from the analysis. Those who missed one appointment but had no relapse detected during the preceding and following appointments were considered to have negative parasitemia for the missed appointment and were not withdrawn from the study. Finally, patients (i) treated with PQ, (ii) misdiagnosed on D0, or (iii) diagnosed with another malaria species during the follow-up were also excluded.
General baseline data were also recorded in the patients’ files: (i) sex, (ii) age, (iii) history of malaria in the last 3 months, (iv) onset of symptoms, (v) weight, (vi) history of travel during the 4 weeks preceding the consultation, and (vii) antimalarial chemoprophylaxis.
Classification of treatment responses.
Treatment responses were classified according to the WHO guidelines (5), as early treatment failure (ETF), late clinical failure (LCF), late parasitological failure (LPF), or adequate clinical and parasitological response (ACPR). Treatment failures (TFs) included ETF, LCF, and LPF.
In order to properly characterize parasite resistance, biological analyses were conducted on samples from all patients who had experienced recurrent parasitemia, including (i) patients presenting TF during the standard or extended follow-up and (ii) patients not followed but diagnosed with TF outside the protocol.
Measurement of antimalarial drug concentration.
CQ+dCQ plasma concentrations ([CQ]) were measured at the day of treatment failure (DF) by liquid chromatography combined with tandem mass spectrometry (TSQ Quantum Ultra; Thermo Fisher, France) as previously reported by Hodel et al., with minor modifications (28). Using OASE 96-well microplates (Waters, France), 100 μl of plasma was mixed with acetonitrile. Proteins and phospholipids were eliminated by positive pressure using the 96-Positive Pressure system (Waters, France). Eluents were evaporated at room temperature. Dry residues were dissolved in 100 μl of mobile phase, and 10 μl was injected into the system. For both molecules, the method was linear between 10 and 1,000 ng/ml. For homemade and external controls from the WorldWide Antimalarial Resistance Network, coefficients of variation were below 10% and bias values were ±10%.
A plasma concentration greater than 100 ng/ml was considered adequate up to D35 (8). When the measurement was conducted before D28, results were compared to drug concentrations observed in patients with adequate clinical and parasitological responses on the same day of follow-up. Then TFs were classified as (i) TF due to subtherapeutic concentration if [CQ] is <100 ng/ml, (ii) TF due to resistance if [CQ] is >100ng/ml, and (iii) “unclassified” if no drug concentration was available.
DNA extraction.
Parasite DNA was extracted from 200 μl of blood using QIAamp genomic DNA kits according to the manufacturer’s instructions (Qiagen, Courtaboeuf, France).
Microsatellite characterization.
The genetic background of parasites was compared between D0 and DF using a set of six microsatellite loci (3.27, 8.504, 11.162, 13.239, MS8, and MS9) (29–31). This panel was selected because of its high polymorphism in the general parasite population of French Guiana (0.62 < expected heterozygosity [HE] < 0.68 [data not shown]). Microsatellites were analyzed by nested PCR following previously described procedures and with the primers listed in Table S1 in the supplemental material. The homologous genetic profile (based on the allelic sizes) between D0 and DF suggested a recrudescence of resistant forms, while heterogeneous profiles suggested a new infection. However, whatever the genetic profile, relapses could not be excluded.
Analysis of pvmdr1 and pvcrt-o as putative molecular markers of CQR.
The pvmdr1 and pvcrt-o gene sequences and gene copy numbers were analyzed on all isolates associated with a recurrence collected on D0 and DF according to the previously described methods (26, 27). PCR products were visualized using 2% agarose gel electrophoresis before a double-strand Sanger sequencing. The generally accepted Sanger sequencing limit in case of a mixed genotype is around 10% for the minor genotype (32). Sequences were analyzed with Geneious 8.1.7 software (Biomatters, Ltd., Auckland, New Zealand). Positive and negative controls were systematically included in each series of genotyping. In the absence of known published genotype for the pvcrt-o gene, a sample set of 28 samples associated with adequate treatment response has also been analyzed in order to compare the results.
Ethical and consent approval.
Data and samples were all obtained as standard medical care for any patient presenting fever on hospital admission in French Guiana. According to the French legislation (article L.1211-2 of the French Public Health Code), biobanking and secondary use for scientific purpose of data and human clinical samples are possible as long as the corresponding patients are informed and have not given any objection. In our study, information was given to every patient through the Cayenne Hospital brochure, and no immediate or delayed patient opposition was reported. In cases involving infants, parents or guardians had to report their opposition to the hospital. Samples received from the National Reference Center (NRC) biobank were approved and registered by the French Ministry for Research and the French Ethics Committee (declaration no. DC-2010-1223, collection Nu2). According to the French legislation, no institutional review board approval was required.
Statistical analysis.
Data were collected with Microsoft Excel 2016 (Microsoft, Redmond, WA, USA). Statistical computing was analyzed with R software (R Foundation, Vienna, Austria). Percentages were calculated according to the total number of patients followed up to D28 with a 95% confidence interval. Medians were associated with range. The Wilcoxon test and Fisher’s test were performed to compare data between ACPR and TF after 28 ± 2 days of follow-up. A P value of <0.05 was considered significant.
Supplementary Material
ACKNOWLEDGMENTS
The National Reference Center for Malaria in French Guiana acknowledges its partners involved in diagnosis and care of malaria in French Guiana, who collaborate for several years in malaria surveillance and allow generation of precious data for public health and malaria control. The authors are grateful to Hervé Bogreau and Antoine Adde for support and advice during the statistical analyses and Marie-Hélène Rodier, Christine Imbert, Blandine Rammaert, and Lucie Sedille for comments on the manuscript.
This work was supported by Santé Publique France (French Ministry of Health) and the French Ministry for National Education, Higher Education and Research. The Regional Health Agency of French Guiana financed C.H. We acknowledge an Investissement d’Avenir grant from the Agence Nationale de la Recherche (CEBA: ANR-10-LABX-25-01).
P.R. is a staff member of the World Health Organization. P.R. alone is responsible for the views expressed in this publication, and they do not necessarily represent the decisions, policy, or views of the World Health Organization.
L.M., E.L., P.R., and F.D. conceived and coordinated the study. F.D., R.N., P.A., G.W., L.E., and M.D. collected clinical data. M.D. and D.B. collected biological data. Y.L., B.V., L.M., and S.P. confirmed the diagnostic data and updated the biobank collection. P.H. and C.H. determined the pharmacological concentration. C.H., L.M., Y.L., and B.V. carried out the molecular genetic studies. C.H., L.M., P.R., and S.P. analyzed the data. C.H. carried out statistical analysis. C.H. and L.M. wrote the manuscript. P.H., S.P., L.E., E.L., P.R., and F.D. reviewed the manuscript. All authors read and approved the final manuscript.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/AAC.02116-18.
REFERENCES
- 1.WHO. 2018. World malaria report. World Health Organization, Geneva, Switzerland. [Google Scholar]
- 2.Carme B. 2005. Substantial increase of malaria in inland areas of eastern French Guiana. Trop Med Int Health 10:154–159. doi: 10.1111/j.1365-3156.2004.01365.x. [DOI] [PubMed] [Google Scholar]
- 3.Musset L, Pelleau S, Girod R, Ardillon V, Carvalho L, Dusfour I, Gomes MS, Djossou F, Legrand E. 2014. Malaria on the Guiana Shield: a review of the situation in French Guiana. Mem Inst Oswaldo Cruz 109:525–533. doi: 10.1590/0074-0276140031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cellule-interrégionale d’Epidémiologie de Guyane. 2019. Situation du paludisme en Guyane: point du 16 Avril 2019. http://invs.santepubliquefrance.fr/Publications-et-outils/Points-epidemiologiques/Tous-les-numeros/Guyane/2019/Situation-epidemiologique-du-paludisme-en-Guyane.-Point-au-16-avril-2019.
- 5.WHO. 2015. Guidelines for the treatment of malaria. World Health Organization, Geneva, Switzerland. [Google Scholar]
- 6.WHO. 2015. Control and elimination of Plasmodium vivax malaria—a technical brief. World Health Organization, Geneva, Switzerland. [Google Scholar]
- 7.Imwong M, Snounou G, Pukrittayakamee S, Tanomsing N, Kim JR, Nandy A, Guthmann JP, Nosten F, Carlton J, Looareesuwan S, Nair S, Sudimack D, Day NP, Anderson TJ, White NJ. 2007. Relapses of Plasmodium vivax infection usually result from activation of heterologous hypnozoites. J Infect Dis 195:927–933. doi: 10.1086/512241. [DOI] [PubMed] [Google Scholar]
- 8.Baird JK, Leksana B, Masbar S, Fryauff DJ, Sutanihardja MA, Suradi, Wignall FS, Hoffman SL. 1997. Diagnosis of resistance to chloroquine by Plasmodium vivax: timing of recurrence and whole blood chloroquine levels. Am J Trop Med Hyg 56:621–626. doi: 10.4269/ajtmh.1997.56.621. [DOI] [PubMed] [Google Scholar]
- 9.WHO. 2009. Methods for surveillance of antimalarial drug efficacy. World Health Organization, Geneva, Switzerland. [Google Scholar]
- 10.Ehrman FC, Ellis JM, Young MD. 1945. Plasmodium Vivax Chesson strain. Science 101:377. doi: 10.1126/science.101.2624.377. [DOI] [PubMed] [Google Scholar]
- 11.Brega S, Meslin B, de Monbrison F, Severini C, Gradoni L, Udomsangpetch R, Sutanto I, Peyron F, Picot S. 2005. Identification of the Plasmodium vivax mdr-like gene (pvmdr1) and analysis of single-nucleotide polymorphisms among isolates from different areas of endemicity. J Infect Dis 191:272–277. doi: 10.1086/426830. [DOI] [PubMed] [Google Scholar]
- 12.Schousboe ML, Ranjitkar S, Rajakaruna RS, Amerasinghe PH, Morales F, Pearce R, Ord R, Leslie T, Rowland M, Gadalla NB, Konradsen F, Bygbjerg IC, Roper C, Alifrangis M. 2015. Multiple origins of mutations in the mdr1 gene—a putative marker of chloroquine resistance in P vivax. PLoS Negl Trop Dis 9:e0004196. doi: 10.1371/journal.pntd.0004196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hupalo DN, Luo Z, Melnikov A, Sutton PL, Rogov P, Escalante A, Vallejo AF, Herrera S, Arévalo-Herrera M, Fan Q, Wang Y, Cui L, Lucas CM, Durand S, Sanchez JF, Baldeviano GC, Lescano AG, Laman M, Barnadas C, Barry A, Mueller I, Kazura JW, Eapen A, Kanagaraj D, Valecha N, Ferreira MU, Roobsoong W, Nguitragool W, Sattabonkot J, Gamboa D, Kosek M, Vinetz JM, González-Cerón L, Birren BW, Neafsey DE, Carlton JM. 2016. Population genomics studies identify signatures of global dispersal and drug resistance in Plasmodium vivax. Nat Genet 48:953–958. doi: 10.1038/ng.3588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sa JM, Nomura T, Neves J, Baird JK, Wellems TE, del Portillo HA. 2005. Plasmodium vivax: allele variants of the mdr1 gene do not associate with chloroquine resistance among isolates from Brazil, Papua, and monkey-adapted strains. Exp Parasitol 109:256–259. doi: 10.1016/j.exppara.2004.12.005. [DOI] [PubMed] [Google Scholar]
- 15.Summers RL, Nash MN, Martin RE. 2012. Know your enemy: understanding the role of PfCRT in drug resistance could lead to new antimalarial tactics. Cell Mol Life Sci 69:1967–1995. doi: 10.1007/s00018-011-0906-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nomura T, Carlton JM, Baird JK, del Portillo HA, Fryauff DJ, Rathore D, Fidock DA, Su X, Collins WE, McCutchan TF, Wootton JC, Wellems TE. 2001. Evidence for different mechanisms of chloroquine resistance in 2 Plasmodium species that cause human malaria. J Infect Dis 183:1653–1661. doi: 10.1086/320707. [DOI] [PubMed] [Google Scholar]
- 17.Melo GC, Monteiro WM, Siqueira AM, Silva SR, Magalhaes BM, Alencar AC, Kuehn A, Portillo HA, Fernandez-Becerra C, Lacerda MV. 2014. Expression levels of pvcrt-o and pvmdr-1 are associated with chloroquine resistance and severe Plasmodium vivax malaria in patients of the Brazilian Amazon. PLoS One 9:e105922. doi: 10.1371/journal.pone.0105922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Phillips EJ, Keystone JS, Kain KC. 1996. Failure of combined chloroquine and high-dose primaquine therapy for Plasmodium vivax malaria acquired in Guyana, South America. Clin Infect Dis 23:1171–1173. doi: 10.1093/clinids/23.5.1171. [DOI] [PubMed] [Google Scholar]
- 19.Gomes MS, Vieira JL, Machado RL, Nacher M, Stefani A, Musset L, Legrand E, Menezes RA, Júnior AA, Sousa AP, Couto VS, Couto ÁA. 2015. Efficacy in the treatment of malaria by Plasmodium vivax in Oiapoque, Brazil, on the border with French Guiana: the importance of control over external factors. Malar J 14:402. doi: 10.1186/s12936-015-0925-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.de Santana Filho FS, Arcanjo AR, Chehuan YM, Costa MR, Martinez-Espinosa FE, Vieira JL, Barbosa MG, Alecrim WD, Alecrim MG. 2007. Chloroquine-resistant Plasmodium vivax, Brazilian Amazon. Emerg Infect Dis 13:1125–1126. doi: 10.3201/eid1307.061386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Marques MM, Costa MR, Santana Filho FS, Vieira JL, Nascimento MT, Brasil LW, Nogueira F, Silveira H, Reyes-Lecca RC, Monteiro WM, Lacerda MV, Alecrim MG. 2014. Plasmodium vivax chloroquine resistance and anemia in the western Brazilian Amazon. Antimicrob Agents Chemother 58:342–347. doi: 10.1128/AAC.02279-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Douine M, Lazrek Y, Blanchet D, Pelleau S, Chanlin R, Corlin F, Hureau L, Volney B, Hiwat H, Vreden S, Djossou F, Demar M, Nacher M, Musset L. 2018. Predictors of antimalarial self-medication in illegal gold miners in French Guiana: a pathway towards artemisinin resistance. J Antimicrob Chemother 73:231–239. doi: 10.1093/jac/dkx343. [DOI] [PubMed] [Google Scholar]
- 23.Anez A, Moscoso M, Laguna A, Garnica C, Melgar V, Cuba M, Gutierrez S, Ascaso C. 2015. Resistance of infection by Plasmodium vivax to chloroquine in Bolivia. Malar J 14:261. doi: 10.1186/s12936-015-0774-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Naing C, Aung K, Win DK, Wah MJ. 2010. Efficacy and safety of chloroquine for treatment in patients with uncomplicated Plasmodium vivax infections in endemic countries. Trans R Soc Trop Med Hyg 104:695–705. doi: 10.1016/j.trstmh.2010.08.009. [DOI] [PubMed] [Google Scholar]
- 25.Hanf M, Stephani A, Basurko C, Nacher M, Carme B. 2009. Determination of the Plasmodium vivax relapse pattern in Camopi, French Guiana. Malar J 8:278. doi: 10.1186/1475-2875-8-278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Faway E, Musset L, Pelleau S, Volney B, Casteras J, Caro V, Menard D, Briolant S, Legrand E. 2016. Plasmodium vivax multidrug resistance-1 gene polymorphism in French Guiana. Malar J 15:540. doi: 10.1186/s12936-016-1595-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Silva SR, Almeida ACG, da Silva GAV, Ramasawmy R, Lopes SCP, Siqueira AM, Costa GL, Sousa TN, Vieira JLF, Lacerda MVG, Monteiro WM, de Melo GC. 2018. Chloroquine resistance is associated to multi-copy pvcrt-o gene in Plasmodium vivax malaria in the Brazilian Amazon. Malar J 17:267. doi: 10.1186/s12936-018-2411-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hodel EM, Zanolari B, Mercier T, Biollaz J, Keiser J, Olliaro P, Genton B, Decosterd LA. 2009. A single LC-tandem mass spectrometry method for the simultaneous determination of 14 antimalarial drugs and their metabolites in human plasma. J Chromatogr B Analyt Technol Biomed Life Sci 877:867–886. doi: 10.1016/j.jchromb.2009.02.006. [DOI] [PubMed] [Google Scholar]
- 29.Ferreira MU, Karunaweera ND, da Silva-Nunes M, da Silva NS, Wirth DF, Hartl DL. 2007. Population structure and transmission dynamics of Plasmodium vivax in rural Amazonia. J Infect Dis 195:1218–1226. doi: 10.1086/512685. [DOI] [PubMed] [Google Scholar]
- 30.Imwong M, Nair S, Pukrittayakamee S, Sudimack D, Williams JT, Mayxay M, Newton PN, Kim JR, Nandy A, Osorio L, Carlton JM, White NJ, Day NP, Anderson TJ. 2007. Contrasting genetic structure in Plasmodium vivax populations from Asia and South America. Int J Parasitol 37:1013–1022. doi: 10.1016/j.ijpara.2007.02.010. [DOI] [PubMed] [Google Scholar]
- 31.Rezende AM, Tarazona-Santos E, Fontes CJ, Souza JM, Couto AD, Carvalho LH, Brito CF. 2010. Microsatellite loci: determining the genetic variability of Plasmodium vivax. Trop Med Int Health 15:718–726. doi: 10.1111/j.1365-3156.2010.02535.x. [DOI] [PubMed] [Google Scholar]
- 32.Rohlin A, Wernersson J, Engwall Y, Wiklund L, Björk J, Nordling M. 2009. Parallel sequencing used in detection of mosaic mutations: comparison with four diagnostic DNA screening techniques. Hum Mutat 30:1012–1020. doi: 10.1002/humu.20980. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.